A Proteomics Insight into Advancements in the Rice–Microbe Interaction
Abstract
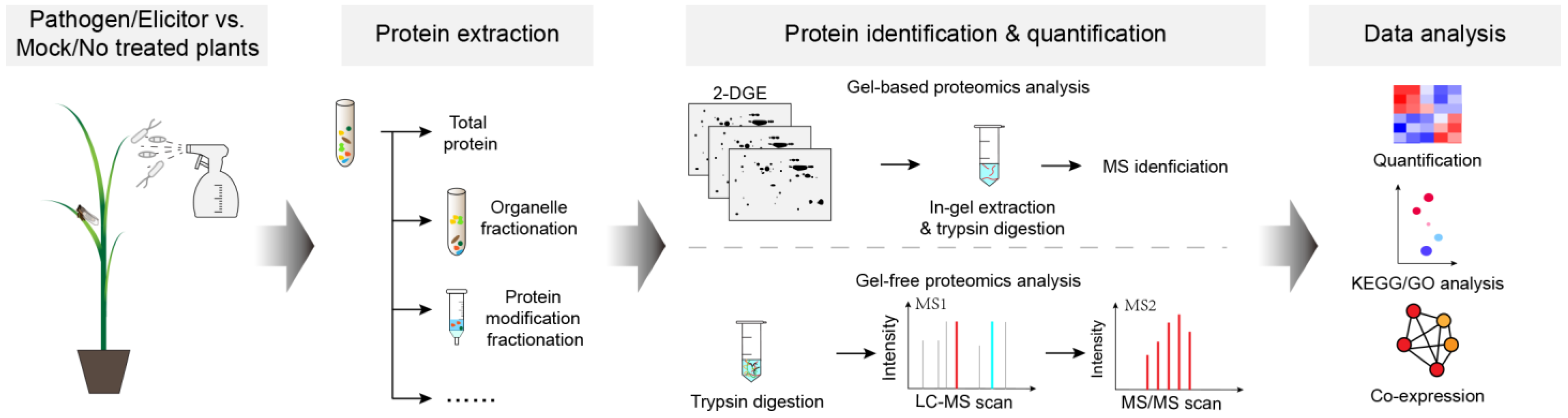
1. Introduction
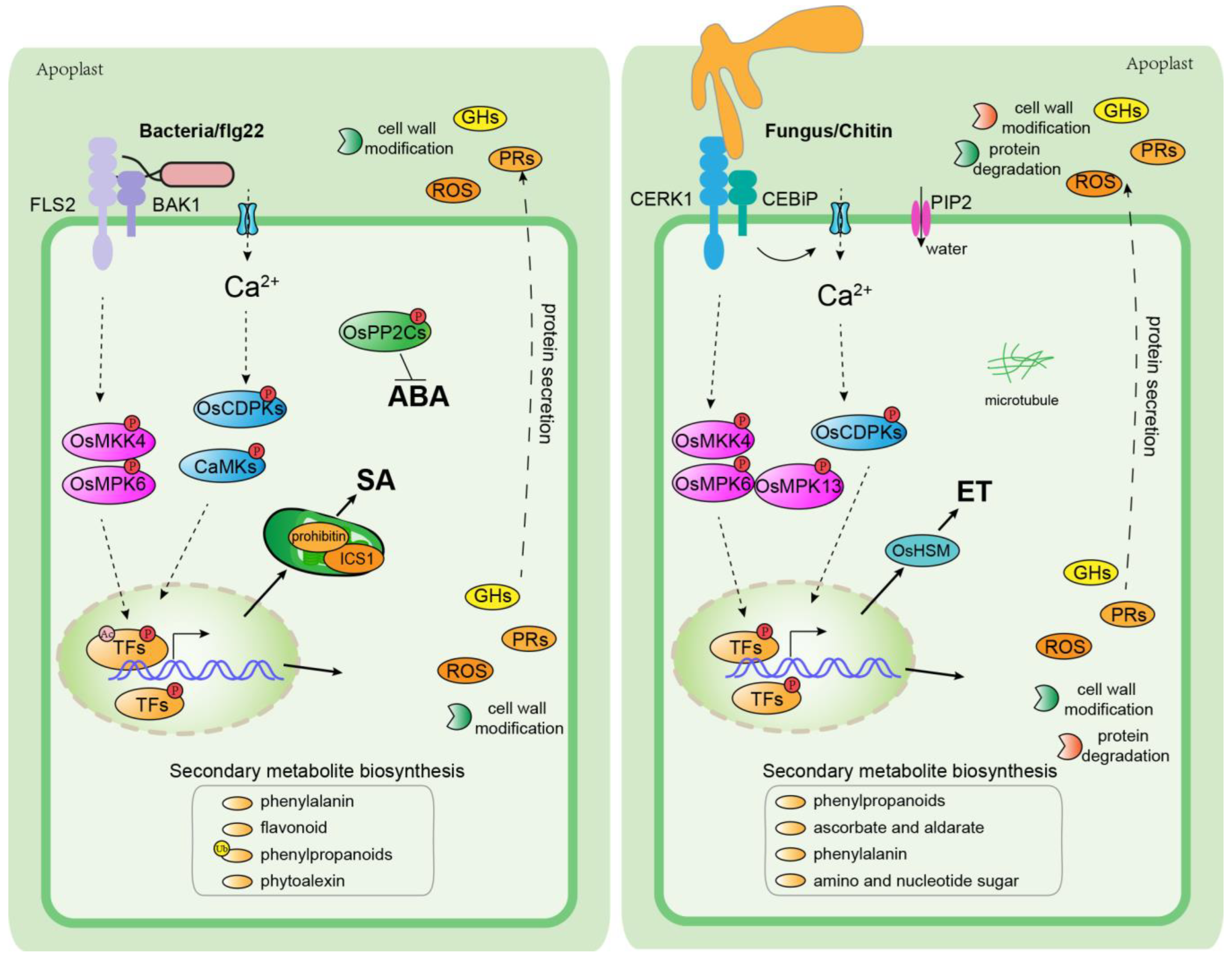
2. Interactions between Rice and Pathogenic Bacteria
3. Interactions between Rice and Growth-Promoting Bacteria
4. Interactions between Rice and Fungal Pathogens
5. Interactions between Rice and Virus Pathogens
6. Summary and Perspectives
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Vo, K.T.X.; Rahman, M.M.; Trinh, K.T.T.; Kim, S.T.; Jeon, J.S. Proteomics and metabolomics studies on the biotic stress responses of rice: An update. Rice 2021, 14, 31. [Google Scholar] [CrossRef]
- Gupta, R.; Wang, Y.; Agrawal, G.K.; Rakwal, R.; Jo, I.H.; Bang, K.H.; Kim, S.T. Time to dig deep into the plant proteome: A hunt for low-abundance proteins. Front. Plant Sci. 2015, 6, 22. [Google Scholar] [CrossRef]
- Biemann, K. Mass spectrometry of peptides and proteins. Annu. Rev. Biochem. 1992, 61, 977–1010. [Google Scholar] [CrossRef]
- Wasinger, V.C.; Cordwell, S.J.; Cerpa-Potjak, A.; Yan, J.X.; Gooley, A.A.; Wilkins, M.R.; Duncan, M.W.; Harris, R.; Williams, K.L.; Humphrey-Smith, I. Progress with gene-product mapping of the Mollicutes: Mycoplasma genitalium. Electrophoresis 1995, 16, 1090–1094. [Google Scholar] [CrossRef]
- Steen, H.; Mann, M. The ABC’s (and XYZ’s) of peptide sequencing. Nat. Rev. Mol. Cell Biol. 2004, 5, 699–711. [Google Scholar] [CrossRef]
- Meleady, P. Two-dimensional gel electrophoresis and 2D-DIGE. Methods Mol. Biol. 2018, 1664, 3–14. [Google Scholar]
- Shiio, Y.; Aebersold, R. Quantitative proteome analysis using isotope-coded affinity tags and mass spectrometry. Nat. Protoc. 2006, 1, 139–145. [Google Scholar] [CrossRef]
- Unwin, R.D. Quantification of proteins by iTRAQ. Methods Mol. Biol. 2010, 658, 205–215. [Google Scholar]
- Zhang, L.; Elias, J.E. Relative protein quantification using tandem mass tag mass spectrometry. Methods Mol. Biol. 2017, 1550, 185–198. [Google Scholar]
- Ong, S.E.; Mann, M. A practical recipe for stable isotope labeling by amino acids in the cell culture (SILAC). Nat. Protoc. 2006, 1, 2650–2660. [Google Scholar] [CrossRef]
- Wang, D.X.; Eraslan, B.; Wieland, T.; Hallstrom, B.; Hopf, T.; Zolg, D.P.; Zecha, J.; Asplund, A.; Li, L.H.; Meng, C.; et al. A deep proteome and transcriptome abundance atlas of 29 healthy human tissues. Mol. Syst. Biol. 2019, 15, 2. [Google Scholar] [CrossRef] [PubMed]
- Omenn, G.S.; Lane, L.; Lundberg, E.K.; Oyerall, C.M.; Deutsch, E.W. Progress on the HUPO Draft Human Proteome: 2017 metrics of the human proteome project. J. Proteome Res. 2017, 16, 4281–4287. [Google Scholar] [CrossRef] [PubMed]
- Baerenfaller, K.; Grossmann, J.; Grobei, M.A.; Hull, R.; Hirsch-Hofmann, M.; Yalovsky, S.; Zimmermann, P.; Grossniklaus, U.; Gruissem, W.; Baginsky, S. Genome-scale proteomics reveals Arabidopsis thaliana gene models and proteome dynamics. Science 2008, 320, 938–941. [Google Scholar] [CrossRef]
- Liu, Y.; Lu, S.; Liu, K.; Wang, S.; Huang, L.; Guo, L. Proteomics: A powerful tool to study plant responses to biotic stress. Plant Methods 2019, 15, 135. [Google Scholar] [CrossRef]
- Jain, A.; Singh, H.B.; Das, S. Deciphering plant-microbe crosstalk through proteomics studies. Microbiol. Res. 2021, 242, 126590. [Google Scholar] [CrossRef]
- Sharma, M.; Sudheer, S.; Usmani, Z.; Rani, R.; Gupta, P. Deciphering the omics of plant-microbe interaction: Perspectives and new insights. Curr. Genom. 2020, 21, 343–362. [Google Scholar] [CrossRef]
- Chen, X.L.; Xie, X.; Wu, L.; Liu, C.; Zeng, L.; Zhou, X.; Luo, F.; Wang, G.L.; Liu, W. Proteomic analysis of ubiquitinated proteins in rice (Oryza sativa) after treatment with pathogen-associated molecular pattern (PAMP) elicitors. Front. Plant Sci. 2018, 9, 1064. [Google Scholar] [CrossRef]
- Tang, B.; Liu, C.; Li, Z.; Zhang, X.; Zhou, S.; Wang, G.L.; Chen, X.L.; Liu, W. Multilayer regulatory landscape during pattern-triggered immunity in rice. Plant Biotechnol. J. 2021, 19, 2629–2645. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Zhang, W.W. Affinity separation and enrichment methods in proteomics analysis. J. Proteom. 2008, 71, 284–303. [Google Scholar] [CrossRef] [PubMed]
- Scherp, P.; Ku, G.; Coleman, L.; Kheterpal, I. Gel-based and gel-free proteomic technologies. Methods Mol. Biol. 2011, 702, 163–190. [Google Scholar] [PubMed]
- Jayaraman, D.; Forshey, K.; Grimsrud, P.A.; Ane, J.M. Leveraging proteomics to understand plant-microbe interactions. Front. Plant Sci. 2012, 3, 44. [Google Scholar] [CrossRef]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Zipfel, C.; Robatzek, S.J. Pathogen-associated molecular pattern-triggered immunity: Veni, vid…? Plant Physiol. 2010, 154, 551–554. [Google Scholar] [CrossRef]
