Performance of Plant-Growth-Promoting Rhizobacteria (PGPR) Isolated from Sandy Soil on Growth of Tomato (Solanum lycopersicum L.)
Abstract
1. Introduction
2. Results
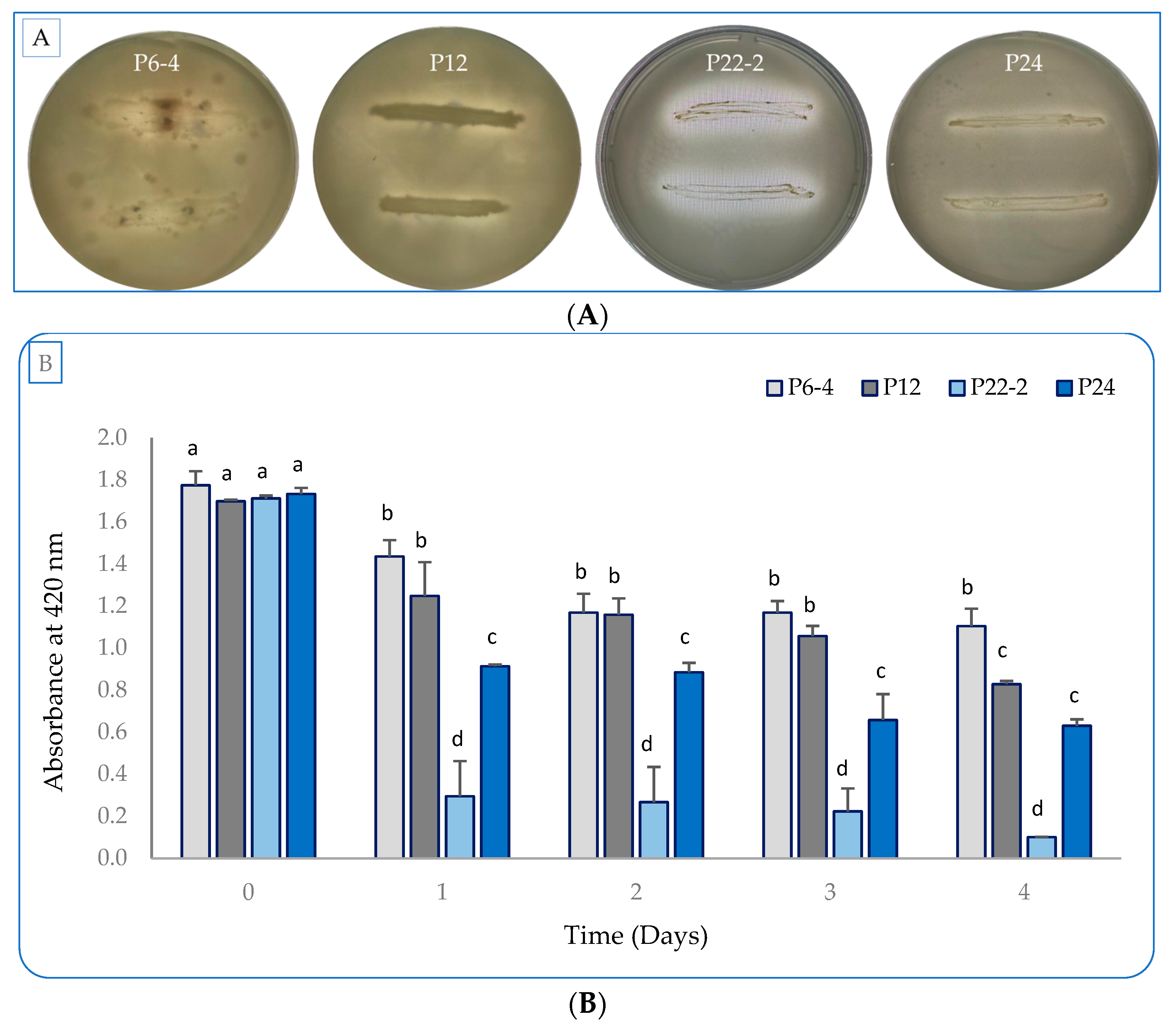
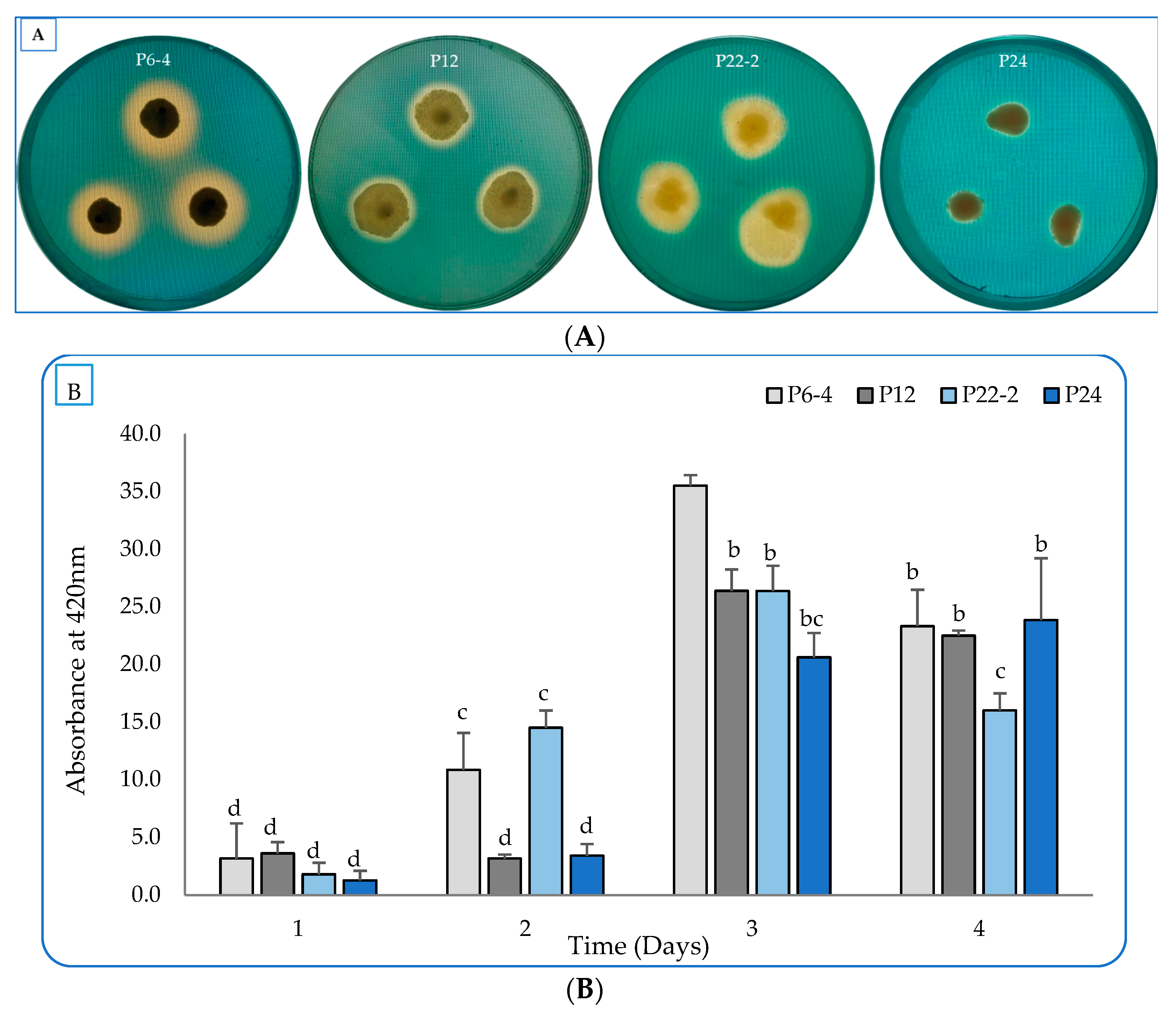
2.1. Evaluation of the Bacterial Plant-Growth-Promoting Characterstics In Vitro
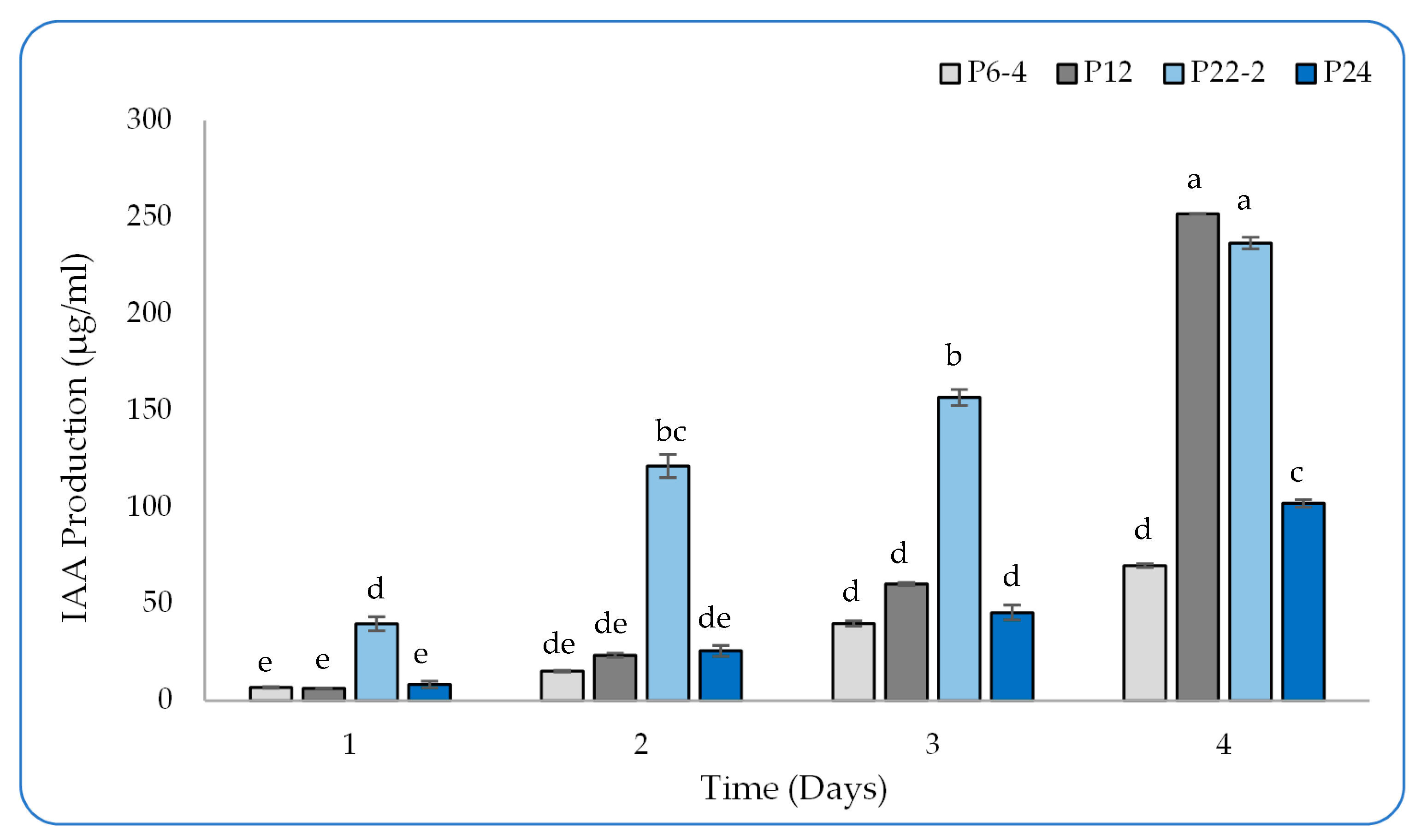
2.2. Detection of IAA Production in the Culture Broth by HPLC
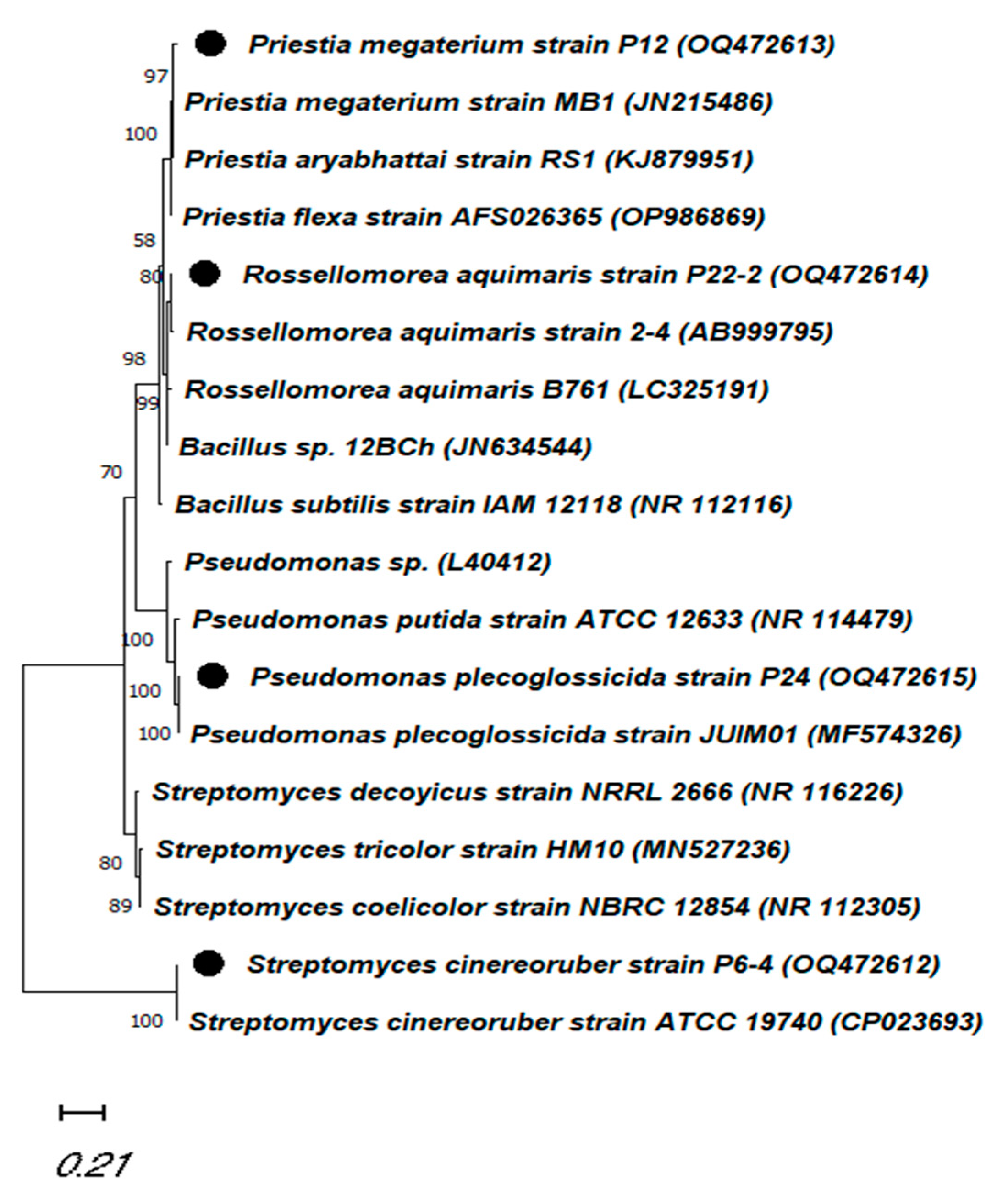
2.3. Identification of the Desired Plant-Growth-Promoting Strains
2.4. Assessing the Isolated PGP Bacteria on Tomato Plant
2.4.1. Vegetative-Growth Traits
2.4.2. Soil and Plant Phosphorus
2.5. Correlations between Studied Traits
3. Discussion
4. Materials and Methods
4.1. Bacterial Isolation and Growth Conditions
4.2. Phosphate Solubilization, IAA, and Siderophore Production of Selected Strains
4.3. IAA Detection in the Culture Broth by HPLC
4.4. Bacterial Identification by 16S rRNA, Accession Numbers, and Phylogenetic-Tree Construction
4.5. Greenhouse Experiment
4.5.1. Experimental Site and Agriculture Conditions
4.5.2. PGPB Treatments under Greenhouse Conditions
4.5.3. Data Collected
The Growth Traits
Phosphorus Analysis
Statistical Analysis and Principal Component Analysis
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rai, S.; Omar, A.F.; Rehan, M.; Al-Turki, A.; Sagar, A.; Ilyas, N.; Sayyed, R.Z.; Hasanuzzaman, M. Crop microbiome: Their role and advances in molecular and omic techniques for the sustenance of agriculture. Planta 2022, 257, 27. [Google Scholar] [CrossRef]
- Bhattacharyya, P.N.; Jha, D.K. Plant growth-promoting rhizobacteria (PGPR): Emergence in agriculture. World J. Microbiol. Biotechnol. 2012, 28, 1327–1350. [Google Scholar] [CrossRef]
- De Souza, R.; Ambrosini, A.; Passaglia, L.M.P. Plant growth-promoting bacteria as inoculants in agricultural soils. Genet. Mol. Biol. 2015, 38, 401–419. [Google Scholar] [CrossRef]
- Katsenios, N.; Andreou, V.; Sparangis, P.; Djordjevic, N.; Giannoglou, M.; Chanioti, S.; Kasimatis, C.-N.; Kakabouki, I.; Leonidakis, D.; Danalatos, N.; et al. Assessment of plant growth promoting bacteria strains on growth, yield and quality of sweet corn. Sci. Rep. 2022, 12, 11598. [Google Scholar] [CrossRef]
- Do Figueiredo, M.V.B.; Seldin, L.; de Araujo, F.F.; de Mariano, R.L.R. Plant Growth Promoting Rhizobacteria: Fundamentals and Applications BT—Plant Growth and Health Promoting Bacteria; Maheshwari, D.K., Ed.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 21–43. ISBN 978-3-642-13612-2. [Google Scholar]
- Dos Lopes, M.J.S.; Dias-Filho, M.B.; Gurgel, E.S.C. Successful Plant Growth-Promoting Microbes: Inoculation Methods and Abiotic Factors. Front. Sustain. Food Syst. 2021, 5, 606454. [Google Scholar] [CrossRef]
