Identification and Expression Analysis of Hexokinases Family in Saccharum spontaneum L. under Drought and Cold Stresses
Abstract
1. Introduction
2. Results
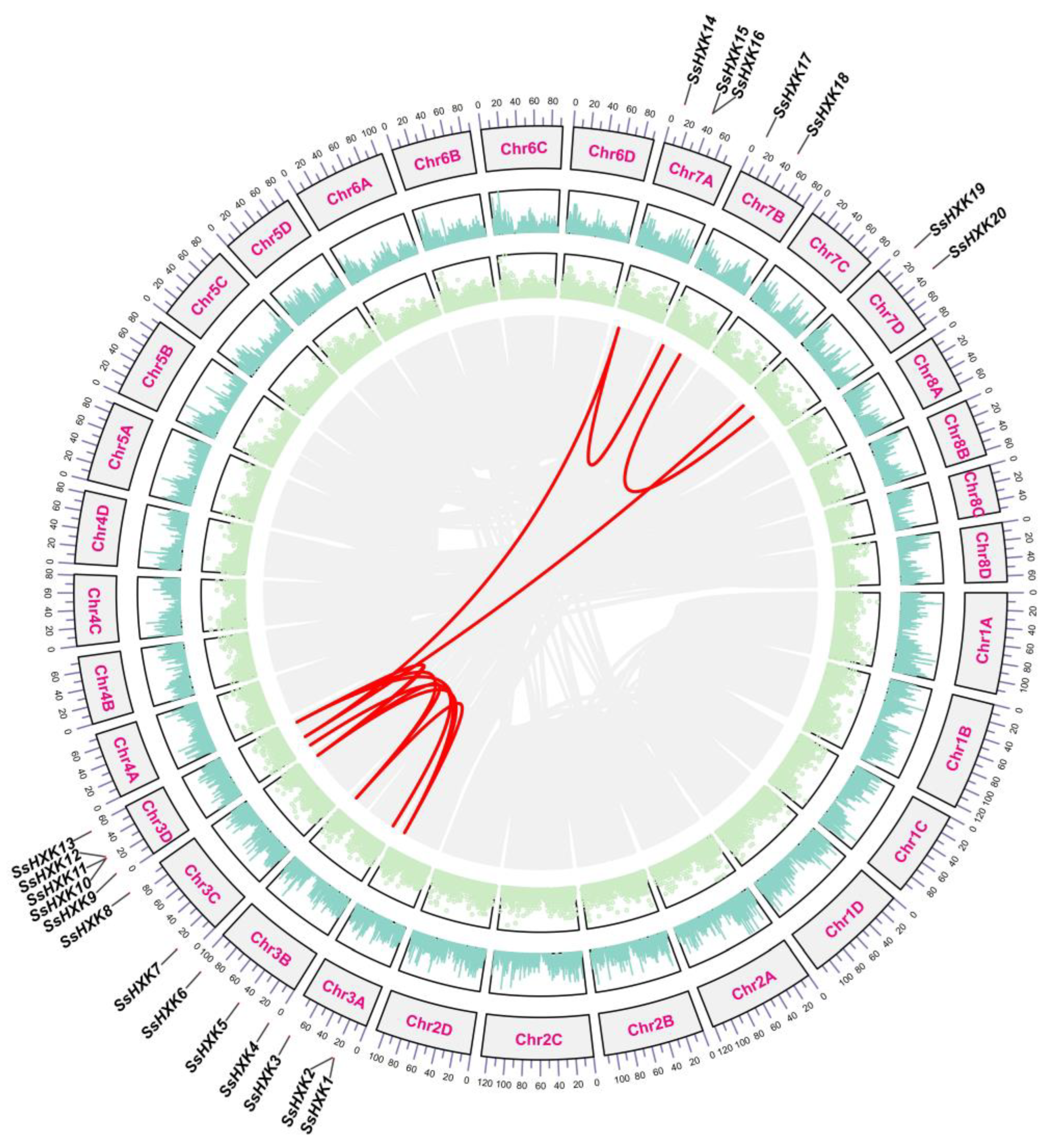
2.1. Identification, Physicochemical Properties and Chromosomal Distribution of SsHXK Genes
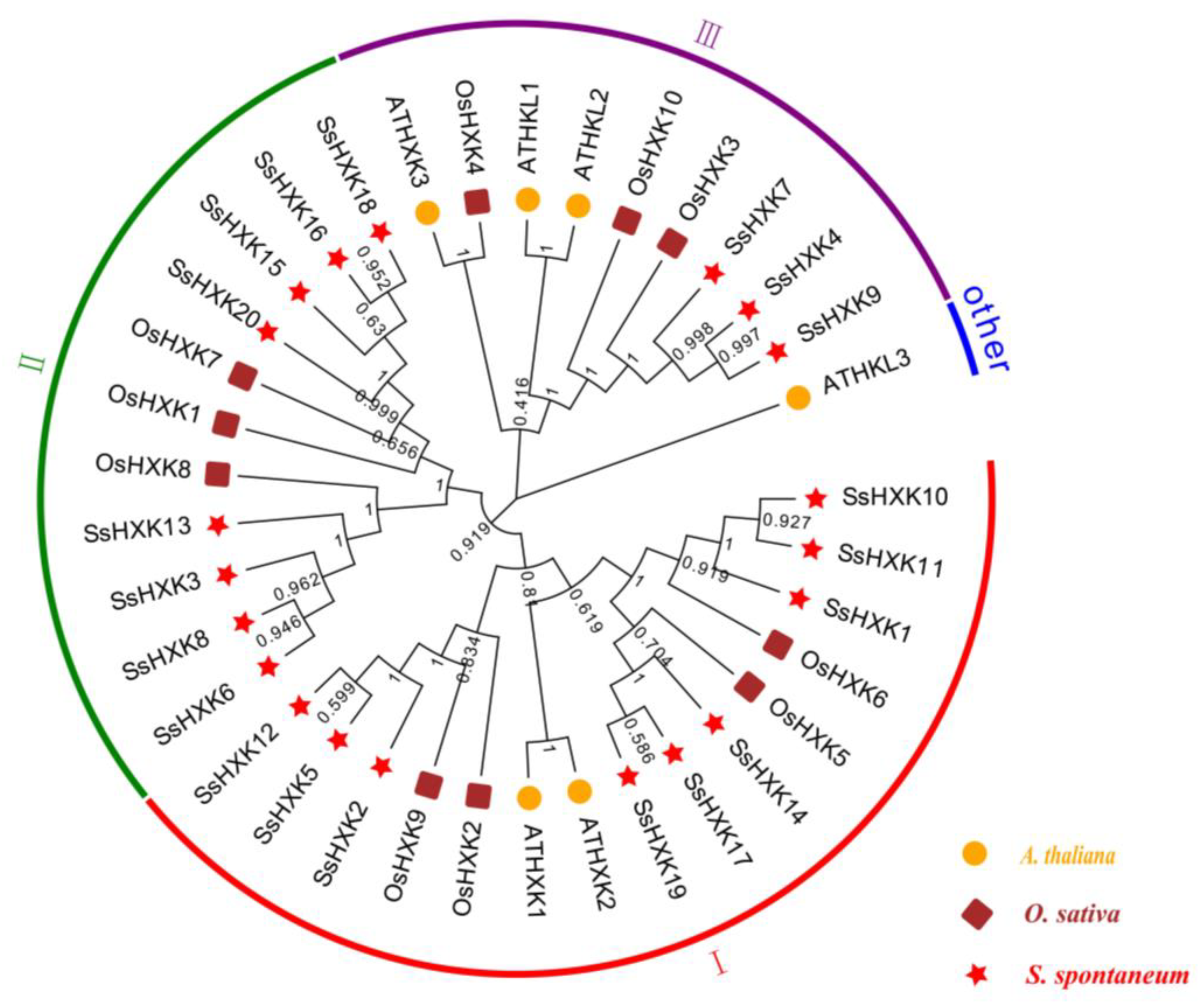
2.2. Phylogenetic Analysis and Synteny Analysis
2.3. Conserved Motifs and Gene Structures of the HXK Gene Family
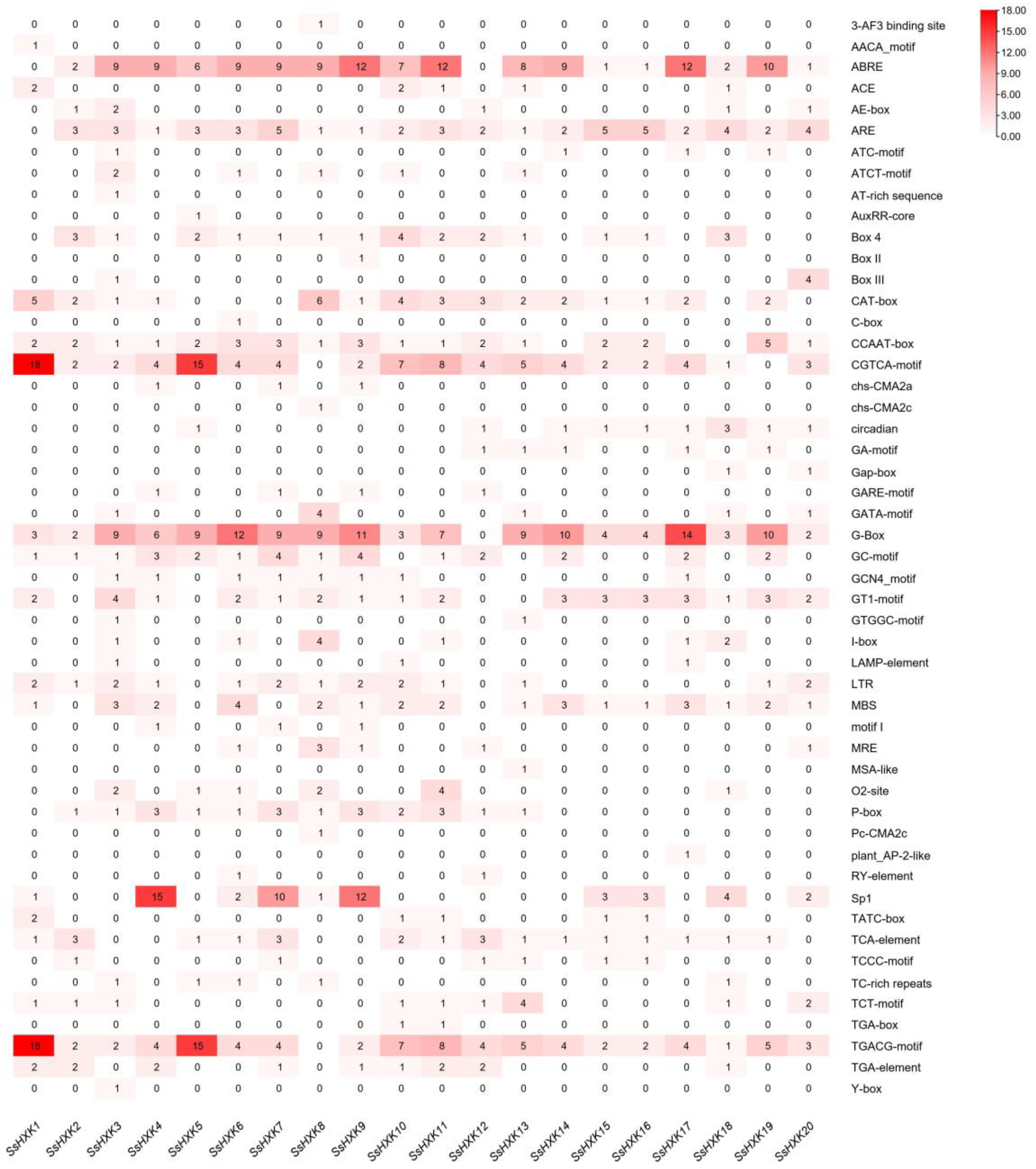
2.4. CREs Analysis of SsHXK Genes
2.5. Expression Pattern of SsHXK Genes during Saccharum Spontaneum Development
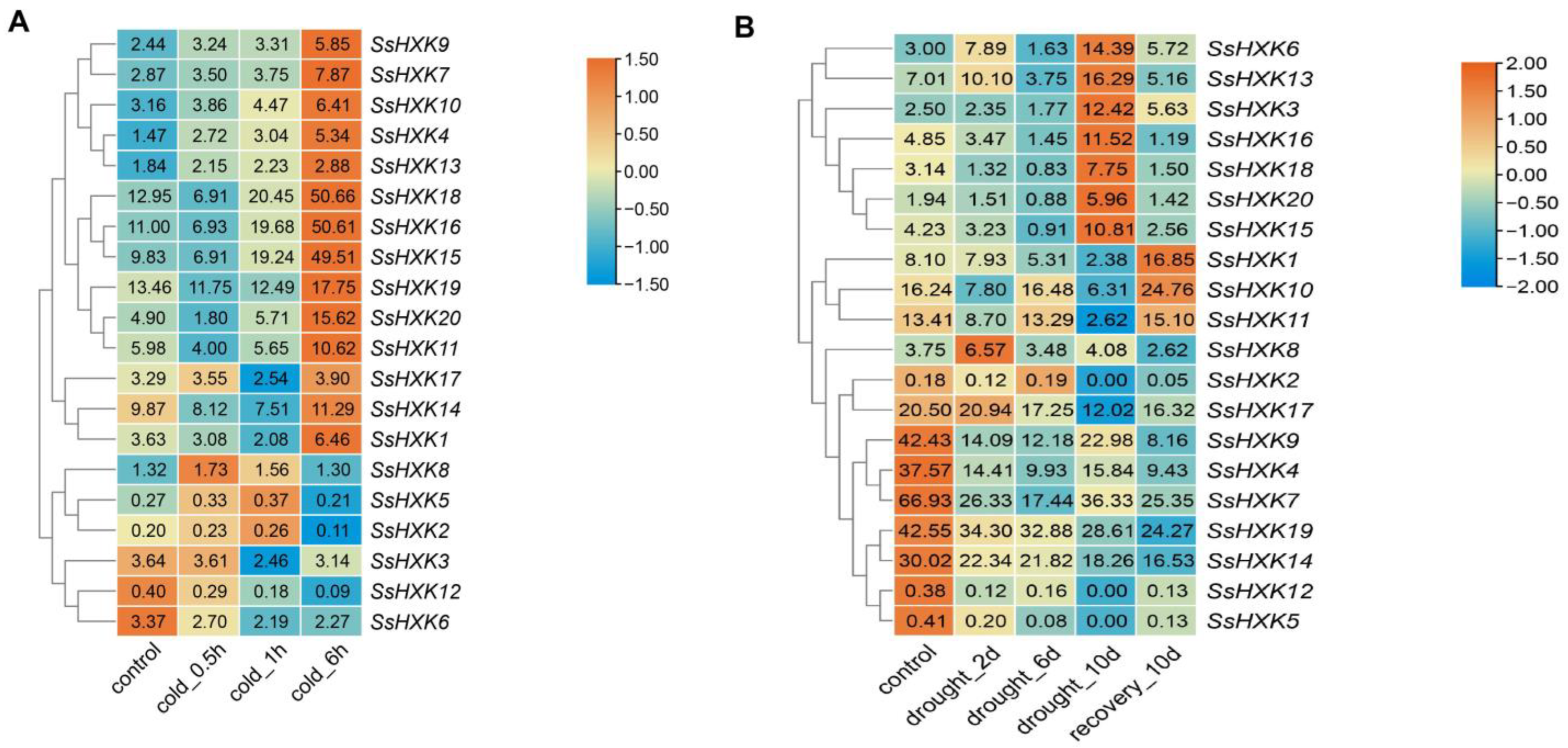
2.6. Expression Pattern of SsHXK Genes under Cold and Drought Treatments
3. Discussion
4. Materials and Methods
4.1. Identification of HXK Gene Family in Sugarcane
4.2. Phylogenetic Analysis and Gene Duplication Analysis of SsHXK Proteins
4.3. Gene Structure and Motif Composition of SsHXK Genes
4.4. Promoter Analysis of SsHXK Genes
4.5. Expression Analysis of SsHXKs Based on RNA-Seq
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhao, B.; Qi, K.; Yi, X.; Chen, G.; Liu, X.; Qi, X.; Zhang, S. Identification of hexokinase family members in pear (Pyrus × bretschneideri) and functional exploration of PbHXK1 in modulating sugar content and plant growth. Gene 2019, 711, 143932. [Google Scholar] [CrossRef] [PubMed]
- Karve, A.; Rauh, B.L.; Xia, X.; Kandasamy, M.; Meagher, R.B.; Sheen, J.; Moore, B.D. Expression and evolutionary features of the hexokinase gene family in Arabidopsis. Planta 2008, 228, 411–425. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.C.; Su, J.; Jin, Y.