Abstract
MADS-box is a class of transcriptional regulators that are ubiquitous in plants and plays important roles in the process of plant growth and development. Identification and analysis of blueberry MADS-box genes can lay a foundation for their function investigations. In the present study, 249 putative MADS-box genes were identified in the blueberry genome. Those MADS-box genes were distributed on 47 out of 48 chromosomes. The phylogenetic and evolutionary analyses showed that blueberry MADS-box genes were divided into 131 type I members and 118 type II members. The type I genes contained an average of 1.89 exons and the type II genes contained an average of 7.83 exons. Motif analysis identified 15 conserved motifs, of which 4 were related to the MADS domain and 3 were related to the K-box domain. A variety of cis-acting elements were found in the promoter region of the blueberry MADS-box gene, indicating that the MADS-box gene responded to various hormones and environmental alterations. A total of 243 collinear gene pairs were identified, most of which had a Ka/Ks value of less than 1. Nine genes belonging to SEP, AP3/PI, and AGL6 subfamilies were screened based on transcriptomic data. The expression patterns of those nine genes were also verified using quantitative PCR, suggesting that VcMADS6, VcMADS35, VcMADS44, VcMADS58, VcMADS125, VcMADS188, and VcMADS212 had potential functions in blueberry fruit ripening. The results of this study provide references for an in-depth understanding of the biological function of the blueberry MADS-box genes and the mechanism of blueberry fruit ripening.
1. Introduction
MADS-box is an important class of transcription regulators that is widely present in eukaryotes. The name of MADS-box originated from four proteins, Saccharomyces cerevisiae transcription factor (MCM1) [1], Arabidopsis flower homeotype gene (AGAMOUS) [2], snapdragon flower homeotype gene (DEFICIENS) [3], and human serum response factor (SRF) [4]. The amino acid sequences of those proteins contain a conserved region of about 60 amino acids, namely, the MADS-box domain [5]. MADS-box genes can be separated into type I and type II according to their sequence features [6]. Type I MADS-box genes in plants are also known as M-type genes and can be further divided into Mα, Mβ, and Mγ subgroups [7,8]. Type II MADS-box genes mainly include a highly conserved MADS (M) region, moderately conserved keratin-like (K) region, relatively conserved intervening (I) region, and variable C-terminal (C) [8,9], which are also called MIKC-type. The MADS domain is involved in DNA binding and protein dimerization. The MADS domain and I domain jointly participate in the process of protein dimerization and affect the dimerization specificity. The K-box domain affects the interaction between MADS-box transcription factors. The C domain plays different functions in the various proteins, which enhance the function of the K-box domain or affect the DNA binding ability of the proteins. The MIKC-type MADS-box genes can be further divided into MIKCc and MIKC* subgroups based on phylogenetic analysis [10]. Both type I and type II MADS-boxes contain a highly conserved MADS domain. The difference between these two types is that they contain divergent MADS domains. Most type II MADS-box genes include three other domains (‘I’, ‘K’, and ‘C’), while most type I MADS-box genes do not [11,12].
MADS-box genes play essential roles in plant growth, development regulation, and signal transduction. Previous studies about MADS-box genes mainly focused on the foundation of Arabidopsis female gametophytes [13], chloroplast formation [14], and embryo [15] and endosperm development [16]. The study interests of type I MADS-box genes were limited, possibly due to scarce transcription abundance and rarely reported functions [17]. By contrast, type II MADS-box genes attracted extensive and deep attention. Type II MADS-box genes were mostly recognized in the floral quartet model and the underlying ABCDE model of organ identity determination [18,19,20]. The expression of five classes of genes A, B, C, D, and E in plants separately or together contributed to floral organ development. The A function is mediated by APETALA1 (AP1) and APETALA2 (AP2), the B function by APETALA3 (AP3) and PISTILLATA (PI), the C function by AGAMOUS (AG), the D function by SHATTERPROOF (SHP) and/or SEEDSTICK (STK), and the E function by SEPALLATA (SEP) [20]. Most of the five classes of genes belong to the MADS-box gene family. Type II MADS-box genes were also found to be involved in the growth and development of plant buds [21], stress resistance [13,22,23], and seed germination [24]. In addition, type II MADS-box genes were associated with fruit ripening [25]. The SHATTERPROOF-like gene (FaSHP) that belongs to the strawberry (Fragaria × ananassa) type II MADS-box was shown to regulate the ripening process of flesh fruit directly or indirectly through other transcription-factor-encoding genes [26]. The expression of MADS-RIN was required for tomato fruit ripening, which provides molecular insights into the developmental regulation of maturation [27].
To date, MADS-box gene families from numerous species were identified, including cabbage [28], tomato [29], sweet potato [30], willow [31], and wheat [32]. However, the blueberry (Vaccinium spp.) MADS-box gene family has not yet been analyzed. Blueberry is cultivated worldwide for its health benefits due to its abundant polyphenolic compound. Fruit ripening is the result of the joint regulation of multiple genes, which involves a series of physiological and biochemical changes. The ripening process of blueberry fruit is a crucial step for the formation and accumulation of fresh fruit flavor and other qualities. MADS-box genes were reported to be related to fruit development and ripening in a variety of plants, but their expression patterns during blueberry fruit ripening are still unclear. In this study, we performed genome-wide identification of the blueberry MADS-box genes. The chromosomal location, gene structure, phylogenetic relationships, and promoter cis-acting elements of MADS-box genes were analyzed. The biochemical features and conserved motifs of MADS-box proteins were also investigated. In addition, we screened MADS-box genes in accordance with the ripening process of blueberry fruit through transcriptomic data. The expression patterns of selected genes were validated using quantitative PCR. We aimed at providing references for understanding the potential function of blueberry MADS-box genes and the possible regulatory roles of MADS-box genes in blueberry fruit ripening.
2. Results
2.1. Identification of MADS-Box Genes in the Blueberry Genome
A total of 249 MADS-box genes (NCBI accession nos. OQ559686–OQ559934) were identified in the blueberry (V. corymbosum cv. ‘Draper’) genome. They were named from VcMADS1 to VcMADS249 according to the chromosome location and physical order on the chromosome (Supplementary Table S1). The amino acid number of MADS-box ranged from 135 (VcMADS98) to 672 (VcMADS195), with an average of 262 amino acids. The maximum and minimum molecular weight was observed at protein VcMADS195 (76.5 kDa) and VcMADS98 (15.5 kDa), respectively. The theoretical pI ranged from 4.56 (VcMADS228) to 10.4 (VcMADS208), with a mean value of 7.92. The instability indexes of MADS-box proteins were from 33.1 to 72.4. The results showed that 91.6% of MADS-box proteins were unstable with instability indexes larger than 40, while only 8.40% of MADS-box proteins had instability indexes less than 40. The predicted results of subcellular localization revealed that 64.7% of MADS-box proteins were localized in the nucleus. In addition, 18.5% and 3.21% of the MADS-box members were in chloroplasts and mitochondria, respectively.