- Backer, R.; Rokem, J.S.; Ilangumaran, G.; Lamont, J.; Praslickova, D.; Ricci, E.; Subramanian, S.; Smith, D.L. Plant Growth-Promoting Rhizobacteria: Context, Mechanisms of Action, and Roadmap to Commercialization of Biostimulants for Sustainable Agriculture. Front. Plant Sci. 2018, 9, 1473. [Google Scholar] [CrossRef] [PubMed]
- Glick, B.R. Plant Growth-Promoting Bacteria: Mechanisms and Applications. Scientifica 2012, 2012, 963401. [Google Scholar] [CrossRef] [PubMed]
- Abdelaal, K.; AlKahtani, M.; Attia, K.; Hafez, Y.; Király, L.; Künstler, A. The Role of Plant Growth-Promoting Bacteria in Alleviating the Adverse Effects of Drought on Plants. Biology 2021, 10, 520. [Google Scholar] [CrossRef]
- Omar, A.F.; Rehan, M.; AL-Turki, A. Alleviating the adverse effects of plant pathogens, drought and salinity stress factors using plant growth promoting bacteria. Not. Bot. Horti Agrobot. Cluj-Napoca 2022, 50, 12911. [Google Scholar] [CrossRef]
- Guang-Can, T.A.O.; Shu-Jun, T.; Miao-Ying, C.A.I.; Guang-Hui, X.I.E. Phosphate-solubilizing and-mineralizing abilities of bacteria isolated from soils. Pedosphere 2008, 18, 515–523. [Google Scholar]
- Rodriguez, H.; Gonzalez, T.; Goire, I.; Bashan, Y. Gluconic acid production and phosphate solubilization by the plant growth-promoting Bacterium azospirillum spp. Naturwissenschaften 2004, 91, 552–555. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, H.; Fraga, R. Phosphate solubilizing bacteria and their role in plant growth promotion. Biotechnol. Adv. 1999, 17, 319–339. [Google Scholar] [CrossRef] [PubMed]
- Omar, A.F.; Abdelmageed, A.H.A.; Al-Turki, A.; Abdelhameid, N.M.; Sayyed, R.Z.; Rehan, M. Exploring the Plant Growth-Promotion of Four Streptomyces Strains from Rhizosphere Soil to Enhance Cucumber Growth and Yield. Plants 2022, 11, 3316. [Google Scholar] [CrossRef]
- Rehan, M.; Barakat, H.; Almami, I.S.; Qureshi, K.A.; Alsohim, A.S. Production and Potential Genetic Pathways of Three Different Siderophore Types in Streptomyces tricolor Strain HM10. Fermentation 2022, 8, 346. [Google Scholar] [CrossRef]
- Dorais, M.; Ehret, D.L.; Papadopoulos, A.P. Tomato (Solanum lycopersicum) health components: From the seed to the consumer. Phytochem. Rev. 2008, 7, 231–250. [Google Scholar] [CrossRef]
- Santa-Cruz, A.; Martinez-Rodriguez, M.M.; Perez-Alfocea, F.; Romero-Aranda, R.; Bolarin, M.C. The rootstock effect on the tomato salinity response depends on the shoot genotype. Plant Sci. 2002, 162, 825–831. [Google Scholar] [CrossRef]