-R.; Zhao, H.Y.; Tian, X.C.; Zhang, C.; Ma, F.W.; Li, M.J.; Ma, B.Q. Genome-wide identification, molecular evolution, and expression divergence of the hexokinase gene family in apple. J. Integr. Agric. 2021, 20, 2112–2125. [Google Scholar] [CrossRef]
- Castro, P.H.; Freitas, S.; Azevedo, H. Plant hexokinase phylogenetic analysis highlights a possible regulation by the posttranslational modifier SUMO. Micropubl. Biol. 2020, 2020, PMC7326332. [Google Scholar]
- Pérez-Díaz, J.; Batista-Silva, W.; Almada, R.; Medeiros, D.B.; Arrivault, S.; Correa, F.; Bastías, A.; Rojas, P.; Beltrán, M.F.; Pozo, M.F.; et al. Prunus Hexokinase 3 genes alter primary C-metabolism and promote drought and salt stress tolerance in Arabidopsis transgenic plants. Sci. Rep. 2021, 11, 7098. [Google Scholar] [CrossRef]
- Sarowar, S.; Lee, J.Y.; Ahn, E.R.; Pai, H.S. A role of hexokinases in plant resistance to oxidative stress and pathogen infection. J. Plant Biol. 2008, 51, 341–346. [Google Scholar] [CrossRef]
- Moore, B.; Zhou, L.; Rolland, F.; Hall, Q.; Cheng, W.H.; Liu, Y.X.; Hwang, I.; Jones, T.; Sheen, J. Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science 2003, 300, 332–336. [Google Scholar] [CrossRef]
- Lugassi, N.; Kelly, G.; Fidel, L.; Yaniv, Y.; Attia, Z.; Levi, A.; Alchanatis, V.; Moshelion, M.; Raveh, E.; Carmi, N.; et al. Expression of Arabidopsis hexokinase in citrus guard cells controls stomatal aperture and reduces transpiration. Front. Plant Sci. 2015, 6, 1114. [Google Scholar] [CrossRef]
- Pego, J.N.V.; Weisbeek, P.J.; Smeekens, S.C.M. Mannose inhibits Arabidopsis germination via a hexokinase-mediated step1. Plant Physiol. 1999, 119, 1017–1024. [Google Scholar] [CrossRef]