2.2. Phylogenetic Analysis and Classification of MADS-Box Genes
To study the phylogenetic relationships of MADS-box genes between blueberry and Arabidopsis, preliminary evolutionary trees were built for 107 Arabidopsis MADS-box proteins and 249 blueberry MADS-box proteins. The results showed that 131 M-type (type I) and 118 MIKC type (type II) MADS-box genes were found in the blueberry genome. Different types of MADS-box genes in blueberry, together with Arabidopsis corresponding type I and type II MADS-box genes, Calam.16G114400.2, and Solyc03g019710.2.1, were taken to construct a phylogenetic tree. The results showed that the blueberry genome consisted of 61 Mα members, 19 Mβ members, and 51 Mγ members (Figure 1). The blueberry MIKC-type MADS-box included 14 MIKC* and 104 MIKCc members, among which MIKCc could be further divided into 13 subgroups (Figure 2). Gene numbers in different subgroups ranged from 1 (AGL15) to 18 (SOC1), which indicated dramatic divergence among the subgroups. The subgroup ANR1 contained 16 genes, followed by the subgroup SEP (14 genes). The subgroups SVP and AP3/PI consisted of 10 and 9 members, respectively. Subgroups AG and FLC both had eight genes, while the subgroups AP1/FUL, AGL6, AGL12, TM8, and TT16 contained fewer members than the other subgroups, which just had six, five, two, four, and three genes, respectively.
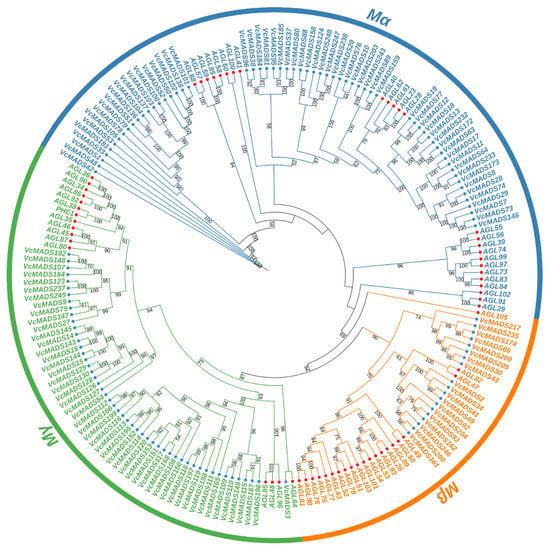
Figure 1.
Phylogenetic tree of type I (M-type) MADS-box proteins from blueberry and Arabidopsis. MADS-box proteins of blueberry and Arabidopsis are marked with blue and red circles, respectively. The digits on the branches represent bootstrap values for 1000 replicates.
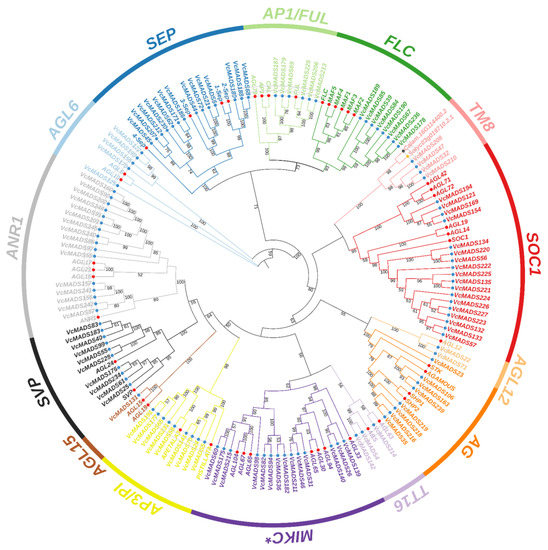
Figure 2.
Phylogenetic tree of type II (MIKC-type) MADS-box proteins from blueberry, Arabidopsis, Calam.16G114400.2, and Solyc03g019710.2.1. MADS-box proteins of blueberry, Arabidopsis, Calam.16G114400.2, and Solyc03g019710.2.1 are marked by blue, red, brown, and orange circles, respectively. The digits on the branches represent bootstrap values for 1000 replicates.
2.3. Chromosomal Locations of MADS-Box Genes
To gain insight into the chromosomal position of the 249 blueberry MADS-box genes, we analyzed their genomic distribution and found that they were unevenly distributed on nearly all chromosomes except chromosome 48 (Figure 3). High MADS-box gene abundance was found on chromosomes 2 (11 genes), 3 (16 genes), 13 (11 genes), 21 (16 genes), and 30 (10 genes), which contained more than 10 MADS-box genes. Whereas, only one MADS-box gene was discovered on each of chromosomes 6, 20, 28, 39, 44, and 46. Further analysis of chromosome location results showed that type I MADS-box genes only existed on eleven chromosomes. MADS-box members from the Mα subfamily were seen on chromosomes 15, 19, 24, and 46, which consisted of two, six, three, and one genes, respectively. Type II MADS-box genes were found on eleven chromosomes, i.e., chromosomes 6, 12, 20, 23, 28, 32, 39–41, 44, and 45. Furthermore, a total of 147 MADS-box genes formed 53 gene clusters distributed on 31 chromosomes, of which 50 gene clusters were composed of the same type of MADS-box genes. Thirty-four gene clusters contained MADS-box genes from the same subfamily, which indicated gene tendency and/or aggregate distribution tendency for blueberry MADS-box genes in the same subgroup.
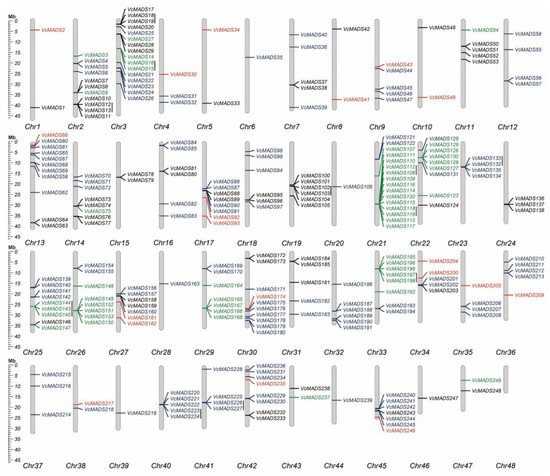
Figure 3.
Chromosome location, distribution, and tandem duplication of MADS-box genes in 48 chromosomes of blueberry. Mα, Mβ, Mγ, and MIKC genes are shown in black, red, green, and purple, respectively. Vertical black lines indicate potential tandem repeat genes.
2.4. Structure and Conserved Motif Analyses of MADS-Box Genes
The length of the MADS-box genes was from 443 to 20,305 bp, with an average number of 5494 base pairs. Regarding the amino acid numbers, VcMADS1 and VcMADS195 were the shortest gene and longest gene, respectively. A significant difference was observed between the type I and type II MADS-box genes. In general, type I MADS-box genes were characterized by a shorter gene length and fewer exons (Figure 4A). The average length of the Mα subfamily genes was 1865 bp. VcMADS203 (12,466 bp) and VcMADS243 (11,980 bp) were significantly longer than the other genes. The Mα subfamily gene contained 1–4 exons, but all genes except VcMADS13, VcMADS18, VcMADS203, and VcMADS243 had only 1–3 exons. It was found that there was a certain relationship between the gene length and the number of exons, and genes with a longer gene length tended to contain more exons, such as VcMADS203 and VcMADS243. In addition, none of the Mα subfamily genes contained a UTR structure, except VcMADS78, which contained two five-prime UTR (untranslated region) structures. The Mβ subfamily gene contained 1–4 exons, and the average length of the gene was 3182 bp, among which VcMADS92 was 13,252 bp. Four genes (VcMADS209, VcMADS205, VcMADS43, and VcMADS30) contained UTR structures, and only VcMADS30 contained a three-prime UTR. The Mγ subfamily genes contained 1–5 exons, but only VcMADS119 and VcMADS165 contained five exons. The average length of the Mγ subfamily genes was 1711 bp and the length of VcMADS195 was 20,305 bp. In contrast to the Mα and Mβ subfamilies, none of the Mγ genes contained UTR structures. The type II genes showed strong conservation within the same subfamily, and the genes in the same subfamily had a similar gene structure and length (Figure 4B). The MIKC* subfamily and AP3/PI subfamily were the two subfamilies with the highest and lowest average numbers of exons among the type II genes, respectively. The MIKC* subfamily genes contained 9.2 exons on average, and the AP3/PI subfamily genes contained 6.8 exons on average. Gene length analysis showed that the AP1/FUL subfamily (16,387 bp) and AP3/PI subfamily (3878 bp) were the subfamilies with the longest and shortest average gene length, respectively. Unlike type I genes, most type II genes contained UTR structures, and in addition to the ANR1 and MIKC* subfamilies, most of the genes belonging to other subfamilies contained both five-prime UTR and three-prime UTR structures. Most type I MADS-box (122, 93.1%) genes contained less than three exons and some genes (9, 6.90%) contained 4 or 5 exons, with an average of 1.89 exons (Figure 4C). In the type II MADS-box genes, the average exon number was 7.83. More than 95 (80.5%) genes contained 7–9 exons, while 15 (12.7%) genes had less than 6 exons and only 8 (6.78%) contained 10–14 exons.