- Dorais, M.; Papadopoulos, A.P.; Gosselin, A. Greenhouse Tomato Fruit Quality. In Horticultural Reviews; John Wiley and Sons: New York, NY, USA, 2000; pp. 239–319. ISBN 9780470650806. [Google Scholar]
- Ministry of Environment, Water and Agriculture. Statistical Yearbook; UN iLibrary: Riyadh, Saudi Arabia, 2020. [Google Scholar]
- Hunt, R.; Causton, D.R.; Shipley, B.; Askew, A.P. A modern tool for classical plant growth analysis. Ann. Bot. 2002, 90, 485–488. [Google Scholar] [CrossRef]
- Bisht, N.; Chauhan, P.S. Excessive and Disproportionate Use of Chemicals Cause Soil Contamination and Nutritional Stress; Larramendy, M.L., Soloneski, S., Eds.; IntechOpen: Rijeka, Saudi Arabia, 2020; Chapter 6; ISBN 978-1-83880-754-2. [Google Scholar]
- Chandini, R.K.; Kumar, R.; Kumar, R.; Prakash, O. The Impact of Chemical Fertilizers on our Environment and Ecosystem. In Research Trends in Environmental Sciences, 2nd ed.; AkiNik Publications: New Delhi, India, 2019; pp. 69–86. ISBN 978-93-5335-062-8. [Google Scholar]
- Savci, S. Investigation of Effect of Chemical Fertilizers on Environment. APCBEE Procedia 2012, 1, 287–292. [Google Scholar] [CrossRef]
- Kumar, S.; Diksha; Sindhu, S.S.; Kumar, R. Biofertilizers: An ecofriendly technology for nutrient recycling and environmental sustainability. Curr. Res. Microb. Sci. 2022, 3, 100094. [Google Scholar] [CrossRef]
- Abdelmageed, A.H.A.; Gruda, N.; Geyer, B.; Hassan, G.M.E. Influence of Bacillus Subtilis FZB24 on the Growth and Development of Tomatoes during Summer in the Arid tropics. In Proceedings of the Gartenbauwissenschaftliche Tagung der DGG und BHGL, Geisenheim, Germany, 26 February 2005. [Google Scholar]
- Ahirwar, N.; Gupta, G.; Singh, V.; Rawlley, R.K.; Ramana, S. Influence on growth and fruit yield of tomato (Lycopersicon esculentum Mill.) plants by inoculation with Pseudomonas fluorescence (SS5): Possible role of plant growth promotion. Int. J. Curr. Microbiol. Appl. Sci. 2015, 4, 720–730. [Google Scholar]
- Puia, C.; Vidican, R.; Szabó, G.; Stoian, V. Potential of biofertilizers to improve performance of local genotype tomatoes. Ital. J. Agron. 2017, 12, 838. [Google Scholar] [CrossRef]
- dos Santos, R.M.; Diaz, P.A.E.; Lobo, L.L.B.; Rigobelo, E.C. Use of Plant Growth-Promoting Rhizobacteria in Maize and Sugarcane: Characteristics and Applications. Front. Sustain. Food Syst. 2020, 4, 136. [Google Scholar] [CrossRef]
- Hayat, R.; Ali, S.; Amara, U.; Khalid, R.; Ahmed, I. Soil beneficial bacteria and their role in plant growth promotion: A review. Ann. Microbiol. 2010, 60, 579–598. [Google Scholar] [CrossRef]
- Bechtaoui, N.; Rabiu, M.K.; Raklami, A.; Oufdou, K.; Hafidi, M.; Jemo, M. Phosphate-Dependent Regulation of Growth and Stresses Management in Plants. Front. Plant Sci. 2021, 12, 679916. [Google Scholar] [CrossRef]