- Karve, R.; Lauria, M.; Virnig, A.; Xia, X.; Rauh, B.L.; Moore, B. Evolutionary lineages and functional diversification of plant hexokinases. Mol. Plant 2010, 3, 334–346. [Google Scholar] [CrossRef]
- Kim, M.; Lim, J.H.; Ahn, C.S.; Park, K.; Kim, G.T.; Kim, W.T.; Pai, H.S. Mitochondria-associated hexokinases play a role in the control of programmed cell death in Nicotiana benthamiana. Plant Cell 2006, 18, 2341–2355. [Google Scholar] [CrossRef] [PubMed]
- Kelly, G.; Moshelion, M.; David-Schwartz, R.; Halperin, O.; Wallach, R.; Attia, Z.; Belausov, E.; Granot, D. Hexokinase mediates stomatal closure. Plant J. 2013, 75, 977–988. [Google Scholar] [CrossRef]
- Dai, N.; Schaffer, A.; Petreikov, M.; Shahak, Y.; Giller, Y.; Ratner, K.; Levine, A.; Granot, D. Overexpression of Arabidopsis hexokinase in tomato plants inhibits growth, reduces photosynthesis, and induces rapid senescence. Plant Cell 1999, 11, 1253–1266. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.G.; Sun, C.H.; Zhang, Q.Y.; An, J.P.; You, C.X.; Hao, Y.J. Glucose sensor MdHXK1 phosphorylates and stabilizes MdbHLH3 to promote anthocyanin biosynthesis in apple. PLoS Genet. 2016, 12, e1006273. [Google Scholar] [CrossRef]
- Sun, M.H.; Ma, Q.J.; Hu, D.G.; Zhu, X.P.; You, C.X.; Shu, H.R.; Hao, Y.J. The Glucose sensor MdHXK1 phosphorylates a tonoplast Na(+)/H(+) exchanger to improve salt tolerance. Plant Physiol. 2018, 176, 2977–2990. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Chen, G.; Zhang, J.; Shen, H.; Kang, J.; Feng, P.; Xie, Q.; Hu, Z. Suppression of a hexokinase gene, SlHXK1, leads to accelerated leaf senescence and stunted plant growth in tomato. Plant Sci. 2020, 298, 110544. [Google Scholar] [CrossRef]