- Cui, H.; Tsuda, K.; Parker, J.E. Effector-triggered immunity: From pathogen perception to robust defense. Annu. Rev. Plant. Biol. 2015, 66, 487–511. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.; Jiang, Z.; Bi, G.; Nomura, K.; Liu, M.; Wang, Y.; Cai, B.; Zhou, J.M.; He, S.Y.; Xin, X.F. Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature 2021, 592, 105–109. [Google Scholar] [CrossRef]
- Ngou, B.P.; Ahn, H.K.; Ding, P.; Jones, J.D. Mutual potentiation of plant immunity by cell-surface and intracellular receptors. Nature 2021, 592, 110–115. [Google Scholar] [CrossRef]
- Durrant, W.E.; Dong, X.J. Systemic acquired resistance. Annu. Rev. Phytopathol. 2004, 42, 185–209. [Google Scholar] [CrossRef]
- Song, W.Y.; Wang, G.L.; Chen, L.L.; Kim, H.S.; Pi, L.Y.; Holsten, T.; Gardner, J.; Wang, B.; Zhai, W.X.; Zhu, L.H. A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 1995, 270, 1804–1806. [Google Scholar] [CrossRef]
- Kaku, H.; Nishizawa, Y.; Ishii-Minami, N.; Akimoto-Tomiyama, C.; Dohmae, N.; Takio, K.; Minami, E.; Shibuya, N.J. Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc. Natl. Acad. Sci. USA 2006, 103, 11086–11091. [Google Scholar] [CrossRef] [PubMed]
- Takai, R.; Isogai, A.; Takayama, S.; Che, F.S. Analysis of flagellin perception mediated by flg22 receptor OsFLS2 in rice. Mol. Plan. Microbe Interact. 2008, 21, 1635–1642. [Google Scholar] [CrossRef]
- Liu, C.H.; Wu, D.Y.; Pollock, J.D. Bioinformatic Challenges of Big Data in Non-Coding RNA Research. Front. Genet. 2012, 3, 178. [Google Scholar] [CrossRef] [PubMed]
- Pruitt, R.N.; Schwessinger, B.; Joe, A.; Thomas, N.; Liu, F.; Albert, M.; Robinson, M.R.; Chan, L.J.; Luu, D.D.; Chen, H.J. The rice immune receptor XA21 recognizes a tyrosine-sulfated protein from a Gram-negative bacterium. Sci. Adv. 2015, 1, e150024. [Google Scholar] [CrossRef]
- Liu, W.; Liu, J.; Triplett, L.; Leach, J.E.; Wang, G.L. Novel insights into rice innate immunity against bacterial and fungal pathogens. Annu. Rev. Phytopathol. 2014, 52, 213–241. [Google Scholar] [CrossRef] [PubMed]
- Shimono, M.; Koga, H.; Akagi, A.; Hayashi, N.; Goto, S.; Sawada, M.; Kurihara, T.; Matsushita, A.; Sugano, S.; Jiang, C.J. Rice WRKY45 plays important roles in fungal and bacterial disease resistance. Mol. Plant Pathol. 2012, 13, 83–94. [Google Scholar] [CrossRef] [PubMed]
- Akamatsu, A.; Wong, H.L.; Fujiwara, M.; Okuda, J.; Nishide, K.; Uno, K.; Imai, K.; Umemura, K.; Kawasaki, T.; Kawano, Y. An OsCEBiP/OsCERK1-OsRacGEF1-OsRac1 module is an essential early component of chitin-induced rice immunity. Cell Host Microbe 2013, 13, 465–476. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Zhai, K.; Xie, Z.; Yang, D.; Zhu, X.; Liu, J.; Wang, X.; Qin, P.; Yang, Y.; Zhang, G.; et al. Epigenetic regulation of antagonistic receptors confers rice blast resistance with yield balance. Science 2017, 355, 962–965. [Google Scholar] [CrossRef]
- Gao, M.; He, Y.; Yin, X.; Zhong, X.; Yan, B.; Wu, Y.; Chen, J.; Li, X.; Zhai, K.; Huang, Y.; et al. Ca2+ sensor-mediated ROS scavenging suppresses rice immunity and is exploited by a fungal effector. Cell 2021, 184, 5391–5404. [Google Scholar] [CrossRef]
- Xu, G.; Yuan, M.; Ai, C.; Liu, L.; Zhuang, E.; Karapetyan, S.; Wang, S.; Dong, X. uORF-mediated translation allows engineered plant disease resistance without fitness costs. Nature 2017, 545, 491–494. [Google Scholar] [CrossRef]
- Gupta, R.; Lee, S.E.; Agrawal, G.K.; Rakwal, R.; Park, S.; Wang, Y.; Kim, S.T. Understanding the plant-pathogen interactions in the context of proteomics-generated apoplastic proteins inventory. Front. Plant Sci. 2015, 6, 352. [Google Scholar] [CrossRef] [PubMed]
- Mahmood, T.; Jan, A.; Kakishima, M.; Komatsu, S.J. Proteomic analysis of bacterial-blight defense-responsive proteins in rice leaf blades. Proteomics 2006, 6, 6053–6065. [Google Scholar] [CrossRef]
- Chen, F.; Yuan, Y.; Li, Q.; He, Z.J. Proteomic analysis of rice plasma membrane reveals proteins involved in early defense response to bacterial blight. Proteomics 2007, 7, 1529–1539. [Google Scholar] [CrossRef]
- Chu, L.Y.; Yan, S.P.; Wang, C.C.; Hu, H.T.; Sun, W.N.; Yan, C.Q.; Chen, J.P.; Yang, L. Pathogenesis-related proteins in somatic hybrid rice induced by bacterial blight. Phytochemistry 2008, 69, 1989–1996. [Google Scholar]
- Chen, X.; Deng, Z.; Yu, C.; Yan, C.; Chen, J. Secretome analysis of rice suspension-cultured cells infected by Xanthomonas oryzae pv. oryza (Xoo). Proteome Sci. 2016, 14, 2. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Dong, Y.; Yu, C.; Fang, X.; Deng, Z.; Yan, C.; Chen, J. Analysis of the proteins secreted from the Oryza meyeriana suspension-cultured cells induced by Xanthomonas oryzae pv. oryzae. PLoS ONE 2016, 11, e0154793. [Google Scholar] [CrossRef]
