Identification of Morphogenesis-Related NDR Kinase Signaling Network and Its Regulation on Cold Tolerance in Maize
Abstract
1. Introduction
2. Results
2.1. Identification of Pivotal Components of Maize MOR Signaling Network
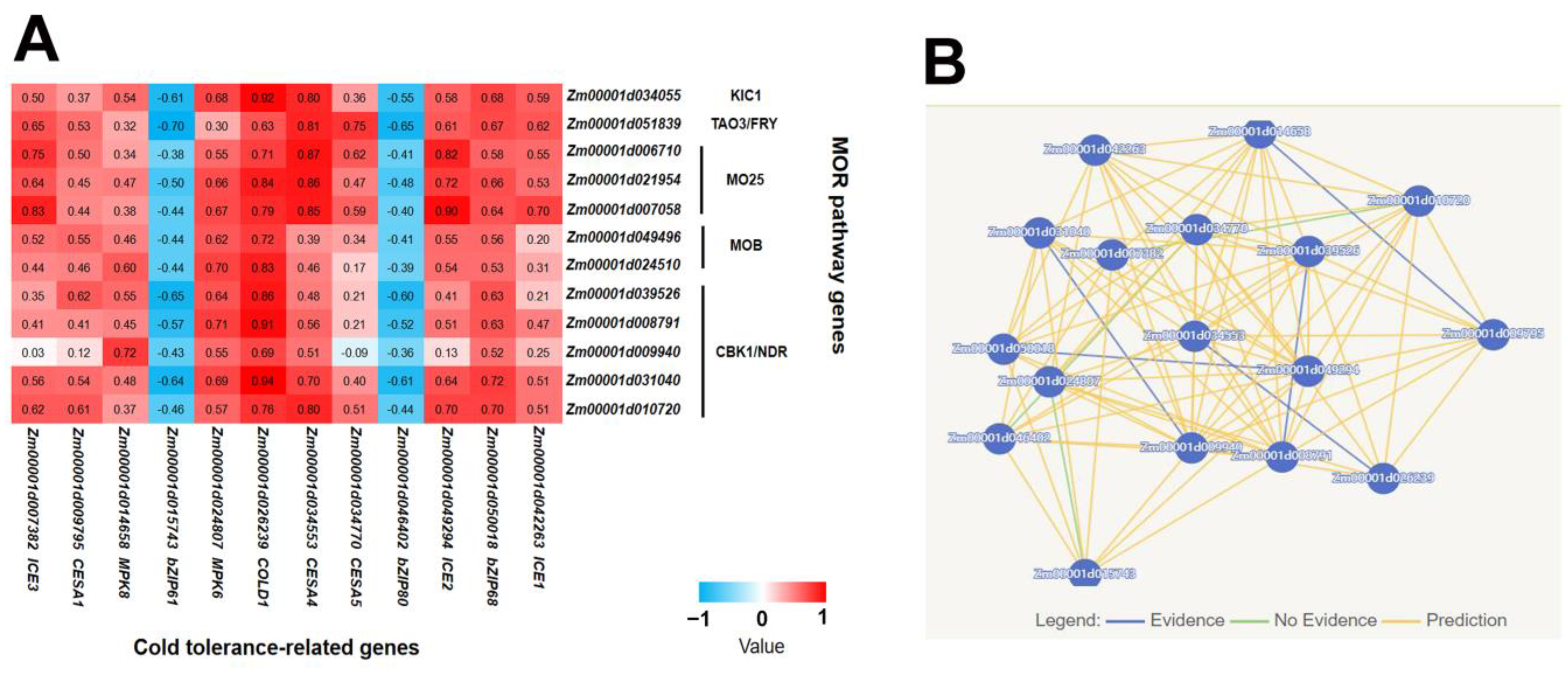
2.2. Co-Expression Analysis of MOR Signaling Network Genes and Cold Tolerance-Related Genes Revealed the Potential Regulatory Mechanism of Cold Tolerance in Maize
2.3. The Expression of Certain MOR Signaling Network Genes in Maize Was Regulated by Cold Stress
2.4. The Mutant of zm00001d010720 Exhibited Heightened Susceptibility to Cold Stress
3. Discussion
3.1. Relevant Components of the MOR Signaling Network Exhibit Conservation in Maize
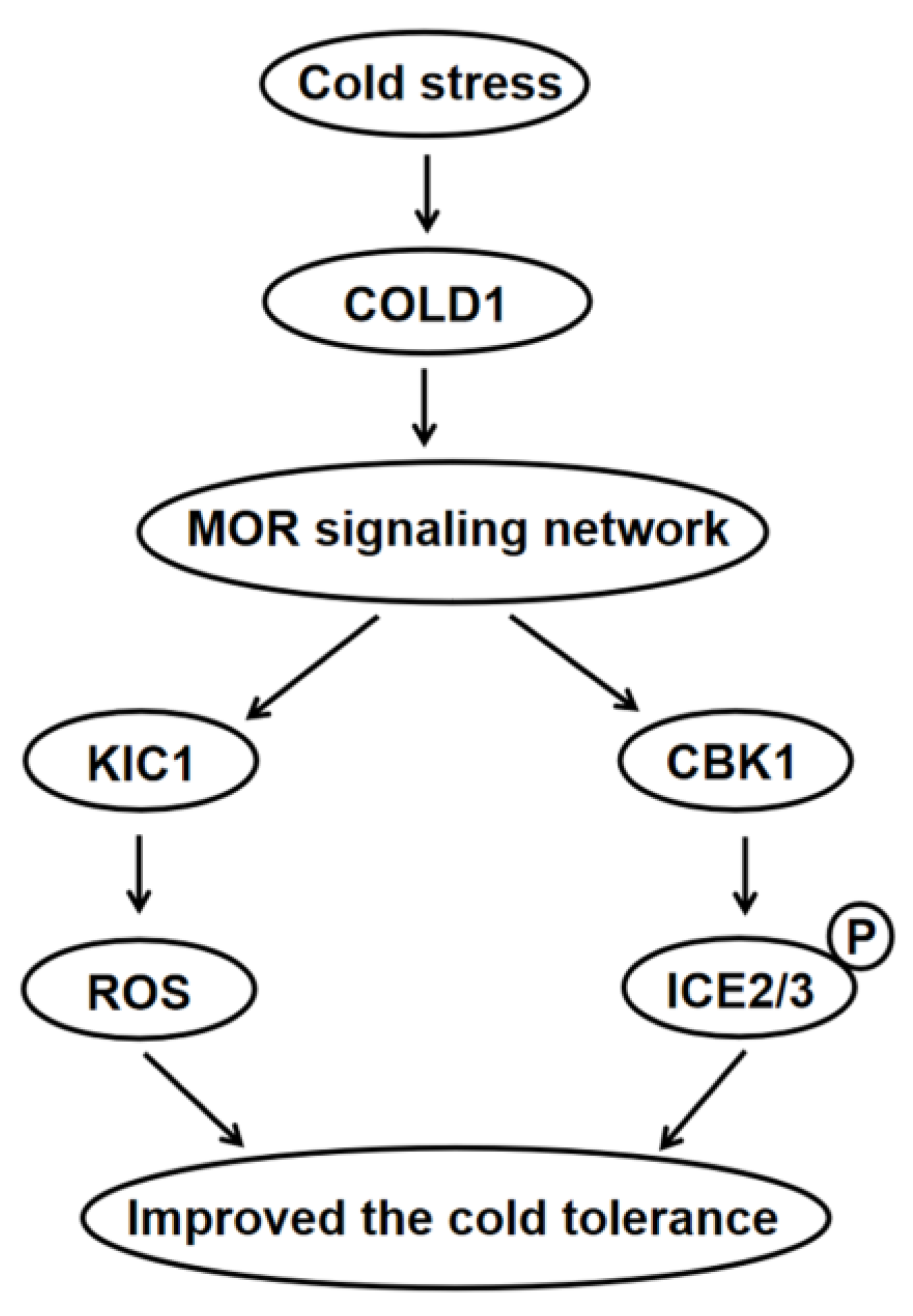
3.2. The MOR Signaling Network Genes Play a Crucial Role in the Regulation of Cold Tolerance in Maize
4. Materials and Methods
4.1. Plant Materials
4.2. Identification of MOR Signaling Network Genes in Maize
4.3. Phylogenetic Analysis
4.4. Co-Expression Analysis
4.5. RNA Extraction and qRT-PCR Analysis
4.6. Identification and Analysis of Mutant zm00001d010720
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zermiani, M.; Begheldo, M.; Nonis, A.; Palme, K.; Mizzi, L.; Morandini, P.; Nonis, A.; Ruperti, B. Identification of the arabidopsis RAM/MOR signalling network: Adding new regulatory players in plant stem cell maintenance and cell polarization. Ann. Bot. 2015, 116, 69–89. [Google Scholar] [CrossRef] [PubMed]
- Bizotto, F.M.; Ceratti, R.S.; Braz, A.S.K.; Masuda, H.P. Evolutionary history of Mo25 gene in plants, a component of RAM/MOR signaling network. Mech. Develop. 2018, 153, 64–73. [Google Scholar] [CrossRef] [PubMed]
- Racki, W.J.; Becam, A.M.; Nasr, F.; Herbert, C.J. CBK1p, a protein similar to the human myotonic dystrophy kinase, is essential for normal morphogenesis in Saccharomyces cerevisiae. Embo. J. 2000, 19, 4524–4532. [Google Scholar] [CrossRef] [PubMed]