- Janská, A.; Marsík, P.; Zelenková, S.; Ovesná, J. Cold stress and acclimation—What is important for metabolic adjustment? Plant Biol. 2010, 12, 395–405. [Google Scholar] [CrossRef]
- Damaris, R.N.; Yang, P. Protein phosphorylation response to abiotic stress in plants. Methods Mol. Biol. 2021, 2358, 17–43. [Google Scholar]
- Iwaniuk, P.; Lozowicka, B. Biochemical compounds and stress markers in lettuce upon exposure to pathogenic Botrytis cinerea and fungicides inhibiting oxidative phosphorylation. Planta 2022, 255, 61. [Google Scholar] [CrossRef]
- Kaczynski, P.; Lozowicka, B.; Wolejko, E.; Iwaniuk, P.; Konecki, R.; Dragowski, W.; Lozowicki, J.; Amanbek, N.; Rusilowska, J.; Pietraszko, A. Complex study of glyphosate and metabolites influence on enzymatic activity and microorganisms association in soil enriched with Pseudomonas fluorescens and sewage sludge. J. Hazard. Mater. 2020, 393, 122443. [Google Scholar] [CrossRef]
- Wang, H.; Xin, H.; Guo, J.; Gao, Y.; Liu, C.; Dai, D.; Tang, L. Genome-wide screening of hexokinase gene family and functional elucidation of HXK2 response to cold stress in Jatropha curcas. Mol. Biol. Rep. 2019, 46, 1649–1660. [Google Scholar] [CrossRef] [PubMed]
- Han, P.; Wei, Y.; Jiang, S.; Chen, Y.; Xu, F.; Wang, H.; Shao, X. N-Acetyl-d-glucosamine inhibition of hexokinase results in downregulation of the phenylpropanoid metabolic pathway and decreased resistance to brown rot in peach fruit. J. Agric. Food Chem. 2022, 70, 3917–3928. [Google Scholar] [CrossRef] [PubMed]
