Turning Garlic into a Modern Crop: State of the Art and Perspectives
Abstract
1. General features and challenges
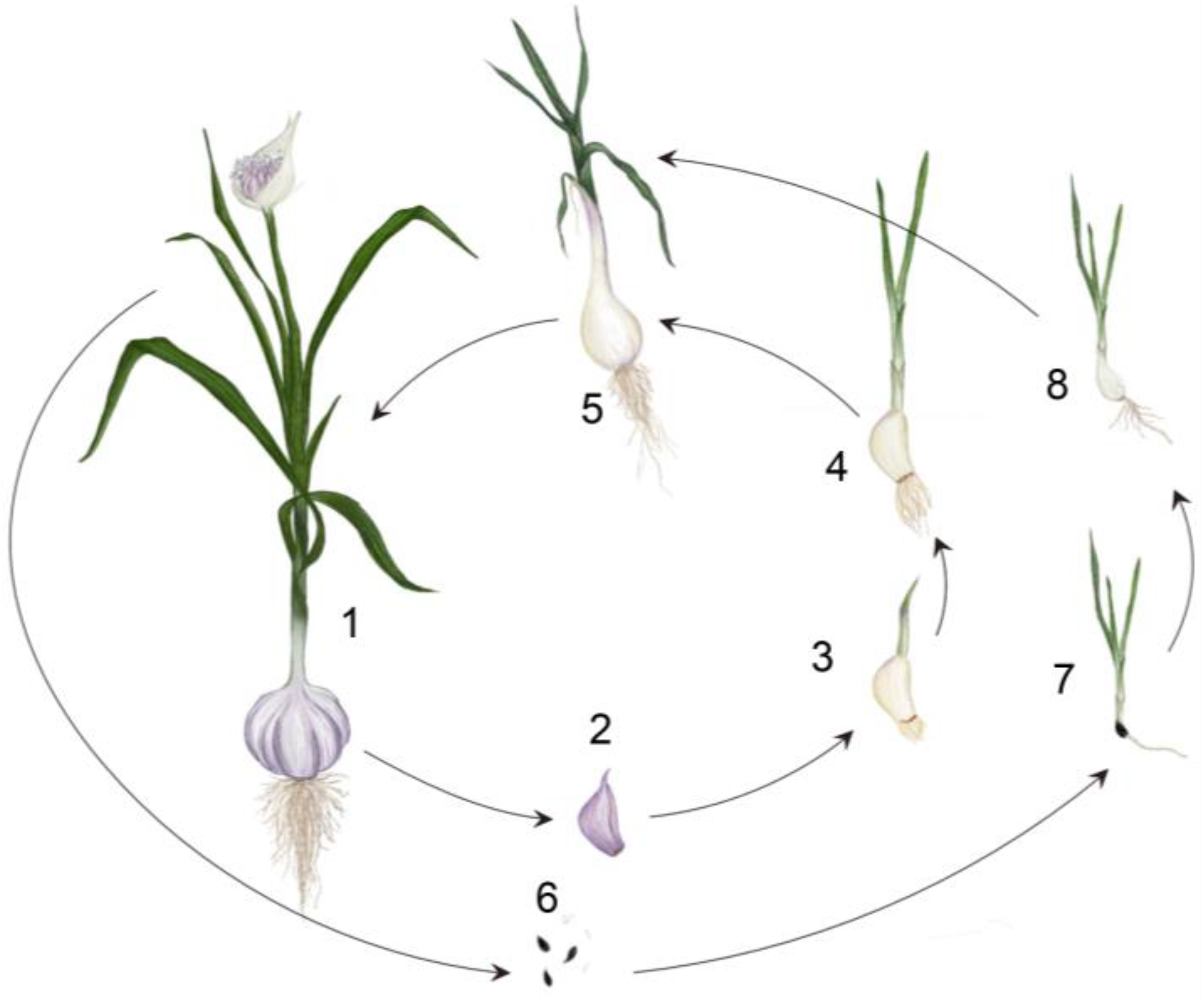
1.1. The Life Cycle of a Garlic Plant: Bulb and Flower Development
1.2. Consequences of Vegetative Propagation and Fertility Restoration
2. The Genetics and Genomics of Garlic: State-of-the-Art
2.1. Molecular Markers and Linkage Maps
2.2. Transcriptomic Approaches in Garlic
2.3. The Genomes of Garlic and Related Species
3. Target Traits for Garlic Breeding
3.1. Yield and Bulb Traits
3.2. Secondary Metabolism as a Target for Garlic Breeding
4. Pathogens of Garlic: Threats, Treatments and Perspectives
4.1. Fungal Pathogens
4.2. Nematodes
4.3. Arthropods: Insects and Mites
4.4. Viruses
4.5. Bacteria and Phytoplasmas
5. Opportunities for the Future of Garlic Breeding
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ricroch, A.; Yockteng, R.; Brown, S.C.; Nadot, S. Evolution of Genome Size across Some Cultivated Allium Species. Genome 2005, 48, 511–520. [Google Scholar] [CrossRef] [PubMed]
- Mes, T.H.; Fritsch, R.M.; Pollner, S.; Bachmann, K. Evolution of the Chloroplast Genome and Polymorphic ITS Regions in Allium Subg. Melanocrommyum. Genome 1999, 42, 237–247. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Dadmohammadi, Y.; Abbaspourrad, A. Flavor Components, Precursors, Formation Mechanisms, Production and Characterization Methods: Garlic, Onion, and Chili Pepper Flavors. Crit. Rev. Food Sci. Nutr. 2022, 62, 8265–8287. [Google Scholar] [CrossRef] [PubMed]
- Borlinghaus, J.; Albrecht, F.; Gruhlke, M.C.H.; Nwachukwu, I.D.; Slusarenko, A.J. Allicin: Chemistry and Biological Properties. Molecules 2014, 19, 12591–12618. [Google Scholar] [CrossRef]
- Mann, L. Anatomy of the Garlic Bulb and Factors Affecting Bulb Development. Hilgardia 1952, 21, 195–251. [Google Scholar] [CrossRef]
- Kamenetsky, R.; Rabinowitch, H.D. Floral Development in Bolting Garlic. Sex. Plant Reprod. 2001, 13, 235–241. [Google Scholar] [CrossRef]
- Winiarczyk, K.; Kosmala, A. Development of the Female Gametophyte in the Sterile Ecotype of the Bolting Allium sativum L. Sci. Hortic. 2009, 121, 353–360. [Google Scholar] [CrossRef]
- Shemesh Mayer, E.; Winiarczyk, K.; Błaszczyk, L.; Kosmala, A.; Rabinowitch, H.D.; Kamenetsky, R. Male Gametogenesis and Sterility in Garlic (Allium sativum L.): Barriers on the Way to Fertilization and Seed Production. Planta 2013, 237, 103–120. [Google Scholar] [CrossRef]
- Mann, L.; Lewis, D. Rest and Dormancy in Garlic. Hilgardia 1956, 26, 161–189. [Google Scholar] [CrossRef]
- Takagi, H. Garlic Allium sativum L. In Onions and Allied Crops: Volume III: Biochemistry, Food Science, and Minor Crops; CRC Press: Boca Raton, FL, USA, 1990; pp. 109–146. [Google Scholar]
- Etoh, T. Studies on the Sterility in Garlic, Allium sativum L. Mem. Fac. Agric. Kagoshima Univ. 1985, 21, 77–132. [Google Scholar]
- Etoh, T. Fertility of the Garlic Clones Collected in Soviet Central Asia. J. Jpn. Soc. Hortic. Sci. 1986, 55, 312–319. [Google Scholar] [CrossRef]
- Kamenetsky, R.; Shafir, I.L.; Zemah, H.; Barzilay, A.; Rabinowitch, H. Environmental Control of Garlic Growth and Florogenesis. J. Am. Soc. Hortic. Sci. 2004, 129, 144–151. [Google Scholar] [CrossRef]
- Guevara-Figueroa, T.; López-Hernández, L.; Lopez, M.; Hurtado, M.D.; Vázquez-Barrios, M.; Guevara-Olvera, L.; González, R.G.; Rivera-Pastrana, D.; Torres-Robles, H.; Mercado-Silva, E. Conditioning Garlic “Seed” Cloves at Low Temperature Modifies Plant Growth, Sugar, Fructan Content, and Sucrose Sucrose Fructosyl Transferase (1-SST) Expression. Sci. Hortic. 2015, 189, 150–158. [Google Scholar] [CrossRef]
- Pooler, M.R.; Simon, P.W. Garlic Flowering in Response to Clone, Photoperiod, Growth Temperature, and Cold Storage. HortScience 1993, 28, 1085–1086. [Google Scholar] [CrossRef]
- Aoba, T.; Takagi, H. Studies on the Bulb Formation in Garlic Plants III. On the Effects of Cooling Treatments of Seed-Bulbs and Day-Length during the Growing Period on Bulbing. J. Jpn. Soc. Hortic. Sci. 1971, 40, 240–245. [Google Scholar] [CrossRef]
- Wu, C.; Wang, M.; Dong, Y.; Cheng, Z.; Meng, H. Growth, Bolting and Yield of Garlic (Allium sativum L.) in Response to Clove Chilling Treatment. Sci. Hortic. 2015, 194, 43–52. [Google Scholar] [CrossRef]
- Mathew, D.; Forer, Y.; Rabinowitch, H.D.; Kamenetsky, R. Effect of Long Photoperiod on the Reproductive and Bulbing Processes in Garlic (Allium sativum L.) Genotypes. Environ. Exp. Bot. 2011, 71, 166–173. [Google Scholar] [CrossRef]
- Shemesh-Mayer, E.; Kamenetsky-Goldstein, R. Traditional and novel approaches in garlic (Allium sativum L.) breeding. In Advances in Plant Breeding Strategies: Vegetable Crops; Springer: Cham, Switzerland, 2021; Volume 8, pp. 3–49. [Google Scholar]
- Peña-Iglesias, A. El Ajo: Virosis, Fisiopatías y Selección Clonal y Sanitaria. I: Parte Teórico-Descriptiva. Bol. Sanid. Veg. Plagas 1988, 14, 461–483. [Google Scholar]