- Wang, Y.; Gupta, R.; Song, W.; Huh, H.-H.; Lee, S.E.; Wu, J.; Agrawal, G.K.; Rakwal, R.; Kang, K.Y.; Park, S.R. Label-free quantitative secretome analysis of Xanthomonas oryzae pv. oryzae highlights the involvement of a novel cysteine protease in its pathogenicity. J. Proteom. 2017, 169, 202–214. [Google Scholar] [CrossRef]
- Hou, Y.; Qiu, J.; Tong, X.; Wei, X.; Nallamilli, B.R.; Wu, W.; Huang, S.; Zhang, J. A comprehensive quantitative phosphoproteome analysis of rice in response to bacterial blight. BMC Plant Biol. 2015, 15, 163. [Google Scholar] [CrossRef]
- Ma, H.; Chen, J.; Zhang, Z.; Ma, L.; Yang, Z.; Zhang, Q.; Li, X.; Xiao, J.; Wang, S. MAPK kinase 10.2 promotes disease resistance and drought tolerance by activating different MAPKs in rice. Plant J. 2017, 92, 557–570. [Google Scholar] [CrossRef]
- Gupta, R.; Min, C.W.; Park, S.R.; Kim, S.T. Label-free proteome data of susceptible and resistant rice cultivars in response to Xanthomonas oryzae pv. oryzae inoculation. Data Br. 2022, 41, 107890. [Google Scholar] [CrossRef]
- Li, D.; Wang, L.; Teng, S.; Zhang, G.; Guo, L.; Mao, Q.; Wang, W.; Li, M.; Chen, L. Proteomics analysis of rice proteins up-regulated in response to bacterial leaf streak disease. J. Plant Biol. 2012, 55, 316–324. [Google Scholar] [CrossRef]
- Chi, F.; Yang, P.; Han, F.; Jing, Y.; Shen, S. Proteomics analysis of rice seedlings infected by Sinorhizobium meliloti 1021. Proteomics 2010, 10, 1861–1874. [Google Scholar] [CrossRef]
- Naher, U.A.; Panhwar, Q.A.; Othman, R.; Shamshuddin, J.; Ismail, M.R.; Zhou, E. Proteomic study on growth promotion of PGPR inoculated aerobic rice (Oryza sativa L.) cultivar MR219-9. Pak. J. Bot. 2018, 50, 1843–1852. [Google Scholar]
- Kandasamy, S.; Loganathan, K.; Muthuraj, R.; Duraisamy, S.; Seetharaman, S.; Thiruvengadam, R.; Ponnusamy, B.; Ramasamy, S. Understanding the molecular basis of plant growth promotional effect of Pseudomonas fluorescens on rice through protein profiling. Proteome Sci. 2009, 7, 47. [Google Scholar] [CrossRef]
- Konishi, H.; Ishiguro, K.; Komatsu, S. A proteomics approach towards understanding blast fungus infection of rice grown under different levels of nitrogen fertilization. Proteomics 2001, 1, 1162–1171. [Google Scholar] [CrossRef]
- Kim, S.T.; Cho, K.S.; Yu, S.; Kim, S.G.; Hong, J.C.; Han, C.D.; Bae, D.W.; Nam, M.H.; Kang, K.Y. Proteomic analysis of differentially expressed proteins induced by rice blast fungus and elicitor in suspension-cultured rice cells. Proteomics 2003, 3, 2368–2378. [Google Scholar] [CrossRef]
- Kim, S.T.; Kang, Y.H.; Wang, Y.; Wu, J.; Park, Z.Y.; Rakwal, R.; Agrawal, G.K.; Lee, S.Y.; Kang, K.Y. Secretome analysis of differentially induced proteins in rice suspension-cultured cells triggered by rice blast fungus and elicitor. Proteomics 2009, 9, 1302–1313. [Google Scholar] [CrossRef]
- Kim, S.G.; Wang, Y.; Lee, K.H.; Park, Z.Y.; Park, J.; Wu, J.; Kwon, S.J.; Lee, Y.H.; Agrawal, G.R.; Rakwal, R.; et al. In-depth insight into in vivo apoplastic secretome of rice-Magnaporthe oryzae interaction. J. Proteom. 2013, 78, 58–71. [Google Scholar] [CrossRef]
- Kim, S.T.; Kim, S.G.; Hwang, D.H.; Kang, S.Y.; Kim, H.J.; Lee, B.H.; Lee, J.J.; Kang, K.Y. Proteomic analysis of pathogen-responsive proteins from rice leaves induced by rice blast fungus, Magnaporthe grisea. Proteomics 2004, 4, 3569–3578. [Google Scholar] [CrossRef]
- Ma, Z.; Wang, L.; Zhao, M.; Gu, S.; Wang, C.; Zhao, J.; Tang, Z.; Gao, H.; Zhang, L.; Fu, L.; et al. iTRAQ proteomics reveals the regulatory response to Magnaporthe oryzae in durable resistant vs. susceptible rice genotypes. PLoS ONE 2020, 15, e0227470. [Google Scholar] [CrossRef]
- Lee, J.; Bricker, T.M.; Lefevre, M.; Pinson, S.R.; Oard, J.H. Proteomic and genetic approaches to identifying defence-related proteins in rice challenged with the fungal pathogen Rhizoctonia solani. Mol. Plant Pathol. 2006, 7, 405–416. [Google Scholar] [CrossRef]
- Ji, Z.; Zeng, Y.; Liang, Y.; Qian, Q.; Yang, C. Proteomic dissection of the rice-Fusarium fujikuroi interaction and the correlation between the proteome and transcriptome under disease stress. BMC Genom. 2019, 20, 91. [Google Scholar] [CrossRef]
- Kim, J.Y.; Wu, J.; Kwon, S.J.; Oh, H.; Lee, S.E.; Kim, S.G.; Wang, Y.; Agrawal, G.K.; Rakwal, R.; Kang, K.Y. Proteomics of rice and Cochliobolus miyabeanus fungal interaction: Insight into proteins at intracellular and extracellular spaces. Proteomics 2014, 14, 2307–2318. [Google Scholar] [CrossRef]
- Ventelon-Debout, M.; Delalande, F.; Brizard, J.P.; Diemer, H.; Van Dorsselaer, A.; Brugidou, C. Proteome analysis of cultivar-specific deregulations of Oryza sativa indica and O. sativa japonica cellular suspensions undergoing Rice yellow mottle virus infection. Proteomics 2004, 4, 216–225. [Google Scholar] [CrossRef]