- Lugassi, N.; Yadav, B.S.; Egbaria, A.; Wolf, D.; Kelly, G.; Neuhaus, E.; Raveh, E.; Carmi, N.; Granot, D. Expression of Arabidopsis hexokinase in tobacco guard cells increases water-use efficiency and confers tolerance to drought and salt stress. Plants 2019, 8, 613. [Google Scholar] [CrossRef]
- Küstner, L.; Fürtauer, L.; Weckwerth, W.; Nägele, T.; Heyer, A.G. Subcellular dynamics of proteins and metabolites under abiotic stress reveal deferred response of the Arabidopsis thaliana hexokinase-1 mutant gin2-1 to high light. Plant J. 2019, 100, 456–472. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, R.; Ou, H.; Gui, Y.; Wei, J.; Zhou, H.; Tan, H.; Li, Y. Comprehensive transcriptome analysis reveals genes in response to water deficit in the leaves of Saccharum narenga (Nees ex Steud.) hack. BMC Plant Biol. 2018, 18, 250. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, X.; Yang, D.; Yin, Z.; Jiang, Y.; Ling, H.; Huang, N.; Zhang, D.; Wu, J.; Liu, L.; et al. A comprehensive identification and expression analysis of VQ motif-containing proteins in sugarcane (Saccharum spontaneum L.) under phytohormone treatment and cold stress. Int. J. Mol. Sci. 2022, 23, 6334. [Google Scholar] [CrossRef] [PubMed]
- Han, M.; Xu, X.; Xiong, Y.; Wei, H.; Yao, K.; Huang, T.; Long, Y.; Su, T. Genome-wide survey and expression analyses of hexokinase family in poplar (Populus trichocarpa). Plants 2022, 11, 2025. [Google Scholar] [CrossRef]