- Conci, V.C.; Canavelli, A.; Lunello, P.; Di Rienzo, J.; Nome, S.F.; Zumelzu, G.; Italia, R. Yield Losses Associated with Virus-Infected Garlic Plants during Five Successive Years. Plant Dis. 2003, 87, 1411–1415. [Google Scholar] [CrossRef]
- Luciani, G.F.; Mary, A.K.; Pellegrini, C.; Curvetto, N.R. Effects of Explants and Growth Regulators in Garlic Callus Formation and Plant Regeneration. Plant Cell Tissue Organ Cult. 2006, 87, 139–143. [Google Scholar] [CrossRef]
- Ramírez-Malagón, R.; Pérez-Moreno, L.; Borodanenko, A.; Salinas-González, G.; Ochoa-Alejo, N. Differential Organ Infection Studies, Potyvirus Elimination, and Field Performance of Virus-Free Garlic Plants Produced by Tissue Culture. Plant Cell Tissue Organ Cult. 2006, 86, 103–110. [Google Scholar] [CrossRef]
- Vieira, R.L.; da Silva, A.L.; Zaffari, G.R.; Steinmacher, D.A.; de Freitas Fraga, H.P.; Guerra, M.P. Efficient Elimination of Virus Complex from Garlic (Allium sativum L.) by Cryotherapy of Shoot Tips. Acta Physiol. Plant. 2015, 37, 1733. [Google Scholar] [CrossRef]
- Kajimura, Y.; Sugiura, T.; Suenaga, K.; Itakura, Y.; Etoh, T. A New Garlic Growing System from Bulbils through Transplanting. J. Hortic. Sci. Biotechnol. 2000, 75, 176–180. [Google Scholar] [CrossRef]
- Keller, E.J.; Zanke, C.D.; Senula, A.; Breuing, A.; Hardeweg, B.; Winkelmann, T. Comparing Costs for Different Conservation Strategies of Garlic (Allium sativum L.) Germplasm in Genebanks. Genet. Resour. Crop Evol. 2013, 60, 913–926. [Google Scholar] [CrossRef]
- Kononkov, P. The Question of Obtaining Garlic Seed. Sadi Ogorod 1953, 8, 38–40. [Google Scholar]
- Hong, C.; Etoh, T. Fertile Clones of Garlic (Allium sativum L.) Abundant around the Tien Shan Mountains. Jpn. J. Breed. 1996, 46, 349–353. [Google Scholar] [CrossRef]
- Jenderek, M. Generative Reproduction of Garlic (Allium sativum). Ses. Nauk. 1998, 57, 141–145. [Google Scholar]
- Etoh, T.; Noma, Y.; Nishitarumizu, Y.; Wakamoto, T. Seed Productivity and Germinability of Various Garlic [Allium sativum L.] Clones Collected in Soviet Central Asia. Mem. Fac. Agric.-Kagoshima Univ. Jpn. 1988, 24, 129–139. [Google Scholar]
- Jenderek, M.; Hannan, R. Seed Producing Ability of Garlic (Allium sativum L.) Clones from Two Public US Collections. In Proceedings of the Third International Symposium on Edible Alliaceae, Athens, GA, USA, 30 October–3 November 2000; pp. 73–75. [Google Scholar]
- Kamenetsky, R.; Shafir, I.L.; Baizerman, M.; Khassanov, F.; Kik, C.; Rabinowitch, H. Garlic (Allium sativum L.) and Its Wild Relatives from Central Asia: Evaluation for Fertility Potential. In Proceedings of the XXVI International Horticultural Congress: Advances in Vegetable Breeding, Toronto, ON, Canada, 11–17 August 2002; pp. 83–91. [Google Scholar]
- Jenderek, M.M.; Hannan, R.M. Variation in Reproductive Characteristics and Seed Production in the USDA Garlic Germplasm Collection. HortScience 2004, 39, 485–488. [Google Scholar] [CrossRef]
- Singh, H.; Khar, A.; Verma, P. Induced Mutagenesis for Genetic Improvement of Allium Genetic Resources: A Comprehensive Review. Genet. Resour. Crop Evol. 2021, 68, 2669–2690. [Google Scholar] [CrossRef]
- Hailu, M.G.; Mawcha, K.T.; Nshimiyimana, S.; Suharsono, S. Garlic Micro-Propagation and Polyploidy Induction in Vitro by Colchicine. Plant Breed. Biotechnol. 2021, 9, 1–19. [Google Scholar] [CrossRef]
- Wen, Y.; Liu, H.; Meng, H.; Qiao, L.; Zhang, G.; Cheng, Z. In Vitro Induction and Phenotypic Variations of Autotetraploid Garlic (Allium sativum L.) With Dwarfism. Front. Plant Sci. 2022, 13, 917910. [Google Scholar] [CrossRef]
- Etoh, T.; Ogura, H. Peroxidase Isozymes in the Leaves of Various Clones of Garlic, Allium sativum L. Mem. Fac. Agric. Kagoshima Univ. 1981, 17, 71–77. [Google Scholar]
- Maaß, H.; Klaas, M. Infraspecific Differentiation of Garlic (Allium sativum L.) by Isozyme and RAPD Markers. Theor. Appl. Genet. 1995, 91, 89–97. [Google Scholar] [CrossRef]
- Ma, K.-H.; Kwag, J.-G.; Zhao, W.; Dixit, A.; Lee, G.-A.; Kim, H.-H.; Chung, I.-M.; Kim, N.-S.; Lee, J.-S.; Ji, J.-J. Isolation and Characteristics of Eight Novel Polymorphic Microsatellite Loci from the Genome of Garlic (Allium sativum L.). Sci. Hortic. 2009, 122, 355–361. [Google Scholar] [CrossRef]
- Zhao, W.; Chung, J.; Lee, G.; Ma, K.; Kim, H.; Kim, K.; Chung, I.; Lee, J.; Kim, N.; Kim, S. Molecular Genetic Diversity and Population Structure of a Selected Core Set in Garlic and Its Relatives Using Novel SSR Markers. Plant Breed. 2011, 130, 46–54. [Google Scholar] [CrossRef]
- Lee, G.-A.; Kwon, S.-J.; Park, Y.-J.; Lee, M.-C.; Kim, H.-H.; Lee, J.-S.; Lee, S.-Y.; Gwag, J.-G.; Kim, C.-K.; Ma, K.-H. Cross-Amplification of SSR Markers Developed from Allium sativum to Other Allium Species. Sci. Hortic. 2011, 128, 401–407. [Google Scholar] [CrossRef]
- Chen, S.; Chang, Y.; Zhou, J.; Cheng, Z.; Meng, H. Genetic Diversity of Garlic (Allium sativum L.) Germplasm by Simple Sequence Repeats. J. Agric. Biotechnol. 2012, 20, 372–381. [Google Scholar]
