Evaluating Introgression Sorghum Germplasm Selected at the Population Level While Exploring Genomic Resources as a Screening Method
Abstract
1. Introduction
2. Results
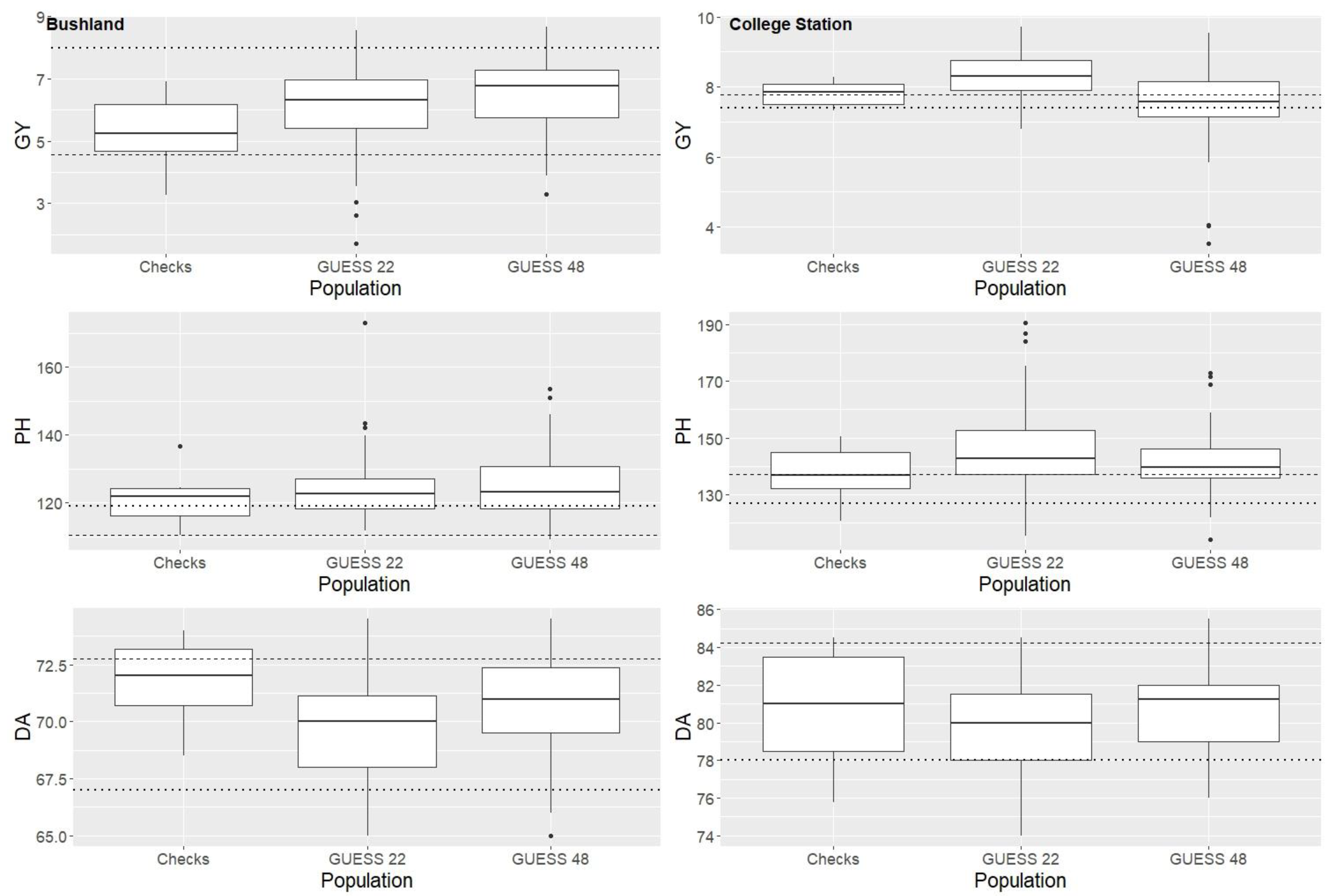
2.1. Statistical Analysis
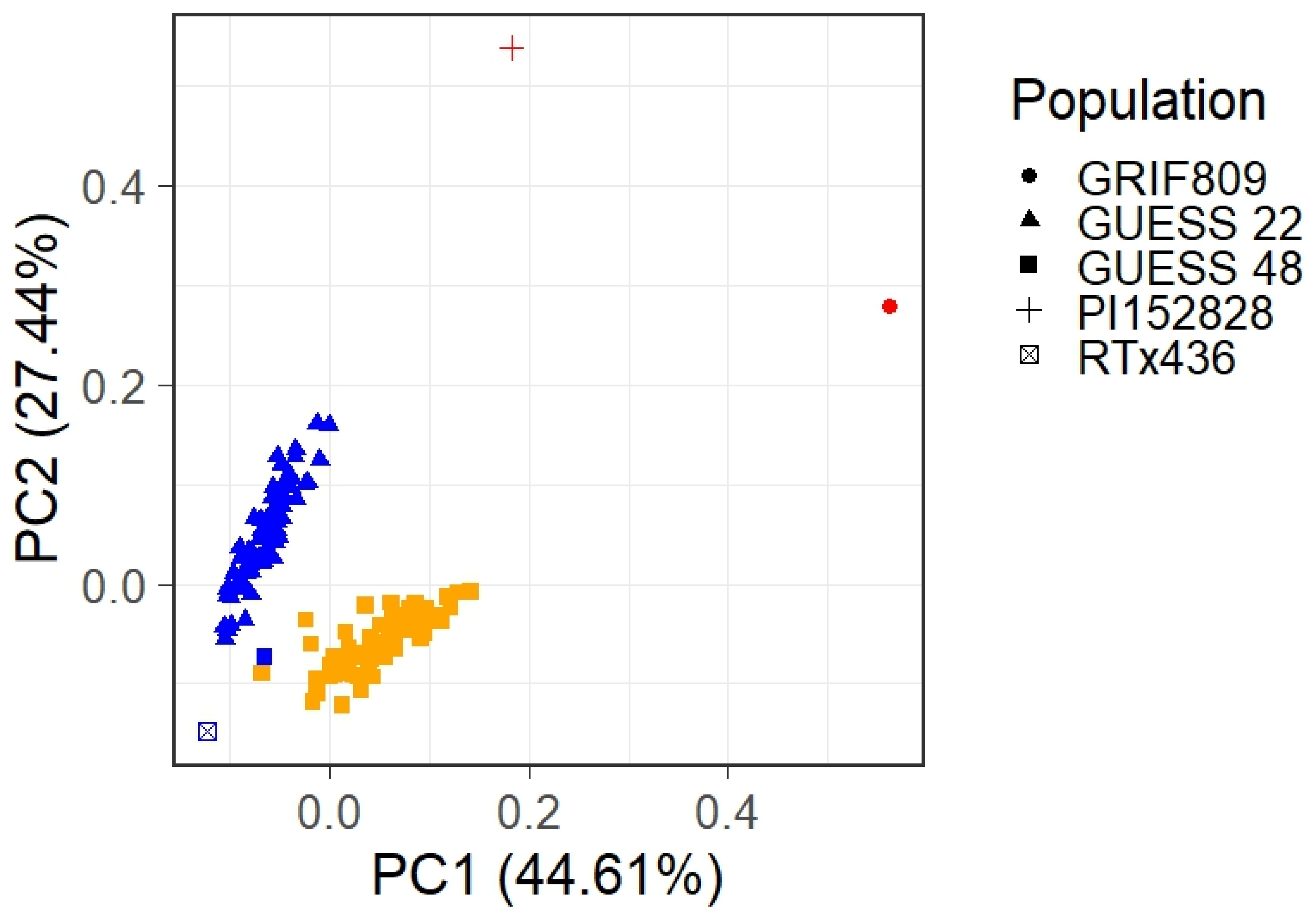
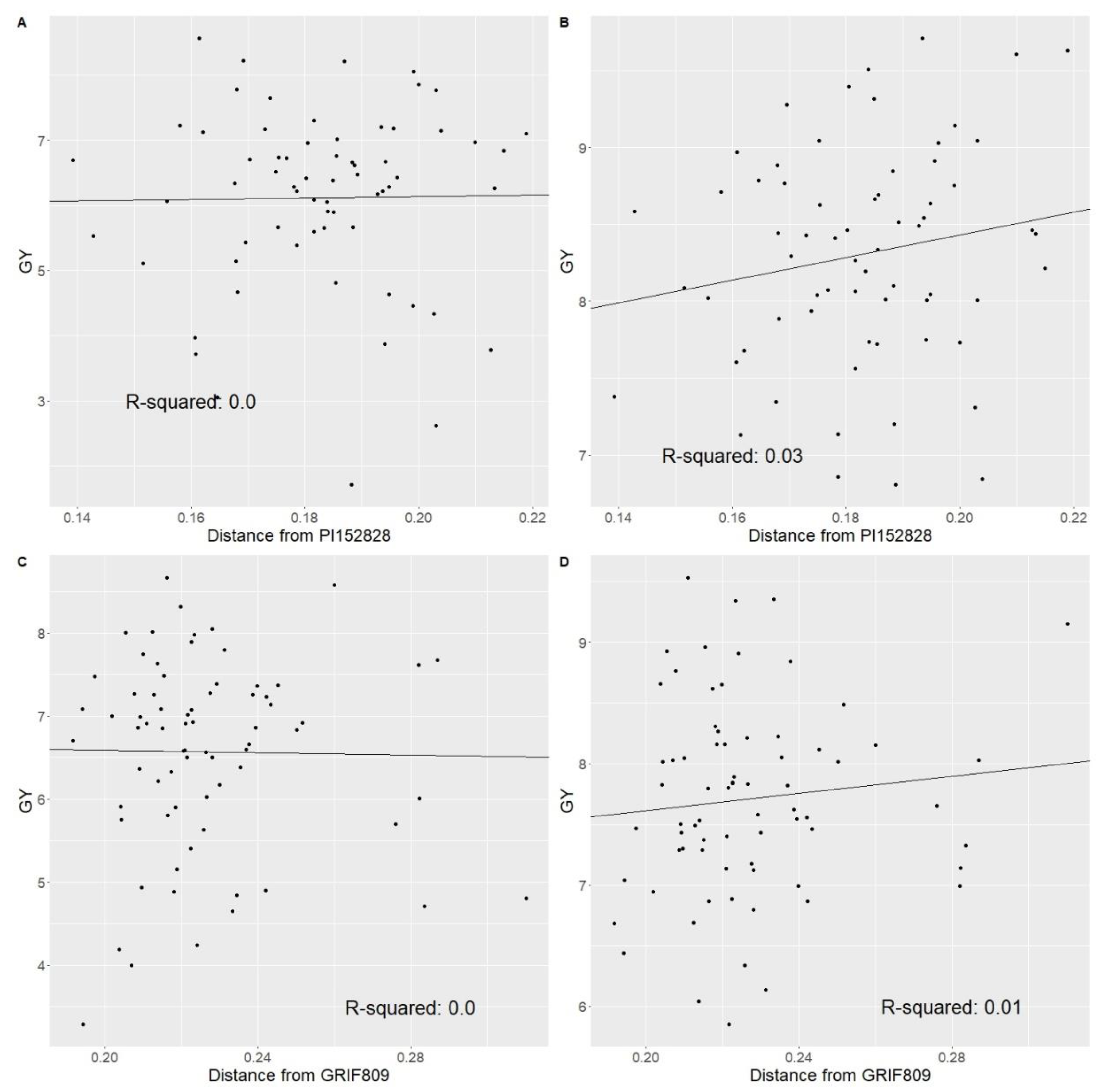
2.2. Population Structure and Genetic Distance
2.3. Genomic Prediction
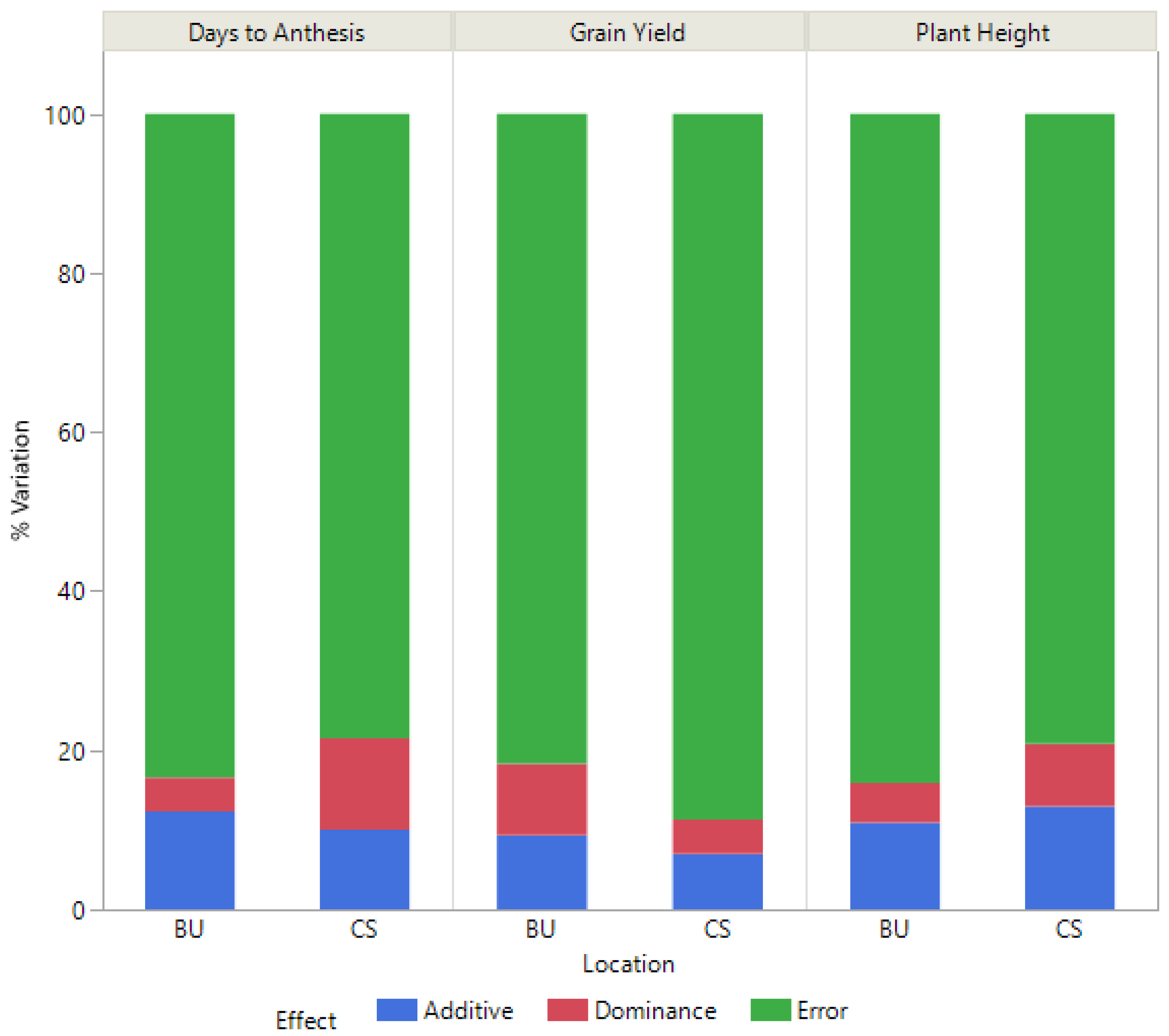
2.3.1. Variance Components
2.3.2. Cross-Validation Scheme CV1
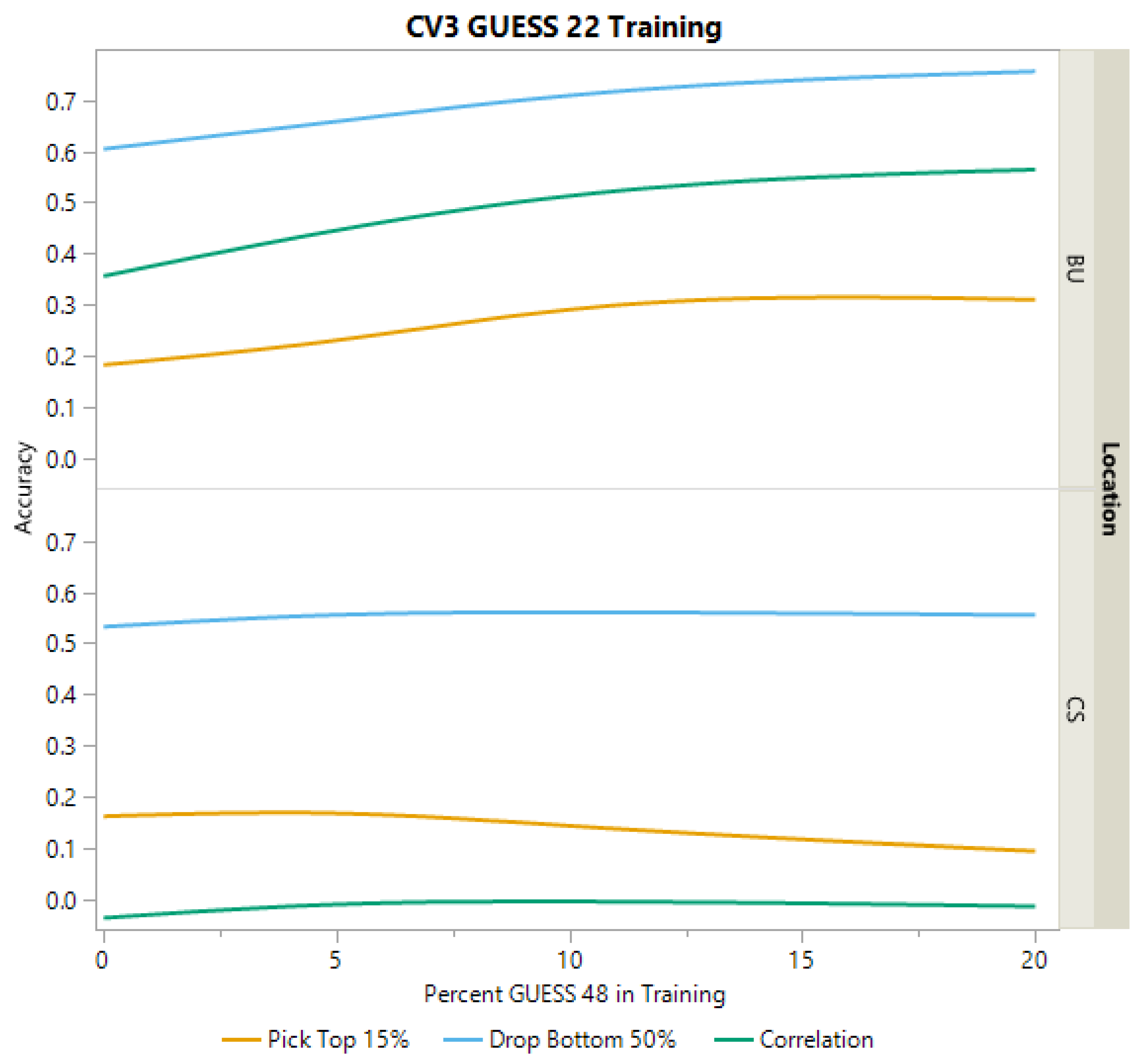
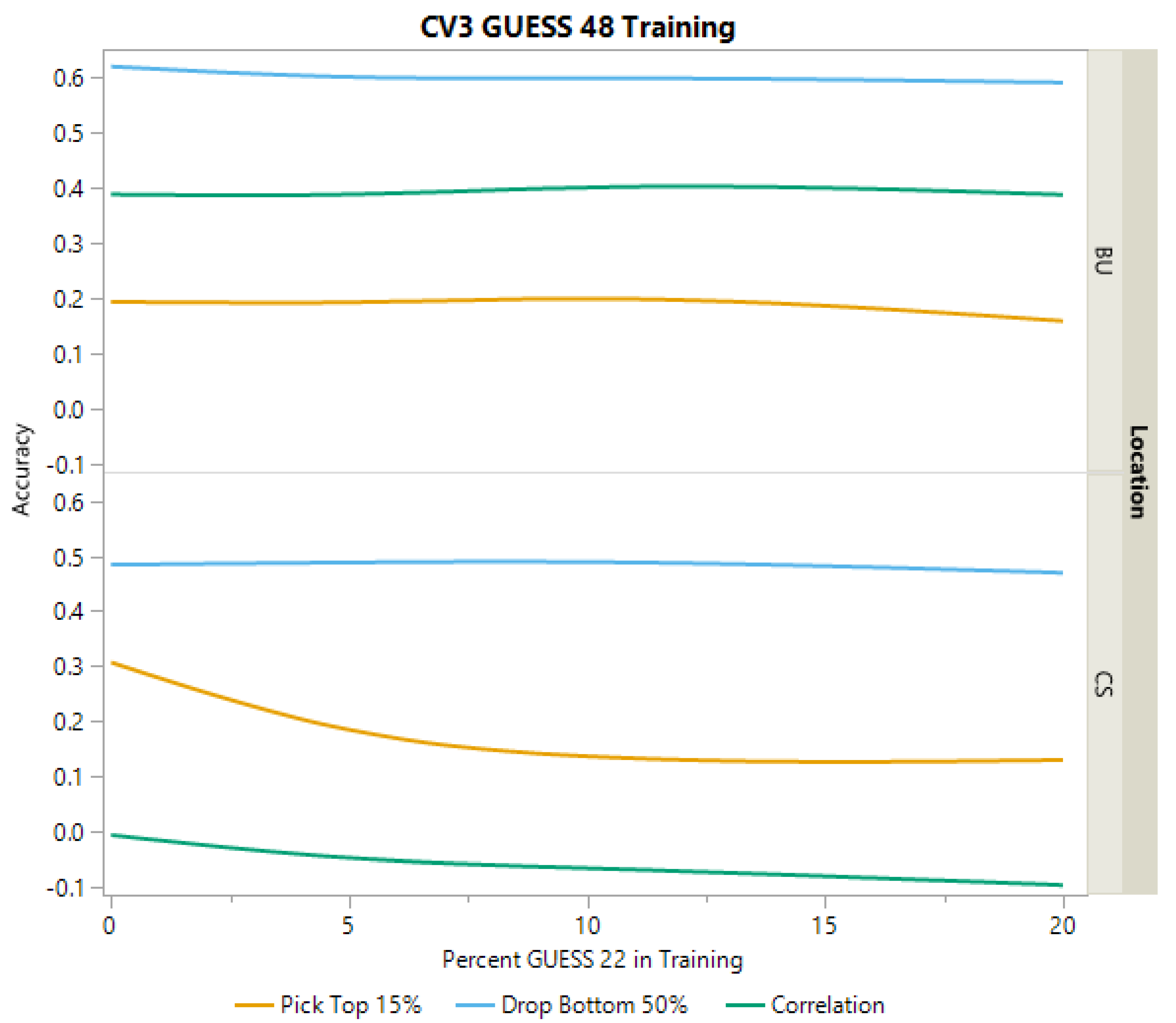
2.3.3. Cross-Validation Scheme CV3
3. Discussion
3.1. BC-NAM Genetic Diversity and Hybrid Performance
3.2. BC-NAM Populations and Genomic Prediction
4. Materials and Methods
4.1. Population Development
4.2. Hybrid Evaluation
4.3. Genotype Data
4.4. Statistical Analysis
4.5. Population Structure Analysis
4.6. Genomic Prediction
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Crozier, D.; Hoffmann, L.; Klein, P.E.; Klein, R.R.; Rooney, W.L. Predicting heterosis in grain sorghum hybrids using sequence-based genetic similarity estimates. J. Crop Improv. 2020, 34, 600–617. [Google Scholar] [CrossRef]