- Bidlingmaier, S.; Weiss, E.L.; Seidel, C.; Drubin, D.G.; Snyder, M. The CBK1p pathway is important for polarized cell growth and cell separation in Saccharomyces cerevisiae. Mol. Cell. Biol. 2001, 21, 2449–2462. [Google Scholar] [CrossRef] [PubMed]
- Weiss, E.L.; Kurischko, C.; Zhang, C.; Shokat, K.; Drubin, D.G.; Luca, F.C. The Saccharomyces cerevisiae MOB2p-CBK1p kinase complex promotes polarized growth and acts with the mitotic exit network to facilitate daughter cell-specific localization of ACE2p transcription factor. J. Cell Biol. 2002, 158, 885–900. [Google Scholar] [CrossRef] [PubMed]
- Nelson, B.; Kurischko, C.; Horecka, J.; Mody, M.; Nair, P.; Pratt, L.; Zougman, A.; McBroom, L.D.B.; Hughes, T.R.; Boone, C.; et al. RAM: A conserved signaling network that regulates Ace2p transcriptional activity and polarized morphogenesis. Mol. Biol. Cell 2003, 14, 3782–3803. [Google Scholar] [CrossRef]
- Jansen, J.M.; Barry, M.F.; Yoo, C.K.; Weiss, E.L. Phosphoregulation of CBK1 is critical for RAM network control of transcription and morphogenesis. J. Cell Biol. 2006, 175, 755–766. [Google Scholar] [CrossRef]
- Maerz, S.; Seiler, S. Tales of RAM and MOR: NDR kinase signaling in fungal morphogenesis. Curr. Opin. Microbiol. 2010, 13, 663–671. [Google Scholar] [CrossRef]
- Saputo, S.; Chabrier-Rosello, Y.; Luca, F.C.; Kumar, A.; Krysan, D.J. The RAM network in pathogenic fungi. Eukaryot. Cell 2012, 11, 708–717. [Google Scholar] [CrossRef]
- Jorgensen, P.; Nelson, B.; Robinson, M.D.; Chen, Y.; Andrews, B.; Tyers, M.; Boone, C. High-Resolution Genetic Mapping with Ordered Arrays of Saccharomyces cerevisiae Deletion Mutants. Genetics 2002, 162, 1091–1099. [Google Scholar] [CrossRef]
- Colman-Lerner, A.; Chin, T.E.; Brent, R. Yeast Cbk1 and Mob2 activate daughter-specific genetic programs to induce asymmetric cell fates. Cell 2001, 107, 739–750. [Google Scholar] [CrossRef] [PubMed]