- Cunha, C.P.; Hoogerheide, E.S.; Zucchi, M.I.; Monteiro, M.; Pinheiro, J.B. New Microsatellite Markers for Garlic, Allium sativum (Alliaceae). Am. J. Bot. 2012, 99, e17–e19. [Google Scholar] [CrossRef]
- Chen, S.; Chen, W.; Shen, X.; Yang, Y.; Qi, F.; Liu, Y.; Meng, H. Analysis of the Genetic Diversity of Garlic (Allium sativum L.) by Simple Sequence Repeat and Inter Simple Sequence Repeat Analysis and Agro-Morphological Traits. Biochem. Syst. Ecol. 2014, 55, 260–267. [Google Scholar] [CrossRef]
- Ovesna, J.; Leišová-Svobodová, L.; Kučera, L. Microsatellite Analysis Indicates the Specific Genetic Basis of Czech Bolting Garlic. Czech J. Genet. Plant Breed. 2014, 50, 226–234. [Google Scholar] [CrossRef]
- Liu, T.; Zeng, L.; Zhu, S.; Chen, X.; Tang, Q.; Mei, S.; Tang, S. Large-Scale Development of Expressed Sequence Tag-Derived Simple Sequence Repeat Markers by Deep Transcriptome Sequencing in Garlic (Allium sativum L.). Mol. Breed. 2015, 35, 204. [Google Scholar] [CrossRef]
- Barboza, K.; Beretta, V.; Kozub, P.C.; Salinas, C.; Morgenfeld, M.M.; Galmarini, C.R.; Cavagnaro, P.F. Microsatellite Analysis and Marker Development in Garlic: Distribution in EST Sequence, Genetic Diversity Analysis, and Marker Transferability across Alliaceae. Mol. Genet. Genom. 2018, 293, 1091–1106. [Google Scholar] [CrossRef] [PubMed]
- Barboza, K.; Salinas, M.C.; Acuña, C.V.; Bannoud, F.; Beretta, V.; Garcia-Lampasona, S.; Burba, J.L.; Galmarini, C.R.; Cavagnaro, P.F. Assessment of Genetic Diversity and Population Structure in a Garlic (Allium sativum L.) Germplasm Collection Varying in Bulb Content of Pyruvate, Phenolics, and Solids. Sci. Hortic. 2020, 261, 108900. [Google Scholar] [CrossRef]
- Li, X.; Qiao, L.; Chen, B.; Zheng, Y.; Zhi, C.; Zhang, S.; Pan, Y.; Cheng, Z. SSR Markers Development and Their Application in Genetic Diversity Evaluation of Garlic (Allium sativum) Germplasm. Plant Divers. 2022, 44, 481–491. [Google Scholar] [CrossRef]
- Jabbes, N.; Geoffriau, E.E.; Le Clerc, V.; Dridi, B.; Hannechi, C. Inter Simple Sequence Repeat Fingerprints for Assess Genetic Diversity of Tunisian Garlic Populations. J. Agric. Sci. 2011, 3, 77–85. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, J.; Chen, Q.; Chang, Y.; Du, J.; Meng, H. Analysis of the Genetic Diversity of Garlic (Allium sativum L.) Germplasm by SRAP. Biochem. Syst. Ecol. 2013, 50, 139–146. [Google Scholar] [CrossRef]
- Havey, M.J.; Ahn, Y.-K. Single Nucleotide Polymorphisms and Indel Markers from the Transcriptome of Garlic. J. Am. Soc. Hortic. Sci. 2016, 141, 62–65. [Google Scholar] [CrossRef]
- Ipek, M.; Sahin, N.; Ipek, A.; Cansev, A.; Simon, P.W. Development and Validation of New SSR Markers from Expressed Regions in the Garlic Genome. Sci. Agric. 2015, 72, 41–46. [Google Scholar] [CrossRef]
- Zewdie, Y.; Havey, M.J.; Prince, J.P.; Jenderek, M.M. The First Genetic Linkages among Expressed Regions of the Garlic Genome. J. Am. Soc. Hortic. Sci. 2005, 130, 569–574. [Google Scholar] [CrossRef]
- Ipek, M.; Ipek, A.; Almquist, S.G.; Simon, P.W. Demonstration of Linkage and Development of the First Low-Density Genetic Map of Garlic, Based on AFLP Markers. Theor. Appl. Genet. 2005, 110, 228–236. [Google Scholar] [CrossRef]
- Elshire, R.J.; Glaubitz, J.C.; Sun, Q.; Poland, J.A.; Kawamoto, K.; Buckler, E.S.; Mitchell, S.E. A Robust, Simple Genotyping-by-Sequencing (GBS) Approach for High Diversity Species. PLoS ONE 2011, 6, e19379. [Google Scholar] [CrossRef]
- Baird, N.A.; Etter, P.D.; Atwood, T.S.; Currey, M.C.; Shiver, A.L.; Lewis, Z.A.; Selker, E.U.; Cresko, W.A.; Johnson, E.A. Rapid SNP Discovery and Genetic Mapping Using Sequenced RAD Markers. PLoS ONE 2008, 3, e3376. [Google Scholar] [CrossRef]
- Jo, J.; Purushotham, P.M.; Han, K.; Lee, H.-R.; Nah, G.; Kang, B.-C. Development of a Genetic Map for Onion (Allium cepa L.) Using Reference-Free Genotyping-by-Sequencing and SNP Assays. Front. Plant Sci. 2017, 8, 1606. [Google Scholar] [CrossRef]
- Choi, Y.; Kim, S.; Lee, J. Construction of an Onion (Allium cepa L.) Genetic Linkage Map Using Genotyping-by-Sequencing Analysis with a Reference Gene Set and Identification of QTLs Controlling Anthocyanin Synthesis and Content. Plants 2020, 9, 616. [Google Scholar] [CrossRef]
- Lee, Y.-R.; Kim, C.W.; Han, J.; Choi, H.J.; Han, K.; Lee, E.S.; Kim, D.-S.; Lee, J.; Siddique, M.I.; Lee, H.-E. Genotyping-by-Sequencing Derived Genetic Linkage Map and Quantitative Trait Loci for Sugar Content in Onion (Allium cepa L.). Plants 2021, 10, 2267. [Google Scholar] [CrossRef]
- Egea, L.A.; Mérida-García, R.; Kilian, A.; Hernandez, P.; Dorado, G. Assessment of Genetic Diversity and Structure of Large Garlic (Allium sativum) Germplasm Bank, by Diversity Arrays Technology “Genotyping-by-Sequencing” Platform (DArTseq). Front. Genet. 2017, 8, 98. [Google Scholar] [CrossRef]