- Rooney, W.; Aydin, S. Genetic Control of a Photoperiod-Sensitive Response in Sorghum bicolor (L.) Moench. Crop Sci. 1999, 39, 397–400. [Google Scholar] [CrossRef]
- Duncan, R.R.; Bramel-Cox, P.J.; Miller, F.R. Contributions of Introduced Sorghum Germplasm to Hybrid Development in the USA. Use Plant Introd. Cultiv. Dev. Part 1 1991, 17, 69–102. [Google Scholar]
- Stephens, J.C. Male Sterility in Sorghum: Its Possible Utilization in Production of Hybrid Seed 1. Agron. J. 1937, 29, 690–696. [Google Scholar] [CrossRef]
- Quinby, J.R.; Martin, J.H. Sorghum Improvement. Adv. Agron. 1954, 6, 305–359. [Google Scholar]
- Stephens, J.C.; Miller, F.R.; Rosenow, D.T. Conversion of Alien Sorghums to Early Combine Genotypes1. Crop Sci. 1967, 7, 396. [Google Scholar] [CrossRef]
- Lin, Y.R.; Schertz, K.F.; Paterson, A.H. Comparative analysis of QTLs affecting plant height and maturity across the Poaceae, in reference to an interspecific sorghum population. Genetics 1995, 141, 391–411. [Google Scholar] [CrossRef]
- Rosenow, D.; Dahlberg, J.; Peterson, G.; Clark, L. Release of 50 converted sorghum lines and 253 partially converted sorghum bulks. Int. Sorghum Millets Newsl. 1995, 19–31. [Google Scholar]