- Alori, E.T.; Glick, B.R.; Babalola, O.O. Microbial Phosphorus Solubilization and Its Potential for Use in Sustainable Agriculture. Front. Microbiol. 2017, 8, 971. [Google Scholar] [CrossRef] [PubMed]
- Prabhu, N.; Borkar, S.; Garg, S. Chapter 11—Phosphate Solubilization by Microorganisms: Overview, Mechanisms, Applications and Advances; Meena, S.N., Naik, M.M.B.T.-A., Eds.; Academic Press: Cambridge, MA, USA, 2019; pp. 161–176. ISBN 978-0-12-817497-5. [Google Scholar]
- Yu, H.; Wu, X.; Zhang, G.; Zhou, F.; Harvey, P.R.; Wang, L.; Fan, S.; Xie, X.; Li, F.; Zhou, H.; et al. Identification of the Phosphorus-Solubilizing Bacteria Strain JP233 and Its Effects on Soil Phosphorus Leaching Loss and Crop Growth. Front. Microbiol. 2022, 13, 892533. [Google Scholar] [CrossRef]
- Lobo, L.L.; de Andrade da Silva, M.S.; Castellane, T.C.; Carvalho, R.F.; Rigobelo, E.C. Effect of Indole-3-Acetic Acid on Tomato Plant Growth. Microorganisms 2022, 10, 2212. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.-F.; Wei, J.-Y.; Chen, H.-W.; Liu, Y.-Y.; Lu, H.-Y.; Chou, J.-Y. Indole-3-acetic acid: A widespread physiological code in interactions of fungi with other organisms. Plant Signal. Behav. 2015, 10, e1048052. [Google Scholar] [CrossRef]
- Labeeuw, L.; Khey, J.; Bramucci, A.R.; Atwal, H.; de la Mata, A.P.; Harynuk, J.; Case, R.J. Indole-3-Acetic Acid Is Produced by Emiliania huxleyi Coccolith-Bearing Cells and Triggers a Physiological Response in Bald Cells. Front. Microbiol. 2016, 7, 828. [Google Scholar] [CrossRef]
- Bhattacharyya, C.; Banerjee, S.; Acharya, U.; Mitra, A.; Mallick, I.; Haldar, A.; Haldar, S.; Ghosh, A.; Ghosh, A. Evaluation of plant growth promotion properties and induction of antioxidative defense mechanism by tea rhizobacteria of Darjeeling, India. Sci. Rep. 2020, 10, 15536. [Google Scholar] [CrossRef]
- Lebrazi, S.; Fadil, M.; Chraibi, M.; Fikri-Benbrahim, K. Screening and optimization of indole-3-acetic acid production by Rhizobium sp. strain using response surface methodology. J. Genet. Eng. Biotechnol. 2020, 18, 21. [Google Scholar] [CrossRef] [PubMed]
- Barbaccia, P.; Gaglio, R.; Dazzi, C.; Miceli, C.; Bella, P.; Lo Papa, G.; Settanni, L. Plant Growth-Promoting Activities of Bacteria Isolated from an Anthropogenic Soil Located in Agrigento Province. Microorganisms 2022, 10, 2167. [Google Scholar] [CrossRef]
- Scavino, A.F.; Pedraza, R.O. The Role of Siderophores in Plant Growth-Promoting Bacteria. In Bacteria in Agrobiology: Crop Productivity; Maheshwari, D., Saraf, M., Aeron, A., Eds.; Springer: Berlin/Heidelberg, German, 2013. [Google Scholar] [CrossRef]