- Gimenez, M.D.; Yañez-Santos, A.M.; Paz, R.C.; Quiroga, M.P.; Marfil, C.F.; Conci, V.C.; García-Lampasona, S.C. Assessment of Genetic and Epigenetic Changes in Virus-Free Garlic (Allium sativum L.) Plants Obtained by Meristem Culture Followed by in Vitro Propagation. Plant Cell Rep. 2016, 35, 129–141. [Google Scholar] [CrossRef]
- Gimenez, M.D.; García Lampasona, S. Before-after Analysis of Genetic and Epigenetic Markers in Garlic: A 13-Year Experiment. Sci. Hortic. 2018, 240, 23–28. [Google Scholar] [CrossRef]
- Burr, B.; Burr, F.A. Recombinant Inbreds for Molecular Mapping in Maize: Theoretical and Practical Considerations. Trends Genet. 1991, 7, 55–60. [Google Scholar] [CrossRef]
- Pollard, D.A. Design and construction of recombinant inbred lines. In Quantitative Trait Loci (QTL); Humana Press: Totowa, NJ, USA, 2012; pp. 31–39. [Google Scholar]
- Arumuganathan, K.; Earle, E.D. Nuclear DNA Content of Some Important Plant Species. Plant Mol. Biol. Rep. 1991, 9, 208–218. [Google Scholar] [CrossRef]
- Kim, D.-W.; Jung, T.-S.; Nam, S.-H.; Kwon, H.-R.; Kim, A.; Chae, S.-H.; Choi, S.-H.; Kim, D.-W.; Kim, R.N.; Park, H.-S. GarlicESTdb: An Online Database and Mining Tool for Garlic EST Sequences. BMC Plant Biol. 2009, 9, 61. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Zhou, S.; Meng, F.; Liu, S. De Novo Assembly and Characterization of the Garlic (Allium sativum) Bud Transcriptome by Illumina Sequencing. Plant Cell Rep. 2012, 31, 1823–1828. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.D.; Ma, G.Q.; Cheng, B.; Li, H.; Liu, S.Q. Identification of Differentially Expressed Genes in Shoot Apex of Garlic (Allium sativum L.) Using Illumina Sequencing. J. Plant Stud. 2013, 2, 136. [Google Scholar] [CrossRef]
- Kamenetsky, R.; Faigenboim, A.; Shemesh Mayer, E.; Ben Michael, T.; Gershberg, C.; Kimhi, S.; Esquira, I.; Rohkin Shalom, S.; Eshel, D.; Rabinowitch, H.D. Integrated Transcriptome Catalogue and Organ-Specific Profiling of Gene Expression in Fertile Garlic (Allium sativum L.). BMC Genom. 2015, 16, 12. [Google Scholar] [CrossRef] [PubMed]
- Shemesh-Mayer, E.; Ben-Michael, T.; Rotem, N.; Rabinowitch, H.D.; Doron-Faigenboim, A.; Kosmala, A.; Perlikowski, D.; Sherman, A.; Kamenetsky, R. Garlic (Allium sativum L.) Fertility: Transcriptome and Proteome Analyses Provide Insight into Flower and Pollen Development. Front. Plant Sci. 2015, 6, 271. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kim, M.-S.; Kim, Y.-M.; Yeom, S.-I.; Cheong, K.; Kim, K.-T.; Jeon, J.; Kim, S.; Kim, D.-S.; Sohn, S.-H. Integrative Structural Annotation of de Novo RNA-Seq Provides an Accurate Reference Gene Set of the Enormous Genome of the Onion (Allium cepa L.). DNA Res. 2015, 22, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Tsukazaki, H.; Yaguchi, S.; Sato, S.; Hirakawa, H.; Katayose, Y.; Kanamori, H.; Kurita, K.; Itoh, T.; Kumagai, M.; Mizuno, S. Development of Transcriptome Shotgun Assembly-Derived Markers in Bunching Onion (Allium fistulosum). Mol. Breed. 2015, 35, 51. [Google Scholar] [CrossRef]
- Zhou, S.-M.; Chen, L.-M.; Liu, S.-Q.; Wang, X.-F.; Sun, X.-D. De Novo Assembly and Annotation of the Chinese Chive (Allium tuberosum Rottler Ex Spr.) Transcriptome Using the Illumina Platform. PLoS ONE 2015, 10, e0133312. [Google Scholar] [CrossRef]
- Rohkin Shalom, S.; Gillett, D.; Zemach, H.; Kimhi, S.; Forer, I.; Zutahy, Y.; Tam, Y.; Teper-Bamnolker, P.; Kamenetsky, R.; Eshel, D. Storage Temperature Controls the Timing of Garlic Bulb Formation via Shoot Apical Meristem Termination. Planta 2015, 242, 951–962. [Google Scholar] [CrossRef]
- Chand, S.K.; Nanda, S.; Rout, E.; Joshi, R.K. Mining, Characterization and Validation of EST Derived Microsatellites from the Transcriptome Database of Allium sativum L. Bioinformation 2015, 11, 145. [Google Scholar] [CrossRef]
- Buso, G.; Paiva, M.; Torres, A.; Resende, F.; Ferreira, M.; Buso, J.; Dusi, A. Genetic Diversity Studies of Brazilian Garlic Cultivars and Quality Control of Garlic-Clover Production. Genet. Mol. Res. 2008, 7, 534–541. [Google Scholar] [CrossRef]
- García-Lampasona, S.; Asprelli, P.; Burba, J.L. Genetic Analysis of a Garlic (Allium sativum L.) Germplasm Collection from Argentina. Sci. Hortic. 2012, 138, 183–189. [Google Scholar] [CrossRef]
- Jo, M.H.; Ham, I.K.; Moe, K.T.; Kwon, S.-W.; Lu, F.-H.; Park, Y.-J.; Kim, W.S.; Kim, M.K.; Kim, T.I.; Lee, E.M. Classification of Genetic Variation in Garlic (‘Allium sativum’ L.) Using SSR Markers. Aust. J. Crop Sci. 2012, 6, 625–631. [Google Scholar]
- Zhu, S.; Tang, S.; Tan, Z.; Yu, Y.; Dai, Q.; Liu, T. Comparative Transcriptomics Provide Insight into the Morphogenesis and Evolution of Fistular Leaves in Allium. BMC Genom. 2017, 18, 60. [Google Scholar] [CrossRef]
- Chaturvedi, A.K.; Shalom, S.R.; Faigenboim-Doron, A.; Teper-Bamnolker, P.; Salam, B.B.; Daus, A.; Kamenetsky, R.; Eshel, D. Differential Carbohydrate Gene Expression during Preplanting Temperature Treatments Controls Meristem Termination and Bulbing in Garlic. Environ. Exp. Bot. 2018, 150, 280–291. [Google Scholar] [CrossRef]