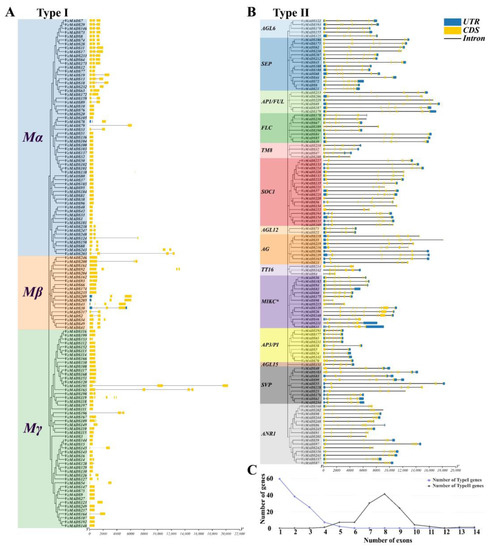
Figure 4.
Gene structure analysis of 249 VcMADS genes. (A) type I MADS-box genes; (B) type II MADS-box genes; (C) exon number and their frequency of MADS-box genes, where blue and black lines indicate type I and type II MADS-box genes, respectively.
We performed a conserved motif analysis for VcMADS proteins with default parameter settings. Fifteen conserved motifs were uncovered in all VcMADS proteins (Table 1). In contrast to the type II genes, motifs 5, 9, and 10 were only found in the type I MADS-box proteins, where motif 9 was only found in the Mα subfamily and motifs 5 and 10 were only found in the Mγ subfamily. In addition, motifs 11, 12, 13, and 14 were mostly present in type I MADS-box proteins. Motif 11 was mainly distributed in the Mγ subfamily, motif 12 was mainly distributed in the Mα and Mγ subfamilies, and motif 14 was only present in the Mγ subfamily in type I MADS-box proteins (Figure 5A). Motifs 6, 8, and 15 were only found in MADS-box type II proteins, in which motifs 6 and 8 were widely distributed in type II proteins. However, FLC subfamily proteins lacked motif 6, MIKC* subfamily proteins lacked motif 8, and only one member contained motif 6. Motif 15 was found only in the SOC1 subfamily proteins (Figure 5B). Motifs 1, 2, 4, and 7 were in the MADS domain region. Motifs 3, 6, and 8 were found in the K-box domain region, of which motifs 6 and 8 were uniquely discovered in type II VcMADS proteins. All MADS-box proteins contained at least one of the four motifs, i.e., motifs 1, 2, 4, and 7, of which motif 2 was the most widely distributed one. Nearly all VcMADS proteins contained motif 2, with the exceptions of VcMADS52 and VcMADS111. Additionally, 90.4% of the proteins contained both motifs 1 and 2, and 69.1% of the proteins contained motifs 1, 2, 4, and 7.

Table 1.
Conserved motifs of blueberry MADS-box proteins.
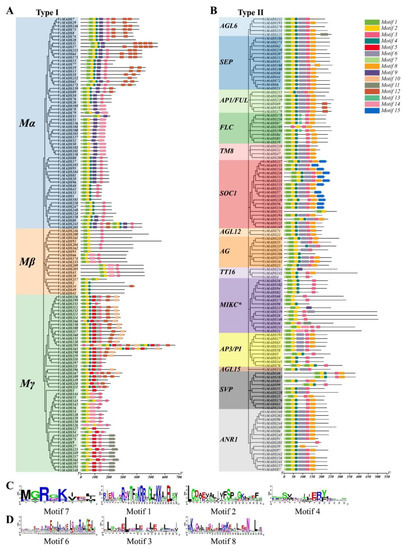
Figure 5.
Conserved motif analysis of 249 VcMADS proteins. (A,B) Conserved motifs of type I and type II MADS-box proteins, respectively. (C) The 4 motifs located in the MADS domain region. (D) The 3 motifs found in the K-box domain region.
We performed sequence alignment and generated sequence logos to check the motif conservation in the MADS and K-box domains. The motifs were ranked according to the position of the protein sequence (Figure 5C,D). The results show that motif 1 contained the most conservative amino acid sites (RQVTFSKRRNGLFKK), while motif 6 showed highly variable sites compared with the other motifs. Highly conservative leucine was found at many sites of motifs 2, 3, 4, and 8. Motif 7 comprised eight amino acids, which was the shortest one among the motifs. The first five amino acids (MGRGK) of motif 7 showed moderate conservation.
2.5. Cis-Acting Element Analysis of MADS-Box Genes
Promoter cis-acting element analysis of MADS-box genes in blueberry showed that 6511 cis-acting elements were identified in the promoter regions of 249 VcMADS genes. We visualized the type and number of cis-acting elements on each gene and clustered them according to gene types and subfamilies (Figure 6A). The results showed that the average number of hormone (gibberellin, methyl jasmonate, and salicylic acid)-responsive elements of type II genes were higher than that of type I genes. However, the average number of auxin-responsive elements and ethylene-responsive elements was lower than that of type I genes. The resistance-related (MYB binding site, oxygen, and wound) response elements of the type II gene were also higher than that of type I genes. In addition, we further investigated the differences in the number of element types in different subfamilies. In type I genes, the average numbers of methyl jasmonate (MeJa)-responsive elements, MYB binding site elements, salicylic acid (SA)-responsive elements, low-temperature-responsive elements, ethylene (ETH)-responsive elements, and meristem regulatory elements in each Mβ subfamily gene was higher than those in the Mα and Mγ subfamilies. Except for oxygen-related elements and ETH-responsive elements, there was no significant difference in the distribution of Mα and Mγ subfamily genes between element types (Figure 6B). We classified and counted the cis-acting elements in the promoter regions. The distribution of elements in the VcMADS genes was analyzed (Figure 6C). The elements can be divided into four categories, namely, light response, hormone response, stress response, and growth and development regulation. Hormone response cis-acting elements was the largest group within them, accounting for 47.2% of the total cis-acting elements. At least one kind of hormone response element was predicted in each gene. The number of cis-acting elements responding to abscisic acid (ABA), SA, andMeJa was higher than other responsive elements. A total of 1797 (27.6%) cis-acting elements associated with light response were found, including light response elements and MYB binding sites related to light response. Furthermore, the elements associated with light response were identified in all VcMADS genes. Stress response elements contained 758 (11.6%) components, which encompassed four subcategories: defense and stress response, low-temperature response, MYB binding site related to drought response, and wound response. There were 821 (12.6%) cis-acting elements that respond to the control of growth and development, including oxygen-related elements, circadian control, the endosperm, the meristem, and the MYB binding site involved in flavonoid biosynthetic genes regulation.