- Schmidt, W.; Thomine, S.; Buckhout, T.J. Editorial: Iron Nutrition and Interactions in Plants. Front. Plant Sci. 2020, 10, 1670. [Google Scholar] [CrossRef] [PubMed]
- Rehan, M.; Alsohim, A.S.; Abidou, H.; Rasheed, Z.; Al Abdulmonem, W. Isolation, Identification, Biocontrol Activity, and Plant Growth Promoting Capability of a Superior Strain HM10. Pol. J. Microbiol. 2021, 70, 245–256. [Google Scholar] [CrossRef] [PubMed]
- Cherif-Silini, H.; Silini, A.; Yahiaoui, B.; Ouzari, I.; Boudabous, A. Phylogenetic and plant-growth-promoting characteristics of Bacillus isolated from the wheat rhizosphere. Ann. Microbiol. 2016, 66, 1087–1097. [Google Scholar] [CrossRef]
- Radhakrishnan, R.; Hashem, A.; Abd_Allah, E.F. Bacillus: A Biological Tool for Crop Improvement through Bio-Molecular Changes in Adverse Environments. Front. Physiol. 2017, 8, 667. [Google Scholar] [CrossRef]
- Singh, P.; Chauhan, P.K.; Upadhyay, S.K.; Singh, R.K.; Dwivedi, P.; Wang, J.; Jain, D.; Jiang, M. Mechanistic Insights and Potential Use of Siderophores Producing Microbes in Rhizosphere for Mitigation of Stress in Plants Grown in Degraded Land. Front. Microbiol. 2022, 13, 898979. [Google Scholar] [CrossRef]
- Singh, T.B.; Sahai, V.; Goyal, D.; Prasad, M.; Yadav, A.; Shrivastav, P.; Ali, A.; Dantu, P.K. Identification, Characterization and Evaluation of Multifaceted Traits of Plant Growth Promoting Rhizobacteria from Soil for Sustainable Approach to Agriculture. Curr. Microbiol. 2020, 77, 3633–3642. [Google Scholar] [CrossRef]
- Phares, C.A.; Amoakwah, E.; Danquah, A.; Afrifa, A.; Beyaw, L.R.; Frimpong, K.A. Biochar and NPK fertilizer co-applied with plant growth promoting bacteria (PGPB) enhanced maize grain yield and nutrient use efficiency of inorganic fertilizer. J. Agric. Food Res. 2022, 10, 100434. [Google Scholar] [CrossRef]
- Kaur, M.; Vyas, P.; Rahi, P.; Sharma, S. Chlorpyrifos- and Carbofuran-Tolerant Phosphate-Solubilising Arthrobacter oxydans and Bacillus flexus Improved Growth and Phosphorus Content in Potato in Pesticide-Amended Soils. Potato Res. 2022, 65, 213–231. [Google Scholar] [CrossRef]
- Mohi-Ud-Din, M.; Hossain, M.A.; Rohman, M.M.; Uddin, M.N.; Haque, M.S.; Ahmed, J.U.; Hossain, A.; Hassan, M.M.; Mostofa, M.G. Multivariate Analysis of Morpho-Physiological Traits Reveals Differential Drought Tolerance Potential of Bread Wheat Genotypes at the Seedling Stage. Plants 2021, 10, 879. [Google Scholar] [CrossRef] [PubMed]
- Zafar, M.M.; Manan, A.; Razzaq, A.; Zulfqar, M.; Saeed, A.; Kashif, M.; Khan, A.I.; Sarfraz, Z.; Mo, H.; Iqbal, M.S.; et al. Exploiting Agronomic and Biochemical Traits to Develop Heat Resilient Cotton Cultivars under Climate Change Scenarios. Agronomy 2021, 11, 1885. [Google Scholar] [CrossRef]