- Li, N.; Qiu, Z.; Lu, X.; Shi, B.; Sun, X.; Tang, X.; Qiao, X. Comparative Transcriptome Analysis of Temperature-Induced Green Discoloration in Garlic. Int. J. Genom. 2018, 2018, 6725728. [Google Scholar] [CrossRef]
- Chen, X.; Liu, X.; Zhu, S.; Tang, S.; Mei, S.; Chen, J.; Li, S.; Liu, M.; Gu, Y.; Dai, Q.; et al. Transcriptome-Referenced Association Study of Clove Shape Traits in Garlic. DNA Res. 2018, 25, 587–596. [Google Scholar] [CrossRef]
- Liu, H.; Wen, Y.; Cui, M.; Qi, X.; Deng, R.; Gao, J.; Cheng, Z. Histological, Physiological and Transcriptomic Analysis Reveal Gibberellin-Induced Axillary Meristem Formation in Garlic (Allium sativum). Plants 2020, 9, 970. [Google Scholar] [CrossRef]
- Sun, X.; Zhu, S.; Li, N.; Cheng, Y.; Zhao, J.; Qiao, X.; Lu, L.; Liu, S.; Wang, Y.; Liu, C.; et al. A Chromosome-Level Genome Assembly of Garlic (Allium sativum) Provides Insights into Genome Evolution and Allicin Biosynthesis. Mol. Plant 2020, 13, 1328–1339. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, C.; Yin, W.; Wei, W.; Wang, Y.; Sa, W.; Liang, J. Single-Molecule Real-Time Sequencing of the Full-Length Transcriptome of Purple Garlic (Allium sativum L. Cv. Leduzipi) and Identification of Serine O-Acetyltransferase Family Proteins Involved in Cysteine Biosynthesis. J. Sci. Food Agric. 2022, 102, 2864–2873. [Google Scholar] [CrossRef] [PubMed]
- Finkers, R.; van Kaauwen, M.; Ament, K.; Burger-Meijer, K.; Egging, R.; Huits, H.; Kodde, L.; Kroon, L.; Shigyo, M.; Sato, S.; et al. Insights from the First Genome Assembly of Onion (Allium cepa). G3 GenesGenomesGenetics 2021, 11, jkab243. [Google Scholar] [CrossRef] [PubMed]
- Liao, N.; Hu, Z.; Miao, J.; Hu, X.; Lyu, X.; Fang, H.; Zhou, Y.-M.; Mahmoud, A.; Deng, G.; Meng, Y.-Q.; et al. Chromosome-Level Genome Assembly of Bunching Onion Illuminates Genome Evolution and Flavor Formation in Allium Crops. Nat. Commun. 2022, 13, 6690. [Google Scholar] [CrossRef] [PubMed]
- Al-Safadi, B.; Arabi, M.; Ayyoubi, Z. Differences in Quantitative and Qualitative Characteristics of Local and Introduced Cultivars and Mutated Lines of Garlic. J. Veg. Crop Prod. 2003, 9, 21–31. [Google Scholar] [CrossRef]
- Mishra, S.S.; Ram, C.; Chakravati, S.K.; Vishwakarma, M.K.; Singh, P.K.; Singh, V.B. Genetic Divergence Studies in Garlic (Allium sativum L.) through Morphological Features. Int. J. Curr. Microbiol. Appl. Sci. 2018, 7, 1013–1020. [Google Scholar] [CrossRef]
- Rout, E.; Nanda, S.; Nayak, S.; Joshi, R.K. Molecular Characterization of NBS Encoding Resistance Genes and Induction Analysis of a Putative Candidate Gene Linked to Fusarium Basal Rot Resistance in Allium sativum. Physiol. Mol. Plant Pathol. 2014, 85, 15–24. [Google Scholar] [CrossRef]
- Martins, N.; Petropoulos, S.; Ferreira, I.C. Chemical Composition and Bioactive Compounds of Garlic (Allium sativum L.) as Affected by Pre-and Post-Harvest Conditions: A Review. Food Chem. 2016, 211, 41–50. [Google Scholar] [CrossRef]
- Khandagale, K.; Krishna, R.; Roylawar, P.; Ade, A.B.; Benke, A.; Shinde, B.; Singh, M.; Gawande, S.J.; Rai, A. Omics Approaches in Allium Research: Progress and Way Ahead. PeerJ 2020, 8, e9824. [Google Scholar] [CrossRef]
- Rocchetti, G.; Zhang, L.; Bocchi, S.; Giuberti, G.; Ak, G.; Elbasan, F.; Yıldıztugay, E.; Ceylan, R.; Picot-Allain, M.C.N.; Mahomoodally, M.F. The Functional Potential of Nine Allium Species Related to Their Untargeted Phytochemical Characterization, Antioxidant Capacity and Enzyme Inhibitory Ability. Food Chem. 2022, 368, 130782. [Google Scholar] [CrossRef]
- Kodera, Y.; Ushijima, M.; Amano, H.; Suzuki, J.; Matsutomo, T. Chemical and Biological Properties of S-1-Propenyl-l-Cysteine in Aged Garlic Extract. Molecules 2017, 22, 570. [Google Scholar] [CrossRef]
- Singiri, J.R.; Swetha, B.; Ben-Natan, A.; Grafi, G. What Worth the Garlic Peel. Int. J. Mol. Sci. 2022, 23, 2126. [Google Scholar] [CrossRef]
- Liu, P.; Weng, R.; Xu, Y.; Feng, Y.; He, L.; Qian, Y.; Qiu, J. Metabolic Changes in Different Tissues of Garlic Plant during Growth. J. Agric. Food Chem. 2020, 68, 12467–12475. [Google Scholar] [CrossRef]
- Eady, C.C.; Kamoi, T.; Kato, M.; Porter, N.G.; Davis, S.; Shaw, M.; Kamoi, A.; Imai, S. Silencing Onion Lachrymatory Factor Synthase Causes a Significant Change in the Sulfur Secondary Metabolite Profile. Plant Physiol. 2008, 147, 2096–2106. [Google Scholar] [CrossRef]
- Kato, M.; Masamura, N.; Shono, J.; Okamoto, D.; Abe, T.; Imai, S. Production and Characterization of Tearless and Non-Pungent Onion. Sci. Rep. 2016, 6, 23779. [Google Scholar] [CrossRef]
- Le, D.; Audenaert, K.; Haesaert, G. Fusarium Basal Rot: Profile of an Increasingly Important Disease in Allium spp. Trop. Plant Pathol. 2021, 46, 241–253. [Google Scholar] [CrossRef]