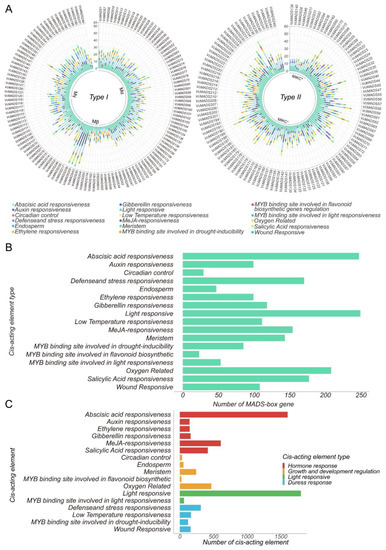
Figure 6.
Cis-acting element prediction in blueberry MADS-box genes. (A) The number of cis-acting elements of type I and type II MADS-box genes. (B) The number of cis-acting elements distributed in MADS-box genes. (C) The total number of cis-acting elements, as well as the category.
2.6. Collinearity and Evolutionary Analyses of MADS-Box Genes
The collinearity results showed that a total of 243 collinear gene pairs with whole-genome replication or segmental replication events were identified, among which 241 duplicate events occurred between different chromosomes and 2 duplicate events occurred on the same chromosome (Figure 7A). The results also indicated that 46.1% of the duplication events were found in type I MADS-box genes, involving 104 type I genes. A total of 103 type II MADS-box genes were discovered that experienced duplication events, accounting for 95.4% of all type II genes. Two or more genes that were located within a 200 kb region on the same chromosome containing were defined as tandem duplication. We found tandem duplication events for 12 VcMADS gene pairs (Figure 3), with 7 duplication events occurring in type I MADS-box genes and 5 duplication events occurring in type II genes. The Ka/Ks values of collinear gene pairs were calculated to unveil the evolutionary selection pressures of VcMADS genes (Figure 7B). The results indicated that most of the collinear VcMADS gene pairs had Ka/Ks values of less than 1, suggesting that most genes may have undergone selective pressure for purification during evolution.
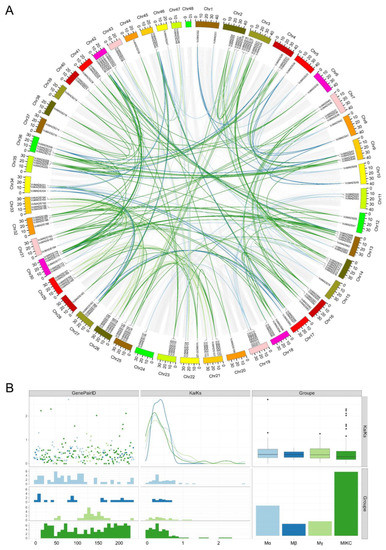
Figure 7.
Collinearity of blueberry MADS-box genes and evolutionary selection pressure analyses. (A) The collinearity of the VcMADS genes. Light blue, dark blue, apricot, and green represent Mα, Mβ, Mγ, and MIKC family gene pairs, respectively. (B) The Ka/Ks distribution of gene pairs. Gene pairs belonging to different subfamilies are shown in light blue, dark blue, apricot, and green, respectively.
2.7. Expression Patterns of MADS-Box Genes during Blueberry Fruit Ripening
Based on the transcriptomic data of blueberry fruits, we found that 137 VcMADS genes showed non-zero FPKM values during six fruit developmental stages. We filtered 27 genes based on an FPKM threshold of 10 from stages S3 to S8 and a heatmap was plotted to show their expression patterns (Figure 8A). Most genes showed lower transcript abundances in stages S3 and/or S4 than those of stages S5, S6, and S8. Generally, genes from the same subfamily displayed similar expression patterns. For instance, AG genes were primarily expressed in the S3 and S4 stages, and FLC genes were commonly expressed from stages S3 to S5. The expression levels of type I MADS-box genes and type II genes differed greatly at the fruit maturation stages. Ten VcMADS genes were highly expressed during fruit ripening, of which seven genes belonged to the SEP subfamily and three genes belonged to the AP1/FUL subfamily. SEP genes and AP1/FUL genes exhibited distinct expression patterns at six stages of fruit ripening. SEP genes showed high expression levels and a general trend consistent with the fruit ripening process. The FPKM values of AP1/FUL subfamily genes increased sharply from stages S5 to S8 and reached summits in stage S5.
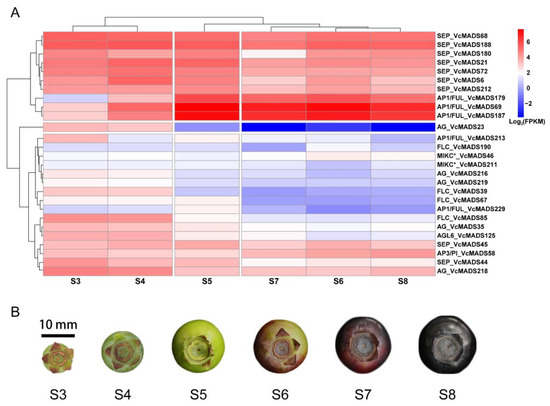
Figure 8.
The heat map of 27 VcMADS genes highly expressed during blueberry fruit ripening. (A) Expression values were quantified as fragments per kilobase per million reads (FPKM) and normalized by taking logarithms. The rows and columns of the heat map were clustered. The row clustering clustered genes with similar expression patterns from S3 to S8, and the column clustering clustered similar stages based on the expression levels of 27 genes in different stages. (B) Six different stages of blueberry during fruit ripening. Blueberry fruit mainly expanded in the S3–S5 stages and changed color in the S6–S8 stages.
Nine genes—namely, VcMADS6, VcMADS44, VcMADS45, VcMADS180, VcMADS188, and VcMADS212 of the SEP subfamily; VcMADS35 of the AG subfamily; VcMADS58 of the AP3/PI subfamily; and VcMADS125 of the AGL6 subfamily—were selected to check relative expression levels using quantitative PCR (qPCR). The expression patterns of those genes at different stages were consistent with those of the transcriptomic data (Figure 9). The results show that six SEP family genes (VcMADS6, VcMADS44, VcMADS45, VcMADS180, VcMADS188, and VcMADS212) were highly expressed during fruit ripening. Among them, the expressions of VcMADS6 and VcMADS45 were higher in the S4 and S5 stages. VcMADS44 had a high expression level in the S3 stage and a low expression level in the S8 stage. The expression level of VcMADS180 did not show dramatic changes except during stage S5. VcMADS188 was upregulated from stages S3 to S5, while it was downregulated in stages S6 and S7. The highest expression level of VcMADS188 was observed at full ripening (S8 stage). The expression level of VcMADS212 at stage S5 was significantly higher than that of other stages. VcMADS58 belongs to the AP3/PI family and is mainly expressed in the S6–S8 stages and the highest expression level was in the S6 stage, with the relative expression reaching 2.27. Compared with other stages, the expression level of VcMADS58 shows a very different expression level alteration. It decreased from stages S3 to S5 and sharply increased to its maximum in stage S6. VcMADS35 belonged to the AG family and its expression level was downregulated from the S3 stage. The lowest expression level of VcMADS35 was seen at full maturity. VcMADS125 belonged to the AGL6 family and was mainly expressed in the S4 stage, with low expression levels in stages S6 and S8.