- Rosenow, D.T.; Dahlberg, J.A.; Stephens, J.C.; Miller, F.R.; Barnes, D.K.; Peterson, G.C.; Johnson, J.W.; Schertz, K.F. Registration of 63 Converted Sorghum Germplasm Lines from the Sorghum Conversion Program. Crop Sci. 1997, 37, 1399–1400. [Google Scholar] [CrossRef]
- Rosenow, D.; Dahlberg, J.; Peterson, G.; Erpelding, J.; Sij, J.; Clark, L.; Hamburger, A.J. Release of 49 Converted Sorghum Germplasm Lines from the Sorghum Conversion Program. Sorghum Res. Rep. 2003, 57–60. [Google Scholar]
- Klein, R.R.; Miller, F.R.; Bean, S.; Klein, P.E. Registration of 40 Converted Germplasm Sources from the Reinstated Sorghum Conversion Program. J. Plant Regist. 2015, 10, 57–61. [Google Scholar] [CrossRef]
- Horne, D.W.; Patil, N.Y.; Klein, R.R.; Miller, F.R.; Hoffmann, L.; Klein, P.E.; Rooney, W.L. Registration of 11 diverse sorghum germplasm lines for grain and silage hybrid production. J. Plant Regist. 2020, 14, 179–188. [Google Scholar] [CrossRef]
- Morris, G.P.; Ramu, P.; Deshpande, S.P.; Hash, C.T.; Shah, T.; Upadhyaya, H.D.; Riera-Lizarazu, O.; Brown, P.J.; Acharya, C.B.; Mitchell, S.E. Population genomic and genome-wide association studies of agroclimatic traits in sorghum. Proc. Natl. Acad. Sci. USA 2013, 110, 453–458. [Google Scholar] [CrossRef]