- Kurischko, C.; Weiss, G.; Ottey, M.; Luca, F.C. A role for the Saccharomyces cerevisiae regulation of ACE2 and polarized morphogenesis signalingnetwork in cell integrity. Genetics 2005, 171, 443–455. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hergovich, A.; Stegert, M.R.; Schmitz, D.; Hemmings, B.A. NDR kinases regulate essential cell processes from yeast to humans. Nat. Rev. Mol. Cell Bio. 2006, 7, 253–264. [Google Scholar] [CrossRef] [PubMed]
- Dan, I.; Watanabe, N.M.; Kusumi, A. The Ste20 group kinases as regulators of MAP kinase cascades. Trends Cell Biol. 2001, 11, 220–230. [Google Scholar] [CrossRef] [PubMed]
- Weiss, E.L. Mitotic exit and separation of mother and daughter cells. Genetics 2012, 192, 1165–1202. [Google Scholar] [CrossRef]
- Wang, D.; Xiao, Y.; Chen, H.; Huang, C.; Chen, P.; Chen, D.; Deng, W.; Wang, J. Combination of genomics, transcriptomics identifies candidate loci related to cold tolerance in Dongxiang wild rice. Plants 2022, 11, 2329. [Google Scholar] [CrossRef]
- Chinnusamy, V.; Zhu, J.; Zhu, J.K. Cold stress regulation of gene expression in plants. Trends Plant Sci. 2007, 12, 444–451. [Google Scholar] [CrossRef]
- Ma, Y.; Dai, X.; Xu, Y.; Luo, W.; Zheng, X.; Zeng, D.; Pan, Y.; Lin, X.; Liu, H.; Zhang, D.; et al. COLD1 confers chilling tolerance in rice. Cell 2015, 160, 1209–1221. [Google Scholar] [CrossRef]
- Ding, Y.; Shi, Y.; Yang, S. Molecular regulation of plant responses to environmental temperatures. Mol. Plant 2020, 13, 544–564. [Google Scholar] [CrossRef]
- Ding, Y.; Yang, S. Surviving and thriving: How plants perceive and respond to temperature stress. Dev. Cell 2022, 57, 947–958. [Google Scholar] [CrossRef]
- Guo, X.; Liu, D.; Chong, K. Cold signaling in plants: Insights into mechanisms and regulation. J. Integr. Plant Biol. 2018, 60, 745–756. [Google Scholar] [CrossRef]
- Shi, Y.; Ding, Y.; Yang, S. Molecular regulation of CBF signaling in cold acclimation. Trends Plant Sci. 2018, 23, 623–637. [Google Scholar] [CrossRef]
- Chinnusamy, V.; Ohta, M.; Kanrar, S.; Lee, B.H.; Hong, X.; Agarwal, M.; Zhu, J.K. ICE1: A regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Gene. Dev. 2003, 17, 1043–1054. [Google Scholar] [CrossRef]