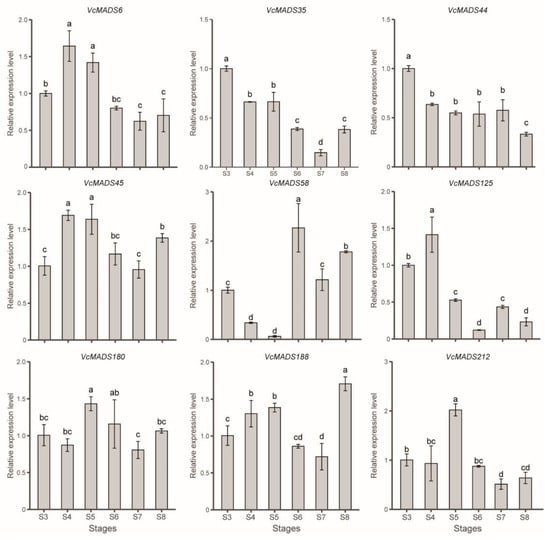
Figure 9.
Relative expression of blueberry MADS-box genes in the fruit developmental stages. Lowercase letters represent significant differences. Different labeled letters for stages indicate significant differences in expression levels (p < 0.05, LSD test).
3. Discussion
3.1. Gene Duplication Led to the Massive Replication of VcMADS Genes
MADS-box genes are ubiquitous in plants and play important roles in plant growth and development, stress response, signal transduction, and other processes. They were systematically identified and analyzed in some species, such as Arabidopsis thaliana (107 genes) [8], Solanum lycopersicum (131 genes) [29], Zea mays (211 genes) [33], and Rhododendron ovatum (77 genes) [34]. A total of 249 VcMADS genes were identified in the genome of blueberry, including 131 type I genes and 118 type II genes. It was found that the number of MADS-box genes in the blueberry was significantly higher than that of previously reported species. This is probably because blueberry has undergone multiple genome-wide doubling events during evolution [35]. Previous studies suggested that the blueberry experienced at least one paleohexaploidization event and two whole-genome duplication (WGD) events during speciation [34,36]. The most recent WGD event occurred about 9.04 million years ago as an independent doubling event [37]. Blueberry and Rhododendron ovatum shared the first two doubling events, but the number of MADS-box genes in Rhododendron ovatum was less than that in blueberry, suggesting that independent doubling events in blueberry evolution led to the massive expansion of VcMADS genes. Furthermore, the collinearity analysis identified 207 VcMADS genes with genome-wide or segmentary replication events, and 24 VcMADS genes with tandem replication events, indicating that small-scale segmentary and tandem replication events also contributed to VcMADS gene expansion in the blueberry genome. Tandem duplication events and segmental duplication events were both significant in the replication of the VcMADS gene, with segmental duplication events possibly acting as the primary driving force.
3.2. Retention and Potential Roles of TM8 Genes in Blueberry
Type II VcMADS genes were clustered into 14 subfamilies, including the TM8 gene subfamily. TM8 genes were absent in a variety of plants, including Arabidopsis and rice [38,39]. Although TM8 genes were present in tomatoes and cucumbers, only some members were found. This is probably because TM8 genes underwent multiple independent losses during the evolution of angiosperms and were not retained when coupling with genome duplication events [39,40]. We found four genes classified into the TM8 subfamily in the blueberry genome, i.e., VcMADS32, VcMADS47, VcMADS208, and VcMADS210. As the ancient subfamily of the MADS-box gene family, few studies on the function of TM8 genes have been conducted, with the exceptions of those involved in the development of female flowers and seeds [38,39]. Four TM8 genes were expressed from stages S5 to S8 during berry ripening, and all reached the maximum during stage S6. This indicated that the TM8 genes acted in pivotal roles in regulating fruit ripening. However, further studies should be conducted to confirm the regulation function of TM8 genes.
3.3. The Possible Mechanism of Extranuclear VcMADS Protein Involved in Gene Regulation
Transcription factors are proteins that bind to specific genes and regulate their transcription, which usually occurs in the eukaryotic nucleus. In this study, 161 VcMADS proteins were predicted to be in the nucleus. There were 86 proteins found outside the nucleus, with 54 of them found in chloroplasts and mitochondria. Two proteins (VcMADS145 and VcMADS160) were predicted to be located at both the nucleus and cytoplasm. Chloroplasts and mitochondria contain DNA and RNA, and both include the machinery necessary for gene transcription. As a result, some VcMADS genes acted in roles in regulating photosynthesis and respiration-related genes. In addition, some transcription factor proteins can shuttle between the nucleus and cytoplasm through the phosphorylation/dephosphorylation nucleocytoplasmic trafficking mechanism [41]. It was reported that brassinosteroid (BR) treatment could recruit the cytoplasmic transcription factor BRASSINAZOLE RESISTANT1 (BZR1, a class of plant-specific transcription factors with noncanonical bHLH domains) into the nucleus and regulate the transcription process of BR response genes [42]. This indicated that the VcMADS proteins located outside the nucleus possibly enter the nucleus when needed for gene regulation. Nevertheless, experimental validation is necessary for testing this hypothesis in future studies.
3.4. Deletion of K-Box Domains in the MIKC* and the FLC Subfamilies
The type II (MIKC) MADS-box proteins generally contain four featured domains, namely, the MADS domain, I domain, K-box domain, and C domain. The MADS domain is the most conserved region in MADS-box proteins, and the K-box domain is moderately conserved in MIKC-type proteins. However, the function of the C domain has not been clearly defined due to its high variability [43]. Therefore, MIKC-type proteins are commonly considered to contain both MADS and K-box domains. The K-box was absent from 18 proteins among the 118 MIKC-type proteins identified in the present study. Four FLC subfamily members and all MIKC* subfamily members did not contain K-box domains. The absence of K-box domains in the MIKC* subfamily was reported in some species, including foxtail millet [23], American beautyberry [44], and litchi [45], which possibly related to MIKC* being a class of genes combining both features of type I (Mδ) and type II genes. Some studies indicated that MIKCc genes may be the most ancient members of MADS-box genes, and type I genes probably evolved from MIKCc genes [10]. This suggested that MIKC* genes may be a class of transition genes retained after the loss of the K-box domain during the evolution of MIKCc genes. FLC genes were thought to be the most frequently lost MADS-box genes in plants [44], which indicated that the missing FLC genes contributed to K-box domain loss in the blueberry genome. The K-box domain contained K1, K2, and K3 subdomains, which corresponded to motifs 6, 3, and 8, respectively. Type II proteins lacking the K-box domain all contained one or two motifs, which is consistent with the idea that the K-box domain was lost during evolution. The contribution of the three subdomains to the K-box domain function was not the same. Some MADS-box proteins gained their K-box domain functions mainly through motifs within them [11]. This indicates that MIKC-type VcMADS proteins with a partial K-box domain possibly functionalize through conserved domains.