- Wang, B.; Hajano, J.U.; Ren, Y.; Lu, C.; Wang, X. iTRAQ-based quantitative proteomics analysis of rice leaves infected by Rice stripe virus reveals several proteins involved in symptom formation. Virol. J. 2015, 12, 99. [Google Scholar] [CrossRef]
- Yang, A.; Yu, L.; Chen, Z.; Zhang, S.; Shi, J.; Zhao, X.; Yang, Y.; Hu, D.; Song, B. Label-free quantitative proteomic analysis of chitosan oligosaccharide-treated rice infected with southern Rice black-streaked dwarf virus. Viruses 2017, 9, 115. [Google Scholar] [CrossRef]
- Wang, Z.; Yu, L.; Jin, L.; Wang, W.; Zhao, Q.; Ran, L.; Li, X.; Chen, Z.; Guo, R.; Wei, Y. Evaluation of rice resistance to southern rice black-streaked dwarf virus and rice ragged stunt virus through combined field tests, quantitative real-time PCR, and proteome analysis. Viruses 2017, 9, 37. [Google Scholar] [CrossRef]
- Yu, L.; Wang, W.; Zeng, S.; Chen, Z.; Yang, A.; Shi, J.; Zhao, X.; Song, B. Label-free quantitative proteomics analysis of Cytosinpeptidemycin responses in southern rice black-streaked dwarf virus-infected rice. Pestic. Biochem. Physiol. 2018, 147, 20–26. [Google Scholar] [CrossRef]
- Min, C.W.; Jang, J.W.; Lee, G.H.; Gupta, R.; Yoon, J.; Park, H.J.; Cho, H.S.; Park, S.R.; Kwon, S.W.; Cho, L.H.; et al. TMT-based quantitative membrane proteomics identified PRRs potentially involved in the perception of MSP1 in rice leaves. J. Proteom. 2022, 267, 104687. [Google Scholar] [CrossRef]
- Ande, S.R.; Nguyen, K.H.; Nyomba, B.G.; Mishra, S.J. Prohibitin in adipose and immune functions. Trends Endocrinol. Metab. 2016, 27, 531–541. [Google Scholar] [CrossRef]
- Snedden, W.A. Characterization of the plant homologue of prohibitin, a gene associated with antiproliferative activity in mammalian cells. Plant Mol. Biol. 1997, 33, 753–756. [Google Scholar] [CrossRef]
- Seguel, A.; Jelenska, J.; Herrera-Vásquez, A.; Marr, S.K.; Joyce, M.B.; Gagesch, K.R.; Shakoor, N.; Jiang, S.C.; Fonseca, A.; Wildermuth, M.C.; et al. PROHIBITIN3 forms complexes with ISOCHORISMATE SYNTHASE1 to regulate stress-induced salicylic acid biosynthesis in Arabidopsis. Plant Physiol. 2018, 176, 2515–2531. [Google Scholar] [CrossRef]
- Chen, K.; Guo, T.; Li, X.M.; Yang, Y.B.; Dong, N.Q.; Shi, C.L.; Ye, W.W.; Shan, J.X.; Lin, H.X. NAL8 encodes a prohibitin that contributes to leaf and spikelet development by regulating mitochondria and chloroplasts stability in rice. BMC Plant Biol. 2019, 19, 395. [Google Scholar] [CrossRef]
- Ahn, C.S.; Lee, J.H.; Reum Hwang, A.; Kim, W.T.; Pai, H.S. Prohibitin is involved in mitochondrial biogenesis in plants. Plant J. 2006, 46, 658–667. [Google Scholar] [CrossRef]
- Zhang, F.; Huang, L.; Zeng, D.; Cruz, C.V.; Li, Z.; Zhou, Y.J. Comparative proteomic analysis reveals novel insights into the interaction between rice and Xanthomonas oryzae pv. oryzae. BMC Plant Biol. 2020, 20, 563. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Qin, G.; Zhang, Y.; Liu, C.; Wei, M.; Cen, Z.; Yan, Y.; Luo, T.; Li, Z.; Liang, H. Bacterial leaf streak 1 encoding a mitogen-activated protein kinase confers resistance to bacterial leaf streak in rice. Plant J. 2021, 107, 1084–1101. [Google Scholar] [CrossRef]
- Kishi-Kaboshi, M.; Okada, K.; Kurimoto, L.; Murakami, S.; Umezawa, T.; Shibuya, N.; Yamane, H.; Miyao, A.; Takatsuji, H.; Takahashi, A.J. A rice fungal MAMP-responsive MAPK cascade regulates metabolic flow to antimicrobial metabolite synthesis. Plant J. 2010, 63, 599–612. [Google Scholar] [CrossRef]
- Guo, T.; Chen, K.; Dong, N.Q.; Shi, C.L.; Ye, W.W.; Gao, J.P.; Shan, J.X.; Lin, H.X. GRAIN SIZE AND NUMBER1 negatively regulates the OsMKKK10-OsMKK4-OsMPK6 cascade to coordinate the trade-off between grain number per panicle and grain size in rice. Plant Cell 2018, 30, 871–888. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Duan, P.; Yu, H.; Zhou, Z.; Zhang, B.; Wang, R.; Li, J.; Zhang, G.; Zhuang, S.; Lyu, J.; et al. Control of grain size and weight by the OsMKKK10-OsMKK4-OsMAPK6 signaling pathway in rice. Mol. Plant. 2018, 11, 860–873. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Min, C.W.; Son, S.; Lee, G.H.; Jang, J.W.; Kwon, S.W.; Park, S.R.; Kim, S.T. Comparative proteome profiling of susceptible and resistant rice cultivars identified an arginase involved in rice defense against Xanthomonas oryzae pv. oryzae. Plant Physiol. Bioch. 2022, 171, 105–114. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Y.; Wang, Y. Apoplastic proteases: Powerful weapons against pathogen infection in plants. Plant Commun. 2020, 1, 100085. [Google Scholar] [CrossRef]
- Rodriguez, P.L. Protein phosphatase 2C (PP2C) function in higher plants. Plant Mol. Biol. 1998, 38, 919–927. [Google Scholar] [CrossRef]
- Liu, Q.; Yan, S.; Huang, W.; Yang, J.; Dong, J.; Zhang, S.; Zhao, J.; Yang, T.; Mao, X.; Zhu, X. NAC transcription factor ONAC066 positively regulates disease resistance by suppressing the ABA signaling pathway in rice. Plant Mol. Biol. 2018, 98, 289–302. [Google Scholar] [CrossRef]