- Zhou, Y.; Srinivasan, S.; Mirnezami, S.V.; Kusmec, A.; Fu, Q.; Attigala, L.; Salas Fernandez, M.G.; Ganapathysubramanian, B.; Schnable, P.S. Semiautomated Feature Extraction from RGB Images for Sorghum Panicle Architecture GWAS. Plant Physiol. 2019, 179, 24–37. [Google Scholar] [CrossRef]
- Brown, P.J.; Rooney, W.L.; Franks, C.; Kresovich, S. Efficient Mapping of Plant Height Quantitative Trait Loci in a Sorghum Association Population With Introgressed Dwarfing Genes. Genetics 2008, 180, 629–637. [Google Scholar] [CrossRef]
- Hayes, C.M.; Burow, G.B.; Brown, P.J.; Thurber, C.; Xin, Z.; Burke, J.J. Natural Variation in Synthesis and Catabolism Genes Influences Dhurrin Content in Sorghum. Plant Genome 2015, 8, plantgenome2014-09. [Google Scholar] [CrossRef]
- Miller, F.; Rooney, L.; Rosenow, D. Registration of Tx2818 through Tx2857 Sorghum bicolor (L.) Moench germplasm lines. Crop Sci. 1990, 30, 1168–1170. [Google Scholar] [CrossRef]
- Rooney, W.L. Registration of Tx2912 through Tx2920 sorghum germplasm lines. (Registrations Of Germplasms). Crop Sci. 2003, 43, 442–444. [Google Scholar] [CrossRef]
- Schertz, K.F. Registration of A2 Tx2753 and B Tx2753 Sorghum Germplasms1 (Reg. No. GP 30 and 31). Crop Sci. 1977, 17, 983. [Google Scholar] [CrossRef]