3.5. Potential Roles of MADS-Box Genes in Fruit Ripening in Blueberry
As an important transcriptional regulator in eukaryotes, MADS-box was widely studied in the process of fruit development. Two FUL genes in tomato, namely, TDR4/FUL1 and MBP7/FUL2, were involved in cell wall modification, cuticle development, and volatile accumulation during fruit ripening [33]. The expression of four SEP genes in bananas caused an increase in ethylene content and affected banana maturation [46]. The expression of SEP4-like genes in strawberry was necessary for berry ripening [37]. The MADS-box gene is a critical component of the network that controls fruit ripening [40]. Blueberry fruit firmness changed dramatically during the fruit ripening stages. We found that the expression patterns of 16 genes potentially accompanied a decline in fruit firmness, among which 9 genes belonged to the SEP gene subfamily with high FPKM values. The results were further validated through quantitative PCR, which means that SEP genes may play an important role in the ripening process of fruits, which is consistent with the results of the previous study [47]. The three AP1/FUL genes were highly expressed from stages S5 to S8. This indicates that the three AP1/FUL genes were also significant for fruit ripening. However, the specific regulatory mechanisms of the MADS-box gene in the blueberry fruit softening process need to be explored further in the future.
4. Materials and Methods
4.1. Identification of MADS-Box Genes in Blueberry
The amino acid sequences of the blueberry cultivar ‘Draper’ was downloaded from the Vaccinium Genome Database (https://www.vaccinium.org/ (accessed on 5 September 2022) [35]. A local protein database was constructed using the MAKEBLASTDB function in the BLAST+ 2.13.0 toolkit. The MADS protein sequences (M-type_MADS and MIKC_MADS types) of Arabidopsis thaliana were downloaded from Plant Transcription Factor Database (http://planttfdb.gao-lab.org/ (accessed on 7 September 2022)) and used to query the blueberry MADS-box homologs. Hit targets with an e-value less than 1 × 10−20 were kept for further analyses. Other parameters of BLASTP were kept as default. In addition, we performed HMMER [48] analysis using seed alignment of the MADS-box domain (PF00319), which was obtained from the Pfam database (https://pfam.xfam.org/ (accessed on 8 September 2022)) [49]. Blueberry MADS-box gene candidates were screened by using an e-value threshold of 1 × 10−10. We merged the outputs from BLAST and HMMER and removed redundant results for downstream analyses. The conserved domains of candidates were verified using the online tool NCBI Conserved Domain Search (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi (accessed on 12 September 2022)). The members that contained a full MADS domain were recognized as blueberry MADS-box genes. The biochemical properties of blueberry MADS-box proteins were analyzed with ExPASy protparam (https://web.expasy.org/protparam/ (accessed on 14 September 2022). Subcellular localization of blueberry MADS-box proteins was predicted using WoLF PSORT (https://wolfpsort.hgc.jp/ (accessed on 15 September 2022).
4.2. Phylogenetic Analysis and Classification of Blueberry MADS-Box Genes
The amino acid sequences of Arabidopsis MADS-box were downloaded from TAIR (https://www.arabidopsis.org/ (accessed on 26 September 2022) and were aligned with the blueberry MADS-box proteins using ClustalW [50] with default parameters. Using the maximum likelihood method, a primary phylogenetic tree was constructed using IQ-TREE (multicore version 2.2.0) [51] with 1000 bootstraps. Based on the classification rules of the Arabidopsis MADS-box, the blueberry MADS-box was divided into type I and type II in the primary tree. Type I and II MADS-boxes in Arabidopsis were used to build the type I and type II phylogeny trees with corresponding blueberry MADS-boxes. In addition, TM8 is a class of genes belonging to the type II MADS-box, but such genes were lost in a variety of plants and are not present in Arabidopsis. To determine the presence of the TM8 gene in blueberry, we added two TM8 genes, namely, Solyc03G019710.2.1 in tomato [29] and Calam.16G1144000.2 in American beautyberry [44], as grouping criteria for TM8 class genes when constructing the type II phylogenetic tree. The online tree plotting tool iTOL (https://itol.embl.de/upload.cgi (accessed on 29 September 2022) [52] was used to complement the phylogenetic tree.
4.3. Chromosomal Location and Conserved Motif Analyses of MADS-Box in Blueberry
The distribution information of the blueberry MADS-box gene on the chromosome was extracted from the genome general feature file. We used the online tool MG2C (http://mg2c.iask.in/mg2c_v2.1/ (accessed on 2 October 2022) to draw the chromosome location of blueberry MADS-box genes. MEME (http://meme-suite.org/ (accessed on 6 October 2022) [53] was used to analyze the conserved motif of blueberry MADS-box genes and the maximum number of conserved motifs was set to 15. The gene structures and conserved motifs of the MADS-box family in the blueberry were visualized using Tbtools [54].
4.4. Cis-Acting Element, Collinearity, and Evolutionary Selection Analysis of Blueberry MADS-Box Genes
We extracted 1500 bp sequences of blueberry MADS-box genes upstream of the start codon using Tbtools. Sequences were submitted to the website PlantCare (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/ (accessed on 15 October 2022) [55] for cis-acting element analyses. MCScanX [43] and Circos [56] were used to estimate the collinearity relationships of MADS-box genes and to plot the collinearity figure, respectively. Furthermore, to understand the selection pressure on the MADS-box genes in blueberry, Ka/Ks values of MADS-box gene pairs were calculated using KaKs_Calculator [57].
4.5. Plant Materials
The southern highbush blueberry (V. corymbosum cv. ‘Star’) used in this study was collected from the blueberry Germplasm Resource Nursery of Zhejiang Normal University. According to Zifkin et al.’s method for identifying the developmental stages of blueberry fruits [58], fruit samples were collected from stages S3 to S8. The berries were immediately frozen in liquid nitrogen and stored at −80 °C for downstream studies. Total RNA from blueberry fruits was extracted using the CTAB method. Synthesis of the first strand of complementary DNA was performed using a reverse transcription kit (Vazyme, Nanjing, China).
4.6. Expression Pattern of MADS-Box Genes during Blueberry Fruit Ripening
Transcript abundance of blueberry fruits at different developmental stages, represented by fragments per kilobase per million reads (FPKM), was obtained from our previous transcriptomic data. MADS-box genes that showed significantly different expression levels between ripening stages were screened. Nine genes were selected for the qPCR assay to check their expression pattern. Primers were designed using NCBI Primer-BLAST and were synthesized by the Sangon Biotech Company (Shanghai, China). The specificity of primers was verified using agarose electrophoresis and dissolution curves. qPCR was performed using the QuantStudio™ 1 real-time quantitative system. The total volume of the reaction system was 10 μL with 5 μL 2× SYBR Green qPCR premix, 2 μL double distillation H2O, 1 μL complementary DNA, and 1 μL for forward and reverse primers. The VcGAPDH gene was chosen as the internal reference gene. The primer sequences used in the current study are shown in Table S2. The procedure for qPCR was 10 min at 95 °C, followed by 40 cycles of 95 °C for 30 s, 57 °C for 30 s, and 72 °C for 30 s. The 2−ΔΔCt method was used to calculate the relative expression, with three biological replicates per sample. R scripts were used to analyze the relative expression and to draw the histogram.
Supplementary Materials
The following supporting information can be downloaded from https://www.mdpi.com/article/10.3390/plants12071424/s1, Table S1. Protein information analysis of MADS box gene family members in blueberry. Table S2. Primers used in this study.
Author Contributions
Conceptualization, Y.Z. and W.G.; methodology, X.W., Q.H. and Z.S.; software, X.W., J.L., W.L. and Y.Z.; validation, X.W., Y.Z. and W.C.; data curation, X.W., Q.H., Z.S., X.L. (Xiaoyi Li) and Y.Z.; writing—original draft preparation, X.W., Q.H. and Y.Z.; writing—review and editing, G.C.B., X.L. (Xiaoyi Li), X.L. (Xiaoying Lu), Y.L. and L.X.; visualization, X.W., Y.Z. and Y.L.; supervision, Y.Z., L.X. and W.G.; project administration, Y.Z. and W.G.; funding acquisition, W.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Key Research Project of Science and Technology Department of Zhejiang Province, grant number 2021C02066-9; the Key Research and Development Program of Science and Technology Department of Zhejiang Province, grant number 2018C02007.