- Fursova, O.V.; Pogorelko, G.V.; Tarasov, V.A. Identification of ICE2, a gene involved in cold acclimation which determines freezing tolerance in Arabidopsis thaliana. Gene 2009, 429, 98–103. [Google Scholar] [CrossRef] [PubMed]
- Tang, K.; Zhao, L.; Ren, Y.; Yang, S.; Zhu, J.K.; Zhao, C. The transcription factor ICE1 functions in cold stress response by binding to the promoters of CBF and COR genes. J. Integr. Plant Biol. 2020, 62, 258–263. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Li, H.; Zhang, X.; Xie, Q.; Gong, Z.; Yang, S. OST1 kinase modulates freezing tolerance by enhancing ICE1 stability in Arabidopsis. Dev. Cell 2015, 32, 278–289. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Ding, Y.; Shi, Y.; Zhang, X.; Zhang, S.; Gong, Z.; Yang, S. MPK3- and MPK6-mediated ICE1 phosphorylation negatively regulates ICE1 stability and freezing tolerance in Arabidopsis. Dev. Cell 2017, 43, 630–642. [Google Scholar] [CrossRef]
- Zhao, C.; Wang, P.; Si, T.; Hsu, C.C.; Wang, L.; Zayed, O.; Yu, Z.; Zhu, Y.; Dong, J.; Tao, W.A.; et al. MAP kinase cascades regulate the cold response by modulating ICE1 protein stability. Dev. Cell 2017, 43, 618–629. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.Y.; Zhang, Z.Y.; Li, J.H.; Li, F.; Liu, H.H.; Yang, W.S.; Chong, K.; Xu, Y.Y. OsMAPK3 phosphorylates OsbHLH002/OsICE1 and inhibits its ubiquitination to activate OsTPP1 and enhances rice chilling tolerance. Dev. Cell 2017, 43, 731–743. [Google Scholar] [CrossRef]
- Gao, R.X.; Hu, M.J.; Zhao, H.M.; Lai, J.S.; Song, W.B. Genetic dissection of ear-related traits using immortalized F2 population in maize. J. Integr. Agr. 2022, 21, 2492–2507. [Google Scholar] [CrossRef]
- Zhao, Y.P.; Zhao, B.B.; Wu, G.X.; Ma, X.J.; Wang, B.B.; Kong, D.X.; Wei, H.B.; Wang, H.Y. Creation of two hyperactive variants of phytochrome B1 for attenuating shade avoidance syndrome in maize. J. Integr. Agr. 2022, 21, 1253–1265. [Google Scholar] [CrossRef]