- Johnson, J.W.; Teetes, G.L.; Rosenow, D.T. Registration of TAM2567 and TAM2568 Greenbug Resistant Sorghum Germplasm Lines1 (Reg. Nos. GP76 and 77). Crop Sci. 1982, 22, 1271. [Google Scholar]
- Jordan, D.R.; Mace, E.S.; Cruickshank, A.W.; Hunt, C.H.; Henzell, R.G. Exploring and exploiting genetic variation from unadapted sorghum germplasm in a breeding program. Crop Sci. 2011, 51, 1444–1457. [Google Scholar] [CrossRef]
- Nice, L.M.; Steffenson, B.J.; Brown-Guedira, G.L.; Akhunov, E.D.; Liu, C.; Kono, T.J.Y.; Morrell, P.L.; Blake, T.K.; Horsley, R.D.; Smith, K.P. Development and genetic characterization of an advanced backcross-nested association mapping (AB-NAM) population of wild× cultivated barley. Genetics 2016, 203, 1453–1467. [Google Scholar] [CrossRef]
- Pham, A.T.; Maurer, A.; Pillen, K.; Brien, C.; Dowling, K.; Berger, B.; Eglinton, J.K.; March, T.J. Genome-wide association of barley plant growth under drought stress using a nested association mapping population. BMC Plant Biol. 2019, 19, 134. [Google Scholar] [CrossRef]