- Rolli, E.; Marasco, R.; Vigani, G.; Ettoumi, B.; Mapelli, F.; Deangelis, M.L.; Gandolfi, C.; Casati, E.; Previtali, F.; Gerbino, R.; et al. Improved plant resistance to drought is promoted by the root-associated microbiome as a water stress-dependent trait. Environ. Microbiol. 2015, 17, 316–331. [Google Scholar] [CrossRef]
- Chaudhry, V.; Runge, P.; Sengupta, P.; Doehlemann, G.; Parker, J.E.; Kemen, E. Shaping the leaf microbiota: Plant–microbe–microbe interactions. J. Exp. Bot. 2021, 72, 36–56. [Google Scholar] [PubMed]
- Stacy, A.; Andrade-Oliveira, V.; McCulloch, J.A.; Hild, B.; Oh, J.H.; Perez-Chaparro, P.J.; Sim, C.K.; Lim, A.I.; Link, V.M.; Enamorado, M.; et al. Infection trains the host for microbiota-enhanced resistance to pathogens. Cell 2021, 184, 615–627.e17. [Google Scholar] [CrossRef] [PubMed]
- Xiang, J.; Ran, J.; Zou, J.; Zhou, X.; Liu, A.; Zhang, X.; Peng, Y.; Tang, N.; Luo, G.; Chen, X. Heat shock factor OsHsfB2b negatively regulates drought and salt tolerance in rice. Plant Cell Rep. 2013, 32, 1795–1806. [Google Scholar] [CrossRef] [PubMed]
- Sesma, A.; Osbourn, A.E. The rice leaf blast pathogen undergoes developmental processes typical of root-infecting fungi. Nature 2004, 431, 582–586. [Google Scholar] [CrossRef]
- Fernandez, J.; Orth, K. Rise of a Cereal Killer: The biology of Magnaporthe oryzae Biotrophic growth. Trends Microbiol. 2018, 26, 582–597. [Google Scholar] [CrossRef]
- Meng, Q.; Gupta, R.; Min, C.W.; Kwon, S.W.; Wang, Y.; Je, B.I.; Kim, Y.J.; Jeon, J.S.; Agrawal, G.K.; Rakwal, R. Proteomics of Rice—Magnaporthe oryzae interaction: What have we learned so far? Front. Plant Sci. 2019, 10, 1383. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.G.; Kim, S.T.; Wang, Y.; Yu, S.; Choi, I.S.; Kim, Y.C.; Kim, W.T.; Agrawal, G.K.; Rakwal, R.; Kang, K.Y. The RNase activity of rice probenazole-induced protein1 (PBZ1) plays a key role in cell death in plants. Mol. Cells 2011, 31, 25–31. [Google Scholar] [CrossRef]
- Lin, S.; Nie, P.; Ding, S.; Zheng, L.; Chen, C.; Feng, R.; Wang, Z.; Wang, L.; Wang, J.; Fang, Z. Quantitative proteomic analysis provides insights into rice defense mechanisms against Magnaporthe oryzae. Int. J. Mol. Sci. 2018, 19, 1950. [Google Scholar] [CrossRef]
- Huang, L.F.; Lin, K.H.; He, S.L.; Chen, J.L.; Jiang, J.Z.; Chen, B.H.; Hou, Y.S.; Chen, R.S.; Hong, C.Y.; Ho, S.L. Multiple patterns of regulation and overexpression of a ribonuclease-like pathogenesis-related protein gene, OsPR10a, conferring disease resistance in rice and Arabidopsis. PLoS ONE 2016, 11, e0156414. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.G.; Kim, S.T.; Wang, Y.; Kim, S.K.; Lee, C.H.; Kim, K.K.; Kim, J.K.; Lee, S.Y.; Kang, K.Y. Overexpression of rice isoflavone reductase-like gene (OsIRL) confers tolerance to reactive oxygen species. Physiol. Plant 2010, 138, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Q.; Li, N.; Dong, L.; Zhang, D.; Fan, S.; Jiang, L.; Wang, X.; Xu, P.; Zhang, S. Overexpression of soybean isoflavone reductase (GmIFR) enhances resistance to Phytophthora sojae in soybean. Front. Plant Sci. 2015, 6, 1024. [Google Scholar] [CrossRef] [PubMed]
- Wrzaczek, M.; Brosché, M.; Salojärvi, J.; Kangasjärvi, S.; Idänheimo, N.; Mersmann, S.; Robatzek, S.; Karpiński, S.; Karpińska, B.; Kangasjärvi, J. Transcriptional regulation of the CRK/DUF26 group of receptor-like protein kinases by ozone and plant hormones in Arabidopsis. BMC Plant Biol. 2010, 10, 95. [Google Scholar] [CrossRef]
- Vaattovaara, A.; Brandt, B.; Rajaraman, S.; Safronov, O.; Veidenberg, A.; Luklová, M.; Kangasjärvi, J.; Löytynoja, A.; Hothorn, M.; Salojärvi, J. Mechanistic insights into the evolution of DUF26-containing proteins in land plants. Commun. Biol. 2019, 2, 56. [Google Scholar] [CrossRef]
- Zhang, L.; Tian, L.H.; Zhao, J.F.; Song, Y.; Zhang, C.J.; Guo, Y. Identification of an apoplastic protein involved in the initial phase of salt stress response in rice root by two-dimensional electrophoresis. Plant Physiol. 2009, 149, 916–928. [Google Scholar] [CrossRef]
- Jiang, J.; Li, J.; Xu, Y.; Han, Y.; Bai, Y.; Zhou, G.; Lou, Y.; Xu, Z.; Chong, K. RNAi knockdown of Oryza sativa root meander curling gene led to altered root development and coiling which were mediated by jasmonic acid signalling in rice. Plant Cell Environ. 2007, 30, 690–699. [Google Scholar] [CrossRef]
- Lourenço, T.F.; Serra, T.S.; Cordeiro, A.M.; Swanson, S.J.; Gilroy, S.; Saibo, N.J.; Oliveira, M.M. The rice E3-ubiquitin ligase HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENE1 modulates the expression of ROOT MEANDER CURLING, a gene involved in root mechanosensing, through the interaction with two ETHYLENE-RESPONSE FACTOR transcription factors. Plant Physiol. 2015, 169, 2275–2287. [Google Scholar]