- Hamad, H.S.; Bleih, E.M.; Gewaily, E.E.; Abou Elataa, A.E.; El Sherbiny, H.A.; Abdelhameid, N.M.; Rehan, M. Cyanobacteria Application Ameliorates Floral Traits and Outcrossing Rate in Diverse Rice Cytoplasmic Male Sterile Lines. Plants 2022, 11, 3411. [Google Scholar] [CrossRef] [PubMed]
- Nautiyal, C.S. An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiol. Lett. 1999, 170, 265–270. [Google Scholar] [CrossRef] [PubMed]
- Gang, S.; Sharma, S.; Saraf, M.; Buck, M.; Schumacher, J. Analysis of Indole-3-acetic Acid (IAA) Production in Klebsiellaby LC-MS/MS and the Salkowski Method. Bio-Protocol 2019, 9, e3230. [Google Scholar] [CrossRef] [PubMed]
- Louden, B.C.; Haarmann, D.; Lynne, A.M. Use of Blue Agar CAS Assay for Siderophore Detection. J. Microbiol. Biol. Educ. 2011, 12, 51–53. [Google Scholar] [CrossRef] [PubMed]
- Arora, N.K.; Verma, M. Modified microplate method for rapid and efficient estimation of siderophore produced by bacteria. 3 Biotech 2017, 7, 381. [Google Scholar] [CrossRef] [PubMed]
- Patel, P.R.; Shaikh, S.S.; Sayyed, R.Z. Modified chrome azurol S method for detection and estimation of siderophores having affinity for metal ions other than iron. Environ. Sustain. 2018, 1, 81–87. [Google Scholar] [CrossRef]
- Payne, S.M. Iron acquisition in microbial pathogenesis. Trends Microbiol. 1993, 1, 66–69. [Google Scholar] [CrossRef] [PubMed]
- Cook, A.E.; Meyers, P.R. Rapid identification of filamentous actinomycetes to the genus level using genus-specific 16S rRNA gene restriction fragment patterns. Int. J. Syst. Evol. Microbiol. 2003, 53, 1907–1915. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Page, A.L. Methods of soil analysis. In Part 2—Chemical and Microbiological Properties; The American Society of Agronomy, Inc., Soil Science Society of America: Madison, WI, USA, 1982. [Google Scholar]
- Rathje Jackson, M.L. Soil chemical analysis. Verlag: Prentice Hall, Inc., Englewood Cliffs, NJ. 1958, 498 S. DM 39.40. Z. Für Pflanz. Düngung Bodenkd. 1959, 85, 251–252. [Google Scholar] [CrossRef]
- Gee, G.W.; Bauder, J.W. Physical and mineralogical methods. Methods Soil Anal. Part 1986, 1, 383–411. [Google Scholar]
- Maynard, D.N.; Hochmuth, G.J. Knott’s Handbook for Vegetable Growers, 5th ed.; John Wiley & Sons: Hoboken, NJ, USA, 2006; pp. 1–621. [Google Scholar] [CrossRef]
- Schwarz, D.; Kläring, H.-P. Allometry to estimate leaf area of tomato. J. Plant Nutr. 2001, 24, 1291–1309. [Google Scholar] [CrossRef]
- Blanco, F.F.; Folegatti, M.V. A new method for estimating the leaf area index of cucumber and tomato plants. Hortic. Bras. 2003, 21, 666–669. [Google Scholar] [CrossRef]