- Jordan, D.R.; Tao, Y.; Godwin, I.D.; Henzell, R.G.; Cooper, M.; McIntyre, C.L. Prediction of hybrid performance in grain sorghum using RFLP markers. Theor. Appl. Genet. 2003, 106, 559–567. [Google Scholar] [CrossRef]
- Maulana, F.; Perumal, R.; Tesso, T. Crop Science Hybrid performance as related to genomic diversity and population structure in public sorghum inbred lines. Crop Sci. 2020, 61, 357–371. [Google Scholar] [CrossRef]
- Fonseca, J.M.O.; Klein, P.E.; Crossa, J.; Pacheco, A.; Perez-Rodriguez, P.; Ramasamy, P.; Klein, R.; Rooney, W.L. Assessing combining abilities, genomic data, and genotype × environment interactions to predict hybrid grain sorghum performance. Plant Genome 2021, 14, e20127. [Google Scholar] [CrossRef]
- Crossa, J.; Pérez, P.; Hickey, J.; Burgueño, J.; Ornella, L.; Ceró N-Rojas, J.; Zhang, X.; Dreisigacker, S.; Babu, R.; Li, Y.; et al. Genomic prediction in CIMMYT maize and wheat breeding programs. Heredity 2014, 112, 48–60. [Google Scholar] [CrossRef]
- Patil, N.Y.; Hoffman, L., Jr.; Winans, N.D.; Perumal, R.; Hayes, C.; Emendack, Y.; Boyles, R.E.; Dahlberg, J.; Klein, R.R.; Klein, P.E.; et al. Registration of Sorghum [Sorghum bicolor (L.) Moench] Backcross-Nested Association Mapping (BC-NAM) Families in a BTx623 or RTx436 Background. J. Plant Regist. 2023; Accepted for publication. [Google Scholar]