- Takeda, T.; Takahashi, M.; Shimizu, M.; Sugihara, Y.; Yamashita, T.; Saitoh, H.; Fujisaki, K.; Ishikawa, K.; Utsushi, H.; Kanzaki, E.; et al. Rice apoplastic CBM1-interacting protein counters blast pathogen invasion by binding conserved carbohydrate binding module 1 motif of fungal proteins. PLoS Pathog. 2022, 18, e1010792. [Google Scholar] [CrossRef]
- Liu, S.; Wang, J.; Han, Z.; Gong, X.; Zhang, H.; Chai, J. Molecular mechanism for fungal cell wall recognition by rice chitin receptor OsCEBiP. Structure 2016, 24, 1192–1200. [Google Scholar] [CrossRef]
- Lin, F.; Manisseri, C.; Fagerström, A.; Peck, M.L.; Vega-Sánchez, M.E.; Williams, B.; Chiniquy, D.M.; Saha, P.; Pattathil, S.; Conlin, B. Cell wall composition and candidate biosynthesis gene expression during rice development. Plant Cell Physiol. 2016, 57, 2058–2075. [Google Scholar] [CrossRef]
- Bacete, L.; Melida, H.; Miedes, E.; Molina, A. Plant cell wall-mediated immunity: Cell wall changes trigger disease resistance responses. Plant J. 2018, 93, 614–636. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wu, J.; Kim, S.G.; Tsuda, K.; Gupta, R.; Park, S.Y.; Kim, S.T.; Kang, K.Y. Magnaporthe oryzae-Secreted Protein MSP1 induces cell death and elicits defense responses in rice. Mol. Plant Microbe Interact. 2016, 29, 299–312. [Google Scholar] [CrossRef] [PubMed]
- Song, D.; Chen, J.; Song, F.; Zheng, Z. A novel rice MAPK gene, OsBIMK2, is involved in disease-resistance responses. Plant Biol. 2006, 8, 587–596. [Google Scholar] [CrossRef]
- Song, F.; Goodman, R.M. Molecular cloning and characterization of a rice phosphoinositide-specific phospholipase C gene, OsPI-PLC1, that is activated in systemic acquired resistance. Physiol. Mol. Plant Pathol. 2002, 61, 31–40. [Google Scholar]
- Li, Y.; Ye, Z.; Nie, Y.; Zhang, J.; Wang, G.L.; Wang, Z. Comparative phosphoproteome analysis of Magnaporthe oryzae-responsive proteins in susceptible and resistant rice cultivars. J. Proteom. 2015, 115, 66–80. [Google Scholar] [CrossRef]
- Hardham, A.R. Microtubules and biotic interactions. Plant J. 2013, 75, 278–289. [Google Scholar] [CrossRef]
- Kobayashi, Y.; Kobayashi, I.; Funaki, Y.; Fujimoto, S.; Takemoto, T.; Kunoh, H. Dynamic reorganization of microfilaments and microtubules is necessary for the expression of non-host resistance in barley coleoptile cells. Plant J. 1997, 11, 525–537. [Google Scholar] [CrossRef]
- Yun, B.W.; Atkinson, H.A.; Gaborit, C.; Greenland, A.; Read, N.D.; Pallas, J.A.; Loake, G.J. Loss of actin cytoskeletal function and EDS1 activity, in combination, severely compromises non-host resistance in Arabidopsis against wheat Powdery mildew. Plant J. 2003, 34, 768–777. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Cha, J.; Choi, C.; Choi, N.; Ji, H.S.; Park, S.R.; Lee, S.; Hwang, D.J. Rice WRKY11 plays a role in pathogen defense and drought tolerance. Rice 2018, 11, 5. [Google Scholar] [CrossRef]
- Zhang, L.; Chen, L.; Dong, H. Plant aquaporins in infection by and immunity against pathogens—A critical review. Front. Plant Sci. 2019, 10, 632. [Google Scholar] [CrossRef]
- Bai, J.; Wang, X.; Yao, X.; Chen, X.; Lu, K.; Hu, Y.; Wang, Z.; Mu, Y.; Zhang, L.; Dong, H. Rice aquaporin OsPIP2;2 is a water-transporting facilitator in relevance to drought-tolerant responses. Plant Direct 2021, 5, e338. [Google Scholar] [CrossRef] [PubMed]
- Tian, S.; Wang, X.; Li, P.; Wang, H.; Ji, H.; Xie, J.; Qiu, Q.; Shen, D.; Dong, H. Plant aquaporin AtPIP1; 4 links apoplastic H2O2 induction to disease immunity pathways. Plant Physiol. 2016, 171, 1635–1650. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Li, Q.T.; Lei, Q.; Feng, C.; Zheng, X.; Zhou, F.; Li, L.; Liu, X.; Wang, Z.; Kong, J. Ectopically expressing MdPIP1; 3, an aquaporin gene, increased fruit size and enhanced drought tolerance of transgenic tomatoes. BMC Plant Biol. 2017, 17, 246. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.T.; Cho, K.S.; Jang, Y.S.; Kang, K.Y. Two-dimensional electrophoretic analysis of rice proteins by polyethylene glycol fractionation for protein arrays. Electrophoresis 2001, 22, 2103–2109. [Google Scholar] [CrossRef] [PubMed]
- Van Bockhaven, J.; Spíchal, L.; Novák, O.; Strnad, M.; Asano, T.; Kikuchi, S.; Höfte, M.; De Vleesschauwer, D. Silicon induces resistance to the brown spot fungus Cochliobolus miyabeanus by preventing the pathogen from hijacking the rice ethylene pathway. New Phytol. 2015, 206, 761–773. [Google Scholar] [CrossRef]
- Wang, Y.; Garrido-Oter, R.; Wu, J.; Winkelmüller, T.M.; Agler, M.; Colby, T.; Nobori, T.; Kemen, E.; Tsuda, K. Site-specific cleavage of bacterial MucD by secreted proteases mediates antibacterial resistance in Arabidopsis. Nat. Commun. 2019, 10, 2853. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Zavaliev, R.; Dong, X. Membrane trafficking in plant immunity. Mol. Plant 2017, 10, 1026–1034. [Google Scholar] [CrossRef]