Data Availability Statement
The RNA-seq datasets presented in this study were deposited in the National Genomics Data Center (NGDC) database and are accessible through GRA accession code CRA010224 (https://bigd.big.ac.cn/gsa/browse/CRA010224, accessed on 19 March 2023).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Passmore, S.; Maine, G.T.; Elble, R.; Christ, C.; Tye, B. Saccharomyces cerevisiae protein involved in plasmid maintenance is necessary for mating of MATα cells. J. Mol. Biol. 1988, 204, 593–606. [Google Scholar] [CrossRef] [PubMed]
- Yanofsky, M.F.; Ma, H.; Bowman, J.L.; Drews, G.N.; Feldmann, K.A.; Meyerowitz, E.M. The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 1990, 346, 35–39. [Google Scholar] [CrossRef]
- Schwarz-Sommer, Z.; Huijser, P.; Nacken, W.; Saedler, H.; Sommer, H. Genetic control of flower development by homeotic genes in Antirrhinum majus. Science 1990, 250, 931–936. [Google Scholar] [CrossRef] [PubMed]
- Norman, C.; Runswick, M.; Pollock, R.; Treisman, R. Isolation and properties of cDNA clones encoding SRF, a transcription factor that binds to the c-fos serum response element. Cell 1988, 55, 989–1003. [Google Scholar] [CrossRef] [PubMed]
- Tröbner, W.; Ramirez, L.; Motte, P.; Hue, I.; Huijser, P.; Lönnig, W.E.; Saedler, H.; Sommer, H.; Schwarz-Sommer, Z. GLOBOSA: A homeotic gene which interacts with DEFICIENS in the control of Antirrhinum floral organogenesis. EMBO J. 1992, 11, 4693–4704. [Google Scholar] [CrossRef]
- Alvarez-Buylla, E.R.; Pelaz, S.; Liljegren, S.J.; Gold, S.E.; Burgeff, C.; Ditta, G.S.; de Pouplana, L.R.; Martínez-Castilla, L.; Yanofsky, M.F. An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proc. Natl. Acad. Sci. USA 2000, 97, 5328–5333. [Google Scholar] [CrossRef]
- De Bodt, S.; Raesm, J.; Florquin, K.; Rombauts, S.; Rouzé, P.; Theissen, G.; van de Peer, Y. Genome wide structural annotation and evolutionary analysis of the type I MADS-box genes in plants. J. Mol. Evol. 2003, 56, 573–586. [Google Scholar] [CrossRef]
- Pařenicová, L.; de Folter, S.; Kieffer, M.; Horner, D.S.; Favalli, C.; Busscher, J.; Cook, H.E.; Ingram, R.M.; Kater, M.M.; Davies, B.; et al. Molecular and phylogenetic analyses of the complete MADS-Box transcription factor family in Arabidopsis: New openings to the MADS world. Plant Cell 2003, 15, 1538–1551. [Google Scholar]
- Becker, A.; Theißen, G. The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol. Phylogenet. Evol. 2003, 29, 464–489. [Google Scholar] [CrossRef]
- Henschel, K.; Kofuji, R.; Hasebe, M.; Saedler, H.; Münster, T.; Theißen, G. Two ancient classes of MIKC-type MADS-box genes are present in the moss physcomitrella patens. Mol. Biol. Evol. 2002, 19, 801–814. [Google Scholar] [CrossRef]
- Kaufmann, K.; Melzer, R.; Theißen, G. MIKC-type MADS-domain proteins: Structural modularity, protein interactions and network evolution in land plants. Gene 2005, 347, 183–198. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yin, X.; Cheng, C.; Wang, H.; Guo, R.; Xu, X.; Zhao, J.; Zheng, Y.; Wang, X. Evolutionary and expression analysis of a MADS-box gene superfamily involved in ovule development of seeded and seedless grapevines. Mol. Genet. Genom. 2015, 290, 825–846. [Google Scholar] [CrossRef]
- Zhao, P.; Zhang, J.; Chen, S.; Wu, J.; Xia, J.; Sun, L.; Ma, S.; Xiang, C. Arabidopsis MADS-box factor AGL16 is a negative regulator of plant response to salt stress by downregulating salt-responsive genes. New Phytol. 2021, 232, 2418–2439. [Google Scholar] [CrossRef] [PubMed]
- Qu, G.; Zheng, T.; Liu, G.; Wang, W.; Zang, L.; Liu, H.; Yang, C. Overexpression of a MADS-box gene from birch (Betula platyphylla) promotes flowering and enhances chloroplast development in transgenic tobacco. PLoS ONE 2013, 8, e63398. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, S.; Spielman, M.; Schulz, R.; Oakey, R.J.; Kelsey, G.; Salazar, A.; Zhang, K.; Pennell, R.; Scott, R.J. Transcriptional profiles underlying parent-of-origin effects in seeds of Arabidopsis thaliana. BMC Plant Biol. 2010, 10, 72. [Google Scholar] [CrossRef]
- Day, R.C.; Herridge, R.P.; Ambrose, B.A.; Macknight, R.C. Transcriptome analysis of proliferating Arabidopsis endosperm reveals biological implications for the control of syncytial division, cytokinin signaling, and gene expression regulation. Plant Physiol. 2008, 148, 1964–1984. [Google Scholar] [CrossRef]
- Masiero, S.; Colombo, L.; Grini, P.E.; Schnittger, A.; Kater, M.M. The emerging importance of type I MADS-Box transcription factors for plant reproduction. Plant Cell 2011, 23, 865–872. [Google Scholar] [CrossRef]
- Causier, B.; Schwarz-Sommer, Z.; Davies, B. Floral organ identity: 20 years of ABCs. Semin. Cell Dev. Biol. 2010, 21, 73–79. [Google Scholar] [CrossRef]
- Litt, A.; Kramer, E.M. The ABC model and the diversification of floral organ identity. Semin. Cell Dev. Biol. 2010, 21, 129–137. [Google Scholar] [CrossRef]
- Theißen, G.; Melzer, R.; Rümpler, F. MADS-domain transcription factors and the floral quartet model of flower development: Linking plant development and evolution. Development 2016, 143, 3259–3271. [Google Scholar] [CrossRef]
- Moser, M.; Asquini, E.; Miolli, G.V.; Weigl, K.; Hanke, M.; Flachowsky, H.; Si-Ammour, A. The MADS-Box gene MdDAM1 controls growth cessation and bud dormancy in apple. Front. Plant Sci. 2020, 11, 1003. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Ma, J.; Luo, D.; Hou, X.; Ma, F.; Zhang, Y.; Meng, Y.; Zhang, H.; Guo, W. CaMADS, a MADS-box transcription factor from pepper, plays an important role in the response to cold, salt, and osmotic stress. Plant Sci. 2019, 280, 164–174. [Google Scholar] [CrossRef]
- Zhao, W.; Zhang, L.; Xu, Z.; Fu, L.; Pang, H.; Ma, Y.; Min, D. Genome-Wide analysis of MADS-Box genes in foxtail millet (Setaria italica L.) and functional assessment of the role of SiMADS51 in the drought stress response. Front. Plant Sci. 2021, 12, 659474. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Zhang, Q.; He, D.; Zhou, Y.; Ni, H.; Tian, D.; Chang, G.; Jing, Y.; Lin, R.; Huang, J.; et al. AGAMOUS-LIKE67 Cooperates with the histone mark reader EBS to modulate seed germination under high temperature. Plant Physiol. 2020, 184, 529–545. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Chen, K.; Grierson, D. A critical evaluation of the role of ethylene and MADS transcription factors in the network controlling fleshy fruit ripening. New Phytol. 2019, 221, 1724–1741. [Google Scholar] [CrossRef]