- Technow, F.; Schrag, T.A.; Schipprack, W.; Bauer, E.; Simianer, H.; Melchinger, A.E. Genome properties and prospects of genomic prediction of hybrid performance in a breeding program of maize. Genetics 2014, 197, 1343–1355. [Google Scholar] [CrossRef]
- Guo, Z.; Tucker, D.M.; Basten, C.J.; Gandhi, H.; Ersoz, E.; Guo, B.; Xu, Z.; Wang, D.; Gay, G. The impact of population structure on genomic prediction in stratified populations. Theor. Appl. Genet. 2014, 127, 749–762. [Google Scholar] [CrossRef]
- Burrell, M.; Sharma, A.; Patil, N.Y.; Collins, S.D.; Anderson, W.F.; Rooney, W.L.; Klein, P.E. Sequencing of an Anthracnose-Resistant Sorghum Genotype and Mapping of a Major QTL Reveal Strong Candidate Genes for Anthracnose Resistance. Crop Sci. 2015, 55, 790–799. [Google Scholar] [CrossRef]
- Patil, N.Y.; Klein, R.R.; Williams, C.L.; Collins, S.D.; Knoll, J.E.; Burrell, A.M.; Anderson, W.F.; Rooney, W.L.; Klein, P.E. Quantitative Trait Loci Associated with Anthracnose Resistance in Sorghum. Crop Sci. 2017, 57, 877–890. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing. 2021. Available online: https://www.r-project.org/ (accessed on 15 March 2022).