- Filyushin, M.A.; Anisimova, O.K.; Kochieva, E.Z.; Shchennikova, A.V. Genome-Wide Identification and Expression of Chitinase Class I Genes in Garlic (Allium sativum L.) Cultivars Resistant and Susceptible to Fusarium Proliferatum. Plants 2021, 10, 720. [Google Scholar] [CrossRef]
- Gálvez, L.; Palmero, D. Fusarium Dry Rot of Garlic Bulbs Caused by Fusarium Proliferatum: A Review. Horticulturae 2022, 8, 628. [Google Scholar] [CrossRef]
- Chand, S.K.; Nanda, S.; Mishra, R.; Joshi, R.K. Multiple Garlic (Allium sativum L.) MicroRNAs Regulate the Immunity against the Basal Rot Fungus Fusarium Oxysporum f. Sp. Cepae. Plant Sci. 2017, 257, 9–21. [Google Scholar] [CrossRef]
- Chand, S.K.; Nanda, S.; Joshi, R.K. Regulation of MiR394 in Response to Fusarium Oxysporum f. Sp. Cepae (FOC) Infection in Garlic (Allium sativum L.). Front. Plant Sci. 2016, 7, 258. [Google Scholar] [CrossRef]
- Gálvez, L.; Palmero, D. Incidence and Etiology of Postharvest Fungal Diseases Associated with Bulb Rot in Garlic (Alllium sativum) in Spain. Foods 2021, 10, 1063. [Google Scholar] [CrossRef]
- Edel-Hermann, V.; Lecomte, C. Current Status of Fusarium Oxysporum Formae Speciales and Races. Phytopathology 2019, 109, 512–530. [Google Scholar] [CrossRef] [PubMed]
- Cramer, C.S. Breeding and Genetics of Fusarium Basal Rot Resistance in Onion. Euphytica 2000, 115, 159–166. [Google Scholar] [CrossRef]
- Molinero-Ruiz, L.; Rubio-Pérez, E.; González-Domínguez, E.; Basallote-Ureba, M.J. Alternative Hosts for Fusarium Spp. Causing Crown and Root Rot of Asparagus in Spain. J. Phytopathol. 2011, 159, 114–116. [Google Scholar] [CrossRef]
- Anisimova, O.K.; Kochieva, E.Z.; Shchennikova, A.V.; Filyushin, M.A. Thaumatin-like Protein (TLP) Genes in Garlic (Allium sativum L.): Genome-Wide Identification, Characterization, and Expression in Response to Fusarium proliferatum Infection. Plants 2022, 11, 748. [Google Scholar] [CrossRef] [PubMed]
- Anisimova, O.K.; Shchennikova, A.V.; Kochieva, E.Z.; Filyushin, M.A. Pathogenesis-Related Genes of PR1, PR2, PR4, and PR5 Families Are Involved in the Response to Fusarium Infection in Garlic (Allium sativum L.). Int. J. Mol. Sci. 2021, 22, 6688. [Google Scholar] [CrossRef] [PubMed]
- Dar, A.A.; Sharma, S.; Mahajan, R.; Mushtaq, M.; Salathia, A.; Ahamad, S.; Sharma, J.P. Overview of Purple Blotch Disease and Understanding Its Management through Chemical, Biological and Genetic Approaches. J. Integr. Agric. 2020, 19, 3013–3024. [Google Scholar] [CrossRef]
- Entwistle, A.R. Allium White Rot and Its Control. Soil Use Manag. 1990, 6, 201–208. [Google Scholar] [CrossRef]
- Gálvez, L.; Gil-Serna, J.; García, M.; Iglesias, C.; Palmero, D. Stemphylium Leaf Blight of Garlic (Allium sativum) in Spain: Taxonomy and in Vitro Fungicide Response. Plant Pathol. J. 2016, 32, 388. [Google Scholar] [CrossRef]
- Lorbeer, J.W.; Seyb, A.M.; de Boer, M.; van den Ende, J.E. Botrytis species on bulb crops. In Botrytis: Biology, Pathology and Control; Elad, Y., Williamson, B., Tudzynski, P., Delen, N., Eds.; Springer: Dordrecht, The Netherlands, 2007; pp. 273–294. ISBN 978-1-4020-2626-3. [Google Scholar]
- Greathead, A. Control of Penicillium Decay of Garlic. Calif. Agric. 1978, 32, 18. [Google Scholar]
- Koike, S.T.; Smith, R.F.; Davis, R.M.; Nunez, J.J.; Voss, R.E. Characterization and Control of Garlic Rust in California. Plant Dis. 2001, 85, 585–591. [Google Scholar] [CrossRef]
- Chand, S.K.; Nanda, S.; Joshi, R.K. Genetics and Molecular Mapping of a Novel Purple Blotch-Resistant Gene ApR1 in Onion (Allium cepa L.) Using STS and SSR Markers. Mol. Breed. 2018, 38, 109. [Google Scholar] [CrossRef]
- Lagunes-Fortiz, E.; Robledo-Paz, A.; Gutiérrez-Espinosa, M.A.; Mascorro-Gallardo, J.O.; Espitia-Rangel, E. Genetic Transformation of Garlic (Allium sativum L.) with Tobacco Chitinase and Glucanase Genes for Tolerance to the Fungus Sclerotium cepivorum. Afr. J. Biotechnol. 2013, 12, 3482–3492. [Google Scholar]
- Kudryavtseva, N.; Havey, M.J.; Black, L.; Hanson, P.; Sokolov, P.; Odintsov, S.; Divashuk, M.; Khrustaleva, L. Cytological Evaluations of Advanced Generations of Interspecific Hybrids between Allium cepa and Allium fistulosum Showing Resistance to Stemphylium vesicarium. Genes 2019, 10, 195. [Google Scholar] [CrossRef]
- Lee, H.-M.; Park, J.-S.; Kim, S.-J.; Kim, S.-G.; Park, Y.-D. Using Transcriptome Analysis to Explore Gray Mold Resistance-Related Genes in Onion (Allium cepa L.). Genes 2022, 13, 542. [Google Scholar] [CrossRef]
- Wako, T.; Yamashita, K.; Tsukazaki, H.; Ohara, T.; Kojima, A.; Yaguchi, S.; Shimazaki, S.; Midorikawa, N.; Sakai, T.; Yamauchi, N.; et al. Screening and Incorporation of Rust Resistance from Allium cepa into Bunching Onion (Allium fistulosum) via Alien Chromosome Addition. Genome 2015, 58, 135–142. [Google Scholar] [CrossRef]
- Poromarto, S.; Widono, S.; Septiriani, D.; Hermawan, K. Decrease in Population of Ditylenchus dipsaci in Garlic Cultivation with the Application of Mycorrhizae and Organic Fertilizers; IOP Publishing: Bristol, UK, 2022; Volume 1114, p. 012062. [Google Scholar]