- Evans, G.C. The Quantitative Analysis of Plant Growth; University of California Press: Berkeley, CA, USA, 1972; Volume 1, ISBN 0520094328. [Google Scholar]
- Olsen, S.R.; Sommers, L.E. Phosphorus. In Methods of Soil Analysis; Agronomy Monographs: Madison, WI, USA, 1983; pp. 403–430. ISBN 9780891189770. [Google Scholar]
- Cottenie, A. Soil and Plant Testing as a Basis of Fertilizer Recommendations; FAO Soils Bulletin (FAO): Via Delle Terme di Caracalla, Italy, 1980; No. 38/2. [Google Scholar]
- Khiddir, S.M. A Statistical Approach in the Use of Parametric Systems Applied to the FAO Framework for Land Evaluation; Rijksuniversiteit Gent. Faculteit van de Wetenschappen: Ghent, Belgium, 1986. [Google Scholar]
- Webster, R. Quantitative and Numerical Methods in Soil Classification and Survey; Clarendon Press: Oxford, UK, 1977. [Google Scholar]
- Addinsoft XLSTAT. Statistical and Data Analysis Solution; Addinsoft XLSTAT: New York, NY, USA, 2019. [Google Scholar]



| Treatments | Plant Height (cm) | No. of Leaves Plant−1 | Root Length (cm) | Leaf DM (%) | Stem DM (%) | Root DM (%) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 21DAT | 45DAT | 21DAT | 45DAT | 21DAT | 45DAT | 21DAT | 45DAT | 21DAT | 45DAT | 21DAT | 45DAT | |
| T1 | 40.00 a | 95.00 bc | 9.67 a | 15.67 bc | 12.67 ab | 21.33 ab | 8.27 b | 12.22 a | 4.84 a | 8.23 a | 9.54 b | 14.20 a |
| T2 | 33.67 ab | 82.33 d | 9.33 a | 13.33 c | 14.27 ab | 18.00 b | 9.86 a | 12.82 a | 4.49 a | 8.58 a | 9.82 b | 11.60 a |
| T3 | 35.33 ab | 101.00 b | 8.67 ab | 17.33 bc | 10.87 b | 19.00 ab | 8.10 b | 11.57 a | 4.69 a | 7.12 a | 8.94 b | 12.09 a |
| T4 | 34.67 ab | 115.67 a | 8.67 ab | 21.67 a | 15.00 a | 26.67 a | 8.38 b | 12.41 a | 5.15 a | 6.77 a | 12.60 a | 12.48 a |
| T5 | 31.50 ab | 102.67 b | 8.00 b | 19.33 ab | 14.50 ab | 25.33 ab | 8.05 b | 12.48 a | 4.60 a | 7.08 a | 10.44 b | 11.65 a |
| T6 | 30.00 b | 87.50 cd | 7.67 b | 15.50 bc | 11.97 ab | 20.50 ab | 8.16 b | 11.52 a | 4.72 a | 7.03 a | 10.24 b | 10.93 a |
| Treatments | Leaf Area (LA) (cm2) | Net Assimilation Rate (NAR) (mg cm2 d−1) | Relative Growth Rate (RGR) (mg g−1 d−1) | |||
|---|---|---|---|---|---|---|
| 21DAT | 45DAT | 21DAT | 45DAT | 21DAT | 45DAT | |
| T1 | 532.71 ab | 5392.97 ab | 0.46 a | 0.48 a | 0.12 b | 0.12 a |
| T2 | 398.45 b | 3183.92 c | 0.35 a | 0.35 a | 0.12 b | 0.13 a |
| T3 | 556.18 ab | 4799.84 abc | 0.43 a | 0.42 a | 0.13 ab | 0.13 a |
| T4 | 770.39 a | 5893.34 ab | 0.47 a | 0.34 a | 0.15 a | 0.11 a |
| T5 | 550.55 ab | 6323.66 a | 0.47 a | 0.42 a | 0.14 ab | 0.14 a |
| T6 | 473.34 b | 4247.94 bc | 0.48 a | 0.47 a | 0.13 ab | 0.13 a |