- Cho, J.I.; Ryoo, N.; Ko, S.; Lee, S.K.; Lee, J.; Jung, K.H.; Lee, Y.H.; Bhoo, S.H.; Winderickx, J.; An, G.; et al. Structure, expression, and functional analysis of the hexokinase gene family in rice (Oryza sativa L.). Planta 2006, 224, 598–611. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, J.; Chen, Y.; Li, R.; Wang, H.; Ding, L.; Wei, J. Isolation, structural analysis, and expression characteristics of the maize (Zea mays L.) hexokinase gene family. Mol. Biol. Rep. 2014, 41, 6157–6166. [Google Scholar] [CrossRef]
- Kim, Y.M.; Heinzel, N.; Giese, J.O.; Koeber, J.; Melzer, M.; Rutten, T.; Von Wiren, N.; Sonnewald, U.; Hajirezaei, M.R. A dual role of tobacco hexokinase 1 in primary metabolism and sugar sensing. Plant Cell Environ. 2013, 36, 1311–1327. [Google Scholar] [CrossRef]
- Dou, L.; Li, Z.; Wang, H.; Li, H.; Xiao, G.; Zhang, X. The hexokinase gene family in cotton: Genome-wide characterization and bioinformatics analysis. Front. Plant Sci. 2022, 13, 882587. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, X.; Geng, S.; Singh, S.K.; Wang, Y.; Pattanaik, S.; Yuan, L. Genome-wide identification of hexokinase gene family in Brassica napus: Structure, phylogenetic analysis, expression, and functional characterization. Planta 2018, 248, 171–182. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.; Liu, X.; Gao, S.; Guo, J.; Su, Y.; Ling, H.; Wang, C.; Li, Z.; Xu, L.; Que, Y. Foreign cry1Ac gene integration and endogenous borer stress-related genes synergistically improve insect resistance in sugarcane. BMC Plant Biol. 2018, 18, 342. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, X.; Tang, H.; Zhang, Q.; Hua, X.; Ma, X.; Zhu, F.; Jones, T.; Zhu, X.; Bowers, J.; et al. Allele-defined genome of the autopolyploid sugarcane Saccharum spontaneum L. Nat. Genet. 