- Bates, D.; Mächler, M.; Bolker, B.M.; Walker, S.C. Fitting Linear Mixed-Effects Models Using lme4. arXiv Prepr. 2014, arXiv:1406.5823. [Google Scholar]
- Wald, A. Tests of statistical hypotheses concerning several parameters when the number of observations is large. Am. Math. Soc. 1943, 54, 426–483. [Google Scholar] [CrossRef]
- Shapiro, S.S.; Wilk, M.B. An Analysis of Variance Test for Normality (Complete Samples). Biometrika 1965, 52, 591. [Google Scholar] [CrossRef]
- Bartlett, M.; Kendall, D. The statistical analysis of variance-heterogeneity and the logarithmic transformation. J. R. Stat. Soc. 1946, 8, 128–138. [Google Scholar] [CrossRef]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef]
- Pérez, P.; De Los Campos, G. Genome-wide regression and prediction with the BGLR statistical package. Genetics 2014, 198, 483–495. [Google Scholar] [CrossRef]
- VanRaden, P.M. Efficient methods to compute genomic predictions. J. Dairy Sci. 2008, 91, 4414–4423. [Google Scholar] [CrossRef]
- Vitezica, Z.G.; Varona, L.; Legarra, A. On the additive and dominant variance and covariance of individuals within the genomic selection scope. Genetics 2013, 195, 1223–1230. [Google Scholar] [CrossRef]
- Amadeu, R.R.; Cellon, C.; Olmstead, J.W.; Garcia, A.A.F.; Resende, M.F.R.; Muñoz, P.R. AGHmatrix: R Package to Construct Relationship Matrices for Autotetraploid and Diploid Species: A Blueberry Example. Plant Genome 2016, 9. [Google Scholar] [CrossRef] [PubMed]
| Variance Component | College Station | |||||
| Grain Yield | Plant Height | Days to Anthesis | ||||
| Estimate | % | Estimate | % | Estimate | % | |
| 0.28 *** | 27.7 | 142.9 *** | 75.4 | 4.91 *** | 70.1 | |
| 0.09 | 8.8 | 1.3 | 0.7 | 0.06 | 0.9 | |
| 0.65 | 63.5 | 45.4 | 23.9 | 2.03 | 29.0 | |
| CVe | 10% | 4.72% | 1.80% | |||
| R | 0.466 | 0.863 | 0.829 | |||
| Variance Component | Bushland | |||||
| Grain Yield | Plant Height | Days to Anthesis | ||||
| Estimate | % | Estimate | % | Estimate | % | |
| 1.19 *** | 54.3 | 41.4 *** | 35.8 | 4.07 *** | 73.41 | |
| 0.15 | 6.6 | 8.9 | 7.7 | 0.002 | 0.03 | |
| 0.86 | 39.1 | 65.4 | 56.5 | 1.47 | 26.56 | |
| CVe | 14.8% | 6.45% | 1.70% | |||
| R | 0.735 | 0.559 | 0.847 | |||
| Test | W | F | p-value | |||
| Shapiro–Wilk | 0.99 | - | 0.0021 ** | |||
| Bartlett | - | 1.2466 | 0.021 * | |||
| College Station | YD Correlation | PH Correlation | DA Correlation | Top 15% YD | Bottom 50% YD |
|---|---|---|---|---|---|
| Mean | 0.179 | 0.364 | 0.633 | 0.132 | 0.599 |
| Standard Error | 0.048 | 0.081 | 0.044 | 0.081 | 0.047 |
| Bushland | |||||
| Mean | 0.485 | 0.143 | 0.257 | 0.332 | 0.703 |
| Standard Error | 0.069 | 0.115 | 0.098 | 0.085 | 0.071 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Winans, N.D.; Klein, R.R.; Fonseca, J.M.O.; Klein, P.E.; Rooney, W.L. Evaluating Introgression Sorghum Germplasm Selected at the Population Level While Exploring Genomic Resources as a Screening Method. Plants 2023, 12, 444. https://doi.org/10.3390/plants12030444
Winans ND, Klein RR, Fonseca JMO, Klein PE, Rooney WL. Evaluating Introgression Sorghum Germplasm Selected at the Population Level While Exploring Genomic Resources as a Screening Method. Plants. 2023; 12(3):444. https://doi.org/10.3390/plants12030444
Chicago/Turabian StyleWinans, Noah D., Robert R. Klein, Jales Mendes Oliveira Fonseca, Patricia E. Klein, and William L. Rooney. 2023. "Evaluating Introgression Sorghum Germplasm Selected at the Population Level While Exploring Genomic Resources as a Screening Method" Plants 12, no. 3: 444. https://doi.org/10.3390/plants12030444
APA StyleWinans, N. D., Klein, R. R., Fonseca, J. M. O., Klein, P. E., & Rooney, W. L. (2023). Evaluating Introgression Sorghum Germplasm Selected at the Population Level While Exploring Genomic Resources as a Screening Method. Plants, 12(3), 444. https://doi.org/10.3390/plants12030444