- Lu, D.; Lin, W.; Gao, X.; Wu, S.; Cheng, C.; Avila, J.; Heese, A.; Devarenne, T.P.; He, P.; Shan, L. Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science 2011, 332, 1439–1442. [Google Scholar] [CrossRef]
- Liszczak, G.; Goldberg, J.M.; Foyn, H.; Petersson, E.J.; Arnesen, T.; Marmorstein, R. Molecular basis for N-terminal acetylation by the heterodimeric NatA complex. Nat. Struct. Mol. Biol. 2013, 20, 1098–1105. [Google Scholar] [CrossRef]
- Jalmi, S.K.; Sinha, A.K. Functional involvement of a mitogen activated protein kinase module, OsMKK3-OsMPK7-OsWRK30 in mediating resistance against Xanthomonas oryzae in rice. Sci. Rep. 2016, 6, 37974. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Chen, J.; Zhao, Y.; Wang, S.; Yuan, M. OsMAPK6 phosphorylates a zinc finger protein OsLIC to promote downstream OsWRKY30 for rice resistance to bacterial blight and leaf streak. J. Integr. Plant Biol. 2022, 64, 1116–1130. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Ni, H.; Chen, Q.; Sun, F.; Zhou, T.; Lan, Y.; Zhou, Y. Comparative proteomic analysis reveals the cross-talk between the responses induced by H2O2 and by long-term Rice black-streaked dwarf virus infection in rice. PLoS ONE 2013, 8, e81640. [Google Scholar]
- Wu, J.; Yang, R.; Yang, Z.; Yao, S.; Zhao, S.; Wang, Y.; Li, P.; Song, X.; Jin, L.; Zhou, T. ROS accumulation and antiviral defence control by microRNA528 in rice. Nat. Plants 2017, 3, 16203. [Google Scholar] [CrossRef] [PubMed]
- Kasajima, I. Difference in oxidative stress tolerance between rice cultivars estimated with chlorophyll fluorescence analysis. BMC Res. Notes 2017, 10, 168. [Google Scholar] [CrossRef]
- Wang, Z.; Li, Y.; Hou, B.; Pronobis, M.I.; Wang, M.; Wang, Y.; Cheng, G.; Weng, W.; Wang, Y.; Tang, Y. An array of 60,000 antibodies for proteome-scale antibody generation and target discovery. Sci. Adv. 2020, 6, eaax2271. [Google Scholar] [CrossRef]
- Zeng, W.; Niu, L.; Wang, Z.; Wang, X.; Wang, Y.; Pan, L.; Lu, Z.; Cui, G.; Weng, W.; Wang, M. Application of an antibody chip for screening differentially expressed proteins during peach ripening and identification of a metabolon in the SAM cycle to generate a peach ethylene biosynthesis model. Hortic. Res. 2020, 7, 31. [Google Scholar] [CrossRef] [PubMed]

| Microbes/Elicitors | Method | Rice Cultivar | Sample | Differentially Accumulated Proteins | References | |
|---|---|---|---|---|---|---|
| Bacterium | Xanthomonas oryzae pv. oryzae (Xoo) | 2-DGE, MALDI-TOF MS | Oryza sativa L. cv. Java 14 | Leaf cytoplasm, membrane protein | PBZ1, PR5, SOD, Peroxiredoxin | [40] |
| 2-DGE, MALDI TOF-TOF MS/MS | Oryza sativa L. japonica cv Nipponbare harboring Xa21-GFP | Leaf plasma membrane protein | PM-associated H+-ATPase, Protein phosphatase, Hypersensitive-induced response protein, Prohibitin | [41] | ||
| 2-DGE, MALDI-TOF-TOF | Oryza meyeriana L. | Leaf total protein | Ascorbate peroxidase, putative Glutathione S-transferase, Mitochondrial chaperonin-60 | [42] | ||
| 2D-DIGE, MALDI-TOF-TOF | O. sativa L. japonica cv Nipponbare | Secreted protein from suspension-cultured cells | Cu/Zn-SOD, Cellulase, CHIT16 | [43] | ||
| 2D-DIGE, MALDI-TOF-TOF | Oryza meyeriana L. | Secreted protein from suspension-cultured cells | Ser/Thr protein phosphatase family protein, Phospholipase C, GDSL-like lipase/acyl hydrolase, OsPDIL1-1, Glucan endo-1,3-beta-glucosidase, Peroxidases, Cu/Zn-SOD, Expansin | [44] | ||
| SDS-PAGE, MudPIT | Oryza sativa L. japonica cv Dongjin | Secreted protein from suspension-cultured cells and leaves | Peroxidase, Peroxiredoxin, Cu/Zn-SOD, Ferrodexin, Glutathione S-transferase, Thioredoxin, Ascorbate peroxidase, Chitinase, Thaumatin-like proteins, Pathogenesis-related bet VI family protein | [45] | ||
| TiO2-MOAC, nLC-MS/MS | Oryza sativa L. cv. IRBB5 IRBB13 | Leaf total protein | PP2Cs, Brassinosteroid insensitive 1-associated receptor kinase 1, OsWRKY72 | [46] | ||
| TMT, LC-MS/MS | Rice introgression line | Leaf total protein | CDPK13, OsMKK4, OsMPK6, OsPR1b | [47] | ||
| nLC-MS/MS | O. sativa L. japonica cv Dongjin, Hwayeong | Low-abundance protein from leaves | CDPKs, PTI-like tyrosin-protein kinase, serine/threonine-protein kinase, OsArg1 | [48] | ||
| Xanthomonas campestris pv. oryzicola (Xoc) | 2-DGE, MALDI-TOF-MS | Oryza sativa L. indica cv. 9311 | Leaf total protein | OsMPK6, Allene oxide synthase 3, receptor-like kinase, L-ascorbate peroxidase 3, PR1-like protein, PR10 | [49] | |
| Sinorhizobium meliloti | 2-DGE, MALDI-TOF-MS | O. sativa L. japonica cv Nipponbare | Root, leaf sheath, leaf total protein | Subtilisin-like proteinase, Exoglucanase, Enolase, Catalase, Auxin-induced protein | [50] | |