2018, 50, 1565–1573. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Zhang, Y.; Zhang, Q.; Wu, R.; Wang, X.; Feng, S.; Chen, S.; Lu, C.; Du, L. Genome-wide identification and characterization of hexokinase genes in Moso Bamboo (Phyllostachys edulis). Front. Plant Sci. 2020, 11, 600. [Google Scholar] [CrossRef]
- Kaltenegger, E.; Leng, S.; Heyl, A. The effects of repeated whole genome duplication events on the evolution of cytokinin signaling pathway. BMC Evol. Biol. 2018, 18, 76. [Google Scholar] [CrossRef]
- Zhang, G.; Wang, F.; Li, J.; Ding, Q.; Zhang, Y.; Li, H.; Zhang, J.; Gao, J. Genome-wide identification and analysis of the VQ motif-containing protein family in Chinese Cabbage (Brassica rapa L. ssp. Pekinensis). Int. J. Mol. Sci. 2015, 16, 28683–28704. [Google Scholar] [CrossRef]
- Song, W.; Zhao, H.; Zhang, X.; Lei, L.; Lai, J. Genome-wide identification of VQ motif-containing proteins and their expression profiles under abiotic stresses in maize. Front. Plant Sci. 2016, 6, 1177. [Google Scholar] [CrossRef]
- Cao, Y.; Meng, D.; Abdullah, M.; Jin, Q.; Lin, Y.; Cai, Y. Genome wide identification, evolutionary, and expression analysis of VQ genes from two pyrus species. Genes 2018, 9, 224. [Google Scholar] [CrossRef]
- Li, Z.; Hua, X.T.; Zhong, W.M.; Yuan, Y.; Wang, Y.J.; Wang, Z.C.; Ming, R.; Zhang, J.S. Genome-wide identification and expression profile analysis of WRKY family genes in the autopolyploid Saccharum spontaneum. Plant Cell Physiol. 2020, 61, 616–630. [Google Scholar] [CrossRef]