- Roberts, P.; Greathead, A. Control of Ditylenchus dipsaci in Infected Garlic Seed Cloves by Nonfumigant Nematicides. J. Nematol. 1986, 18, 66. [Google Scholar] [PubMed]
- Correia, G.S.; de Araujo Filho, J.V.; da Silva, W.R.; Moccellin, R.; Resende, F.V.; Pinheiro, J.B.; da Grinberg, P.S.; Gomes, C.B. Reaction of Garlic Genotypes to Ditylenchus dipsaci and Aspects Related to Productivity in a Naturally Infested Area. Hortic. Bras. 2023, 40, 451–456. [Google Scholar] [CrossRef]
- Koch, M.; Salomon, R. Improvement of garlic via somaclonal variation and virus elimination. In Proceedings of the International Symposium on Alliums for the Tropics, Bangkok, Thailand, 15–19 February 1993; pp. 211–214. [Google Scholar]
- Atighi, M.R.; Verstraeten, B.; De Meyer, T.; Kyndt, T. Genome-wide DNA Hypomethylation Shapes Nematode Pattern-triggered Immunity in Plants. New Phytol. 2020, 227, 545–558. [Google Scholar] [CrossRef]
- Debnath, P.; Karmakar, K. Garlic Mite, Aceria tulipae (Keifer) (Acari: Eriophyoidea)—A Threat for Garlic in West Bengal, India. Int. J. Acarol. 2013, 39, 89–96. [Google Scholar] [CrossRef]
- Dąbrowska, E.; Lewandowski, M.; Koczkodaj, S.; Paduch-Cichal, E. Transmission of Garlic Virus B, Garlic Virus C, Garlic Virus D and Garlic Virus X by Aceria tulipae (Keifer) in Leek. Eur. J. Plant Pathol. 2020, 157, 215–222. [Google Scholar] [CrossRef]
- Kang, S.-G.; Koo, B.-J.; Lee, E.-T.; Chang, M.-U. Allexivirus Transmitted by Eriophyid Mites in Garlic Plants. J. Microbiol. Biotechnol. 2007, 17, 1833–1840. [Google Scholar] [PubMed]
- Yamashita, K.; Sakai, J.; Hanada, K. Characterization of a New Virus from Garlic (Allium sativum L.), Garlic Mite-Borne Mosaic Virus. Jpn. J. Phytopathol. 1996, 62, 483–489. [Google Scholar] [CrossRef]
- Sapáková, E.; Hasíková, L.; Hřivna, L.; Stavělíková, H.; Šefrová, H. Infestation of Different Garlic Varieties by Dry Bulb Mite Aceria tulipae (Keifer) (Acari: Eriophyidae). Acta Univ. Agric. Silvic. Mendel. Brun. 2012, 60, 293–302. [Google Scholar] [CrossRef]
- Karuppaiah, V.; Soumia, P.; Wagh, P.D.; Singh, M. Ephestia cautella (Lepidoptera: Pyralidae): An Emerging Pest on Garlic in Storage. J. Entomol. Zool. Stud. 2018, 6, 2282–2285. [Google Scholar]
- Srinivas, P.S. Pests and their management in onion and garlic. In Trends in Horticultural Entomology; Mani, M., Ed.; Springer Nature: Singapore, 2022; pp. 1177–1187. ISBN 978-981-19034-3-4. [Google Scholar]
- Adams, M.J.; Antoniw, J.F.; Bar-Joseph, M.; Brunt, A.A.; Candresse, T.; Foster, G.D.; Martelli, G.P.; Milne, R.G.; Fauquet, C.M. Virology Division News: The New Plant Virus Family Flexiviridae and Assessment of Molecular Criteria for Species Demarcation. Arch. Virol. 2004, 149, 1045–1060. [Google Scholar] [CrossRef]
- Lunello, P.; Di Rienzo, J.; Conci, V.C. Yield Loss in Garlic Caused by Leek Yellow Stripe Virus Argentinean Isolate. Plant Dis. 2007, 91, 153–158. [Google Scholar] [CrossRef]
- Chen, J.; Chen, J.P.; Adams, M. Characterisation of Some Carla-and Potyviruses from Bulb Crops in China. Arch. Virol. 2002, 147, 419–428. [Google Scholar] [CrossRef]
- Fajardo, T.V.; Nishijima, M.; Buso, J.A.; Torres, A.C.; Ávila, A.C.; Resende, R.O. Garlic Viral Complex: Identification of Potyviruses and Carlavirus in Central Brazil. Fitopatol. Bras. 2001, 26, 619–626. [Google Scholar] [CrossRef]
- Valli, A.; García, J.A.; López-Moya, J.J. Potyviridae. eLS 2015, 1–10. [Google Scholar] [CrossRef]
- Turina, M.; Kormelink, R.; Resende, R.O. Resistance to Tospoviruses in Vegetable Crops: Epidemiological and Molecular Aspects. Annu. Rev. Phytopathol. 2016, 54, 347–371. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, L.A.; Oliveira, A.S.; Melo, F.L.; Ardisson-Araujo, D.M.; Resende, F.V.; Resende, R.O.; Ribeiro, B.M. A New Virus Found in Garlic Virus Complex Is a Member of Possible Novel Genus of the Family Betaflexiviridae (Order Tymovirales). PeerJ 2019, 7, e6285. [Google Scholar] [CrossRef] [PubMed]
- Tsuneyoshi, T.; Matsumi, T.; Deng, T.; Sako, I.; Sumi, S. Differentiation of Allium Carlaviruses Isolated from Different Parts of the World Based on the Viral Coat Protein Sequence. Arch. Virol. 1998, 143, 1093–1107. [Google Scholar] [CrossRef]
- Katis, N.I.; Maliogka, V.I.; Dovas, C.I. Chapter 5—Viruses of the genus Allium in the Mediterranean region. In Advances in Virus Research; Loebenstein, G., Lecoq, H., Eds.; Academic Press: Cambridge, MA, USA, 2012; Volume 84, pp. 163–208. ISBN 0065-3527. [Google Scholar]
- Bag, S.; Schwartz, H.F.; Cramer, C.S.; Havey, M.J.; Pappu, H.R. Iris Yellow Spot Virus (Tospovirus: Bunyaviridae): From Obscurity to Research Priority. Mol. Plant Pathol. 2015, 16, 224–237. [Google Scholar] [CrossRef]
- Prajapati, M.R.; Manav, A.; Singh, J.; Kumar, P.; Kumar, A.; Kumar, R.; Prakash, S.; Baranwal, V.K. Identification and Characterization of a Garlic Virus E Genome in Garlic (Allium sativum L.) Using High-Throughput Sequencing from India. Plants 2022, 11, 224. [Google Scholar] [CrossRef]