| Stenotrophomonas maltophilla and bacillus | 2-DGE, nLC-MS/MS | Oryza sativa L. indica cv. MR219-9 | Leaf sheath total protein | Malate dehydrogenase, HSFB2B, Triosephosphateisomerase | [51] | |
| Pseudomonas fluorescens | 2-DGE, nLC-MS/MS | Oryza sativa L. indica cv. CO43 | Leaf sheath total protein | Thioredoxin, Nucleotide Diphosphate kinase, putative glutathione S-transferase | [52] | |
| Fungus | Magnaporthe oryzae (Mo) | 2-DGE, N-terminal, and internal amino acid sequence analysis | Oryza sativa L. cv Hitomebore | Leaf sheath total protein | Oxygen-evolving enhancer protein 2, Fe-SOD, Cu/Zn-SOD, Thaumatin-like protein | [53] |
| 2-DGE, N-terminal, and internal amino acid sequence analysis | Oryza sativa L. japonica cv. Jinheung | Leaf total protein | PBZ1, SalT, β-Glucosidase, OsIRL, PR10 | [54] | ||
| 2-DGE, MALDI-TOF-MS | Oryza sativa L. japonica cv. Jinheung | Suspension-cultured cell secreted protein | Chitinases, DUF26s, α-Amylases, Germin A | [55] | ||
| 2-DGE, MALDI-TOF-TOF, nESI-LC-MS/MS | Oryza sativa L. japonica cv. Jinheung | Leaf-secreted protein | Xylanase inhibitors, GH family proteins, DUF26s, PR5s, chitinases, PR1s, Proteases, Peroxidases | [56] | ||
| 2-DGE, MALDI-TOF-MS | Oryza sativa L. japonica cv. Jinheung | Leaf total protein | PBZ, PR10, β-1,3-glucanase1, β-1,3-glucanase2, TLP, RLK, POX22.3 | [57] | ||
| iTRAQ, LC-ESI-MS/MS | Oryza sativa L. cv Gangyuan8, Lijiangxintuanheigu | Leaf total protein | Defense proteins (PR1s, PR2s, PR3s, PR8s, PR10s, PR14s, PR15s), redox-oxygen-species-related proteins (Peroxidases, apxs), receptor kinases (DUF26s, RLCKs, LRRs, CDPK, MAPK) | [58] | ||
| Rhizoctonia solani | 2-DGE, ESI-Q-TOF MS | Oryza sativa L. cv Labelle, LSBR-5 | Leaf sheath total protein | β-1,3-glucanase, Stomatal ascorbate Peroxidase, Chitinase, 14-3-3 like protein | [59] | |
| Fusarium fujikuroi | TMT labeling, LC-MS/MS | Oryza sativa L. cv Nipponbare, 9311 | Seedling total protein | PIP2, Peroxidase, PR1, SBT3.8, Monodehydroascorbate reductase, Salicylic acid-binding protein2 | [60] | |
| Cochliobolus miyabeanus | 2-DGE, MALDI-TOF-TOF, nESI-LC-MS/MS | Oryza sativa L. japonica cv. Jinheung | Leaf total and secreted proteins | β-1,3-glucanase, Chitinase, Cu/Zn-SOD, Glutathione reductase, Thioredoxin, Protein disulfide isomerase | [61] | |
| Virus | Rice yellow mottle virus (RYMV) | 2-DGE, MALDI-TOF-MS, nLC-MS/MS | Oryza sativa L. cv IR64, Azucena | Suspension-cultured cell total protein | Cu/Zn-SOD, α-amylases, HSP70s, Ethylene-inducible protein, PR10a, | [62] |
| Rice stripe virus (RSV) | iTRAQ, LC-MS/MS | Oryza sativa L. cv Aichiasahi | Leaf total protein | PR1, PR10, Ascorbate peroxidase 1, Thioredoxin, Cu/Zn-SOD, Mn-SOD, Peroxidases | [63] | |
| Rice black-streaked dwarf virus (RBSDV) | SDS-PAGE, nLC-MS/MS | O. sativa L. japonica cv Nipponbare | Leaf total protein | PP2A, Glycolate oxidase1, Glycolate oxidase 5, Peroxidases, Catalase, Nucleoside diphosphate kinase | [64] | |
| SDS-PAGE, nLC-MS/MS | Oryza sativa L. cv Z1, L2186, FYXZ | Leaf total protein | Dolichyl-diphosphooligosaccharide-protein glycosyltransferase, Hypoxia upregulated protein, Membrane-attack complex | [65] | ||
| Southern rice black-streaked dwarf virus (SRBSDV) | SDS-PAGE, nLC-MS/MS | O. sativa L. japonica cv Nipponbare | Leaf total protein | PR5, PR10, PODs, SODs, CAT | [66] | |
| Elicitors | Chitin, flg22 | Affinity enrichment of ubiquitinated peptide, LC-MS/MS | O. sativa L. japonica cv Nipponbare | Seedlings | ubiquitination system, protein transportation, ligand recognition, membrane trafficking, redox reactions, phenylpropanoid metabolic | [17] |
| Chitin, flg22 | Total, affinity enrichment of ubiquitinated and acetylated peptides, LC-MS/MS | O. sativa L. japonica cv Nipponbare | Seedlings | Enzymes involved in secondary metabolite biosynthesis, WRKY30 | [18] | |
| MSP1 (PTI-inducing protein secreted from M. oryzae) | TMT labeling, LC-MS/MS | O. sativa L. japonica cv Dongjin | Leaf total protein | RLKs, BAK1, MAPK, CRT | [67] | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, L.; Wang, D.; Gupta, R.; Kim, S.T.; Wang, Y. A Proteomics Insight into Advancements in the Rice–Microbe Interaction. Plants 2023, 12, 1079. https://doi.org/10.3390/plants12051079
Wei L, Wang D, Gupta R, Kim ST, Wang Y. A Proteomics Insight into Advancements in the Rice–Microbe Interaction. Plants. 2023; 12(5):1079. https://doi.org/10.3390/plants12051079
Chicago/Turabian StyleWei, Lirong, Dacheng Wang, Ravi Gupta, Sun Tae Kim, and Yiming Wang. 2023. "A Proteomics Insight into Advancements in the Rice–Microbe Interaction" Plants 12, no. 5: 1079. https://doi.org/10.3390/plants12051079
APA StyleWei, L., Wang, D., Gupta, R., Kim, S. T., & Wang, Y. (2023). A Proteomics Insight into Advancements in the Rice–Microbe Interaction. Plants, 12(5), 1079. https://doi.org/10.3390/plants12051079