- Zhu, J.-K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247. [Google Scholar] [CrossRef] [PubMed]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 2007, 58, 221–227. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, Y.; Zhang, Z.; Zhu, J.; Yu, J. KaKs_Calculator 2.0: A toolkit incorporating gamma-series methods and sliding window strategies. Genom. Proteom. Bioinform. 2010, 8, 77–80. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.-Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Jiang, Y.; Liu, X.; Cheng, H.; Han, Y.; Zhang, D.; Wu, J.; Liu, L.; Yan, M.; Que, Y.; et al. Identification and Expression Analysis of Hexokinases Family in Saccharum spontaneum L. under Drought and Cold Stresses. Plants 2023, 12, 1215. https://doi.org/10.3390/plants12061215
Liu Y, Jiang Y, Liu X, Cheng H, Han Y, Zhang D, Wu J, Liu L, Yan M, Que Y, et al. Identification and Expression Analysis of Hexokinases Family in Saccharum spontaneum L. under Drought and Cold Stresses. Plants. 2023; 12(6):1215. https://doi.org/10.3390/plants12061215
Chicago/Turabian StyleLiu, Ying, Yaolan Jiang, Xiaolan Liu, Hefen Cheng, Yuekun Han, Dawei Zhang, Jinfeng Wu, Lili Liu, Mingli Yan, Youxiong Que, and et al. 2023. "Identification and Expression Analysis of Hexokinases Family in Saccharum spontaneum L. under Drought and Cold Stresses" Plants 12, no. 6: 1215. https://doi.org/10.3390/plants12061215
APA StyleLiu, Y., Jiang, Y., Liu, X., Cheng, H., Han, Y., Zhang, D., Wu, J., Liu, L., Yan, M., Que, Y., & Zhou, D. (2023). Identification and Expression Analysis of Hexokinases Family in Saccharum spontaneum L. under Drought and Cold Stresses. Plants, 12(6), 1215. https://doi.org/10.3390/plants12061215