- Roumagnac, P.; Gagnevin, L.; Gardan, L.; Sutra, L.; Manceau, C.; Dickstein, E.R.; Jones, J.B.; Rott, P.; Pruvost, O. Polyphasic Characterization of Xanthomonads Isolated from Onion, Garlic and Welsh Onion (Allium spp.) and Their Relatedness to Different Xanthomonas Species. Int. J. Syst. Evol. Microbiol. 2004, 54, 15–24. [Google Scholar] [CrossRef]
- Conci, V.C.; Gomez, G.G.; Docampo, D.M.; Conci, L.R. Phytoplasma Associated with Symptoms of ‘Tristeza Del Ajo’ (Garlic Decline) in Garlic (Allium sativum L.). J. Phytopathol. 1998, 146, 473–477. [Google Scholar] [CrossRef]
- Parthasarathy, S.; Rajamanickam, S.; Muthamilan, M. Allium Diseases: A Global Perspective. Innov. Farming 2016, 1, 171–178. [Google Scholar]
- Gardan, L.; Bella, P.; Meyer, J.-M.; Christen, R.; Rott, P.; Achouak, W.; Samson, R. Pseudomonas salomonii sp. Nov., Pathogenic on Garlic, and Pseudomonas palleroniana sp. Nov., Isolated from Rice. Int. J. Syst. Evol. Microbiol. 2002, 52, 2065–2074. [Google Scholar]
- Li, B.; Yu, R.R.; Yu, S.H.; Qiu, W.; Fang, Y.; Xie, G.L. First Report on Bacterial Heart Rot of Garlic Caused by Pseudomonas fluorescens in China. Plant Pathol J. 2009, 25, 91–94. [Google Scholar] [CrossRef]
| Crop | Cultivated ha 1 | Tons Produced | Yield (Tons/ha) | Highest Producers |
|---|---|---|---|---|
| Garlic | 1,659,236 | 28,204,854 | 17.00 | China, India, Bangladesh, Egypt |
| Leeks and other alliaceous vegetables | 134,168 | 2,213,183 | 16.49 | Indonesia, Turkey, Belgium, France |
| Onions and shallots, dry | 5,778,767 | 106,592,008 | 18.44 | India, China, Egypt, United States |
| Onions and shallots, green | 215,933 | 4,665,525 | 21.60 | China, Mali, Japan, Republic of Korea |
| Reference | Sequencer | Software | Sample | Results |
|---|---|---|---|---|
| Kim et al., 2009 [67] | Sanger | Leaf and stem tissues | 21,595 ESTs | |
| Sun et al., 2012 [68] | Illumina HiSeq 2000 | SOAPdenovo | Dormant and sprouting vegetative buds | 127,933 unigenes |
| Sun et al., 2013 [69] | Illumina HiSeq 2000 | See [68] | 45,363 DEGs * | |
| Kamenetsky et al., 2015 [70] | Illumina MiSeq | Trinity | Inflorescence, flower, leaf, clove, roots and basal plate | 239,116 (‘extensive’), or 102,042 contigs (‘abundant’ transcriptome) |
| Shemesh-Mayer et al., 2015 [71] | Illumina Hiseq 2000 | Bowtie, DESeq | Flower buds at 3 developmental stages | 16,271 DEGs * |
| Liu et al., 2015 [46] | Illumina HiSeq 2500 | Trinity, MISA | 10 days old plants, 45 days old | 135,360 unigenes; 1506 SSR markers |
| Havey and Ahn, 2016 [52] | Sanger and Roche 454-FLX | SOAPdenovo-trans | Leaf, pseudostem and root tissues | 35,936 contigs; 14,879 SNP and indel markers |
| Zhu et al., 2017 [80] | Illumina HiSeq 2500 | Trinity | Leaf tissue | 132,225 unigenes |
| Chaturvedi et al., 2018 [81] | Illumina HiSeq 2000 | Bowtie, DESeq | Internal buds and storage leaves at two temperatures | 8303 (internal buds) and 14,147 DEGs * (storage leaves) |
| Li et al., 2018 [82] | Illumina HiSeq 4000 | Trinity, CD-HIT | Cloves stored at 4°C for 0, 10, 15 and 40 days | 49,280 unigenes; 5923 DEGs |
| Chen et al., 2018 [83] | PacBio RSII (Iso-Seq CCS) and Illumina HiSeq 2500 | proovread, CD-HIT | Developing bulb | 36,321 transcripts |
| Liu et al., 2020 [84] | Illumina HiSeq 2500 | Trinity, edgeR | Stem (control and treated with GA3) | 159 DEGs * |
| Sun et al., 2020 [85] | Illumina HiSeq 2500 | Trinity, DEGseq | Sprouts, bulbs, flowers, roots, pseudostems and leaves | 34,439 transcripts with constitutive (28,394) or specific (964) expression (out of 57,561 genes predicted in the genome) |
| Wang et al., 2022 [86] | PacBio Sequel (CCS) | Quiver, CD-HIT-EST | Lower bulb, aerial bulb, scape, leaf, clove, basal plate and roots | 36,571 high-quality consensus reads |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Parreño, R.; Rodríguez-Alcocer, E.; Martínez-Guardiola, C.; Carrasco, L.; Castillo, P.; Arbona, V.; Jover-Gil, S.; Candela, H. Turning Garlic into a Modern Crop: State of the Art and Perspectives. Plants 2023, 12, 1212. https://doi.org/10.3390/plants12061212
Parreño R, Rodríguez-Alcocer E, Martínez-Guardiola C, Carrasco L, Castillo P, Arbona V, Jover-Gil S, Candela H. Turning Garlic into a Modern Crop: State of the Art and Perspectives. Plants. 2023; 12(6):1212. https://doi.org/10.3390/plants12061212
Chicago/Turabian StyleParreño, Ricardo, Eva Rodríguez-Alcocer, César Martínez-Guardiola, Lucía Carrasco, Purificación Castillo, Vicent Arbona, Sara Jover-Gil, and Héctor Candela. 2023. "Turning Garlic into a Modern Crop: State of the Art and Perspectives" Plants 12, no. 6: 1212. https://doi.org/10.3390/plants12061212
APA StyleParreño, R., Rodríguez-Alcocer, E., Martínez-Guardiola, C., Carrasco, L., Castillo, P., Arbona, V., Jover-Gil, S., & Candela, H. (2023). Turning Garlic into a Modern Crop: State of the Art and Perspectives. Plants, 12(6), 1212. https://doi.org/10.3390/plants12061212