- Daminato, M.; Guzzo, F.; Casadoro, G. A SHATTERPROOF-like gene controls ripening in non-climacteric strawberries, and auxin and abscisic acid antagonistically affect its expression. J. Exp. Bot. 2013, 64, 3775–3786. [Google Scholar] [CrossRef]
- Vrebalov, J.; Ruezinsky, D.; Padmanabhan, V.; White, R.; Medrano, D.; Drake, R.; Schuch, W.; Giovannoni, J. A MADS-Box gene necessary for fruit ripening at the tomato ripening-inhibitor (Rin) locus. Science 2002, 296, 343–346. [Google Scholar] [CrossRef]
- Duan, W.; Song, X.; Liu, T.; Huang, Z.; Ren, J.; Hou, X.; Li, Y. Genome-wide analysis of the MADS-box gene family in Brassica rapa (Chinese cabbage). Mol. Genet. Genom. 2015, 290, 239–255. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, J.; Hu, Z.; Guo, X.; Tian, S.; Chen, G. Genome-Wide analysis of the MADS-Box transcription factor family in Solanum lycopersicum. Int. J. Mol. Sci. 2019, 20, 2961. [Google Scholar] [CrossRef]
- Shao, Z.; He, M.; Zeng, Z.; Chen, Y.; Hanna, A.; Zhu, H. Genome-Wide identification and expression analysis of the MADS-Box gene family in sweet potato [Ipomoea batatas (L.) Lam]. Front. Genet. 2021, 12, 750137. [Google Scholar] [CrossRef]
- Qu, Y.; Bi, C.; He, B.; Ye, N.; Yin, T.; Xu, L. Genome-wide identification and characterization of the MADS-box gene family in Salix suchowensis. PeerJ 2019, 7, e8019. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Yang, Y.; Luo, W.; Yang, C.; Ding, P.; Liu, Y.; Qiao, L.; Chang, Z.; Geng, H.; Wang, P.; et al. Genome-wide identification and analysis of the MADS-box gene family in bread wheat (Triticum aestivum L.). PLoS ONE 2017, 12, e181443. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Chen, Z.; Xu, L.; Zhang, L.; Zou, Q. Genome-Wide analysis of the MADS-Box gene family in maize: Gene structure, evolution, and relationships. Genes 2021, 12, 1956. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Fang, H.; Wen, X.; Zhang, L. Phylogenetic and expression analysis of MADS-box genes in Rhododendron ovatum. Chin. Bull. Bot. 2022. [Google Scholar] [CrossRef]
- Colle, M.; Leisner, C.P.; Wai, C.M.; Ou, S.; Bird, K.A.; Wang, J.; Wisecaver, J.H.; Yocca, A.E.; Alger, E.I.; Tang, H.; et al. Haplotype-phased genome and evolution of phytonutrient pathways of tetraploid blueberry. Gigascience 2019, 8, giz012. [Google Scholar] [CrossRef]
- Yang, F.; Nie, S.; Liu, H.; Shi, T.; Tian, X.; Zhou, S.; Bao, Y.; Jia, K.; Guo, J.; Zhao, W.; et al. Chromosome-level genome assembly of a parent species of widely cultivated azaleas. Nat. Commun. 2020, 11, 5269. [Google Scholar] [CrossRef]
- Wang, Y.; Nie, F.; Shahid, M.Q.; Baloch, F.S. Molecular footprints of selection effects and whole genome duplication (WGD) events in three blueberry species: Detected by transcriptome dataset. BMC Plant Biol. 2020, 20, 250. [Google Scholar] [CrossRef]
- Daminato, M.; Masiero, S.; Resentini, F.; Lovisetto, A.; Casadoro, G. Characterization of TM8, a MADS-box gene expressed in tomato flowers. BMC Plant Biol. 2014, 14, 319. [Google Scholar] [CrossRef]
- Coenen, H.; Viaene, T.; Vandenbussche, M.; Geuten, K. TM8 represses developmental timing in Nicotiana benthamiana and has functionally diversified in angiosperms. BMC Plant Biol. 2018, 18, 129. [Google Scholar] [CrossRef]
- Gramzow, L.; En, G.U.N.T. Phylogenomics reveals surprising sets of essential and dispensable clades of MIKC(c)-group MADS-box genes in flowering plants. J. Exp. Zool. Part B Mol. Dev. Evol. 2015, 324, 353–362. [Google Scholar] [CrossRef]
- Fu, X.; Liang, C.; Li, F.; Wang, L.; Wu, X.; Lu, A.; Xiao, G.; Zhang, G. The rules and functions of nucleocytoplasmic shuttling proteins. Int. J. Mol. Sci. 2018, 19, 1445. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Wang, R.; Liu, M.; Yuan, W.; Zhao, Z.; Liu, X.; Peng, Y.; Yang, X.; Sun, Y.; Tang, W. Nucleocytoplasmic trafficking and turnover mechanisms of BRASSINAZOLE RESISTANT1 in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2021, 118, e2101838118. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Alhindi, T.; Al-Abdallat, A.M. Genome-Wide identification and analysis of the MADS-Box gene family in American beautyberry (Callicarpa americana). Plants 2021, 10, 1805. [Google Scholar] [CrossRef] [PubMed]
- Guan, H.; Wang, H.; Huang, J.; Liu, M.; Chen, T.; Shan, X.; Chen, H.; Shen, J. Genome-Wide identification and expression analysis of MADS-Box family genes in Litchi (Litchi chinensis Sonn.) and their involvement in floral sex determination. Plants 2021, 10, 2142. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Liu, M.; Wang, J.; Zhang, J.; Miao, H.; Wang, Z.; Jia, C.; Zhang, J.; Xu, B.; Jin, Z. Transcription factor MaMADS36 plays a central role in regulating banana fruit ripening. J. Exp. Bot. 2021, 72, 7078–7091. [Google Scholar] [CrossRef]
- Seymour, G.B.; Ryder, C.D.; Cevik, V.; Hammond, J.P.; Popovich, A.; King, G.J.; Vrebalov, J.; Giovannoni, J.J.; Manning, K. A SEPALLATA gene is involved in the development and ripening of strawberry (Fragaria × ananassa Duch.) fruit, a non-climacteric tissue. J. Exp. Bot. 2011, 62, 1179–1188. [Google Scholar] [CrossRef]
- Mistry, J.; Finn, R.D.; Eddy, S.R.; Bateman, A.; Punta, M. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 2013, 41, e121. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Zhang, Z. KaKs_Calculator 3.0: Calculating selective pressure on coding and non-coding sequences. Genom. Proteom. Bioinform. 2021, 20, 536–540. [Google Scholar] [CrossRef]
- Zifkin, M.; Jin, A.; Ozga, J.A.; Zaharia, L.I.; Schernthaner, J.P.; Gesell, A.; Abrams, S.R.; Kennedy, J.A.; Constabel, C.P. Gene expression and metabolite profiling of developing highbush blueberry fruit indicates transcriptional regulation of flavonoid metabolism and activation of abscisic acid metabolism. Plant Physiol. 2012, 158, 200–224. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).