| Treatment | Plant P-Uptake (mg Plant−1) | Soil-P conc. (mg kg−1) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Leaf | Stem | Root | Total | |||||||
| 21 DAT | 45 DAT | 21 DAT | 45 DAT | 21 DAT | 45 DAT | 21 DAT | 45 DAT | 21 DAT | 45 DAT | |
| T1 | 8.42 b | 98.04 a | 1.71 ab | 26.89 abc | 0.39 ab | 3.35 a | 10.52 b | 130.89 ab | 12.00 a | 24.10 a |
| T2 | 8.71 b | 69.52 b | 1.27 b | 22.27 c | 0.31 b | 1.59 c | 10.29 b | 93.38 c | 9.50 d | 21.77 c |
| T3 | 17.09 ab | 92.76 ab | 1.79 ab | 35.33 ab | 0.46 ab | 1.95 c | 19.34 ab | 130.04 ab | 10.70 c | 22.78 b |
| T4 | 12.46 ab | 105.61 a | 2.62 a | 37.50 a | 0.58 a | 2.24 bc | 15.66 ab | 145.35 a | 11.40 b | 22.35 bc |
| T5 | 18.54 a | 95.94 a | 2.21 ab | 35.77 ab | 0.28 b | 2.96 ab | 21.03 a | 134.67 a | 10.70 c | 22.67 b |
| T6 | 19.46 a | 69.67 b | 2.29 ab | 24.57 bc | 0.38 ab | 2.09 bc | 22.14 a | 96.11 bc | 10.60 c | 24.76 a |
| Properties | Value | |
|---|---|---|
| Soil | Water | |
| Physical properties | ||
| Sand (%) | 93.4 | - |
| Silt (%) | 3.9 | - |
| Clay (%) | 2.7 | - |
| Texture | Sand | - |
| Chemical properties | ||
| 1 pH | 7.82 | 7.27 |
| 2 EC (dS m−1) | 1.04 | 0.95 |
| 3 Nutrients (ppm) | ||
| Total N | 175 | - |
| Available P | 1.26 | - |
| Available K | 74.0 | 42.0 |
| 4 Dissolved ions (meq L−1) | ||
| Dissolved anions (meq L−1) | ||
| Cl− | 8.7 | 8.2 |
| HCO3−1 + CO3−2 | 2.1 | 1.7 |
| Dissolved cations (meq L−1) | ||
| Na+ | 7.3 | 7.1 |
| Ca++ | 2.8 | 1.8 |
| Mg++ | 0.9 | 0.9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rehan, M.; Al-Turki, A.; Abdelmageed, A.H.A.; Abdelhameid, N.M.; Omar, A.F. Performance of Plant-Growth-Promoting Rhizobacteria (PGPR) Isolated from Sandy Soil on Growth of Tomato (Solanum lycopersicum L.). Plants 2023, 12, 1588. https://doi.org/10.3390/plants12081588
Rehan M, Al-Turki A, Abdelmageed AHA, Abdelhameid NM, Omar AF. Performance of Plant-Growth-Promoting Rhizobacteria (PGPR) Isolated from Sandy Soil on Growth of Tomato (Solanum lycopersicum L.). Plants. 2023; 12(8):1588. https://doi.org/10.3390/plants12081588
Chicago/Turabian StyleRehan, Medhat, Ahmad Al-Turki, Adil H. A. Abdelmageed, Noha M. Abdelhameid, and Ayman F. Omar. 2023. "Performance of Plant-Growth-Promoting Rhizobacteria (PGPR) Isolated from Sandy Soil on Growth of Tomato (Solanum lycopersicum L.)" Plants 12, no. 8: 1588. https://doi.org/10.3390/plants12081588
APA StyleRehan, M., Al-Turki, A., Abdelmageed, A. H. A., Abdelhameid, N. M., & Omar, A. F. (2023). Performance of Plant-Growth-Promoting Rhizobacteria (PGPR) Isolated from Sandy Soil on Growth of Tomato (Solanum lycopersicum L.). Plants, 12(8), 1588. https://doi.org/10.3390/plants12081588