- Xie, S.D.; Tian, R.; Zhang, J.J.; Liu, H.M.; Li, Y.P.; Hu, Y.F.; Yu, G.W.; Huang, Y.B.; Liu, Y.H. Dek219 encodes the DICER-LIKE1 protein that affects chromatin accessibility and kernel development in maize. J. Integr. Agr. 2023, 22, 2961–2980. [Google Scholar] [CrossRef]
- Allen, D.J.; Ort, D.R. Impacts of chilling temperatures on photosynthesis in warm-climate plants. Trends Plant Sci. 2001, 6, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, B.; Rasmussen, A.; Porter, J.R. Temperatures and the growth and development of maize and rice: A review. Global Change Biol. 2014, 20, 408–417. [Google Scholar] [CrossRef]
- Riva-Roveda, L.; Escale, B.; Giauffret, C.; Périlleux, C. Maize plants can enter a standby mode to cope with chilling stress. BMC Plant Biol. 2016, 16, 212. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Zhao, C.; Niu, Y.; Chao, W.; He, W.; Wang, Y.; Mao, T.; Bai, X. Understanding and comprehensive evaluation of cold resistance in the seedlings of multiple maize genotypes. Plants 2022, 11, 1881. [Google Scholar] [CrossRef]
- Ben-Haj-Salah, H.; Tardieu, F. Temperature affects expansion rate of maize leaves without change in spatial distribution of cell length. Plant Physiol. 1995, 109, 861–870. [Google Scholar] [CrossRef]
- Warrington, I.J.; Kanemasu, E.T. Corn growth response to temperature and photoperiod II. Leaf-initiation and leaf-appearance rates. Agron. J. 1983, 75, 755–761. [Google Scholar] [CrossRef]
- Hund, A.; Fracheboud, Y.; Soldati, A.; Stamp, P. Cold tolerance of maize seedlings as determined by root morphology and photosynthetic traits. Eur. J. Agron. 2008, 28, 178–185. [Google Scholar] [CrossRef]
- Nguyen, H.T.; Leipner, J.; Stamp, P.; Guerra-Peraza, O. Low temperature stress in maize (Zea mays L.) induces genes involved in photosynthesis and signal transduction as studied by suppression subtractive hybridization. Plant Physiol. Bioch. 2009, 47, 116–122. [Google Scholar] [CrossRef]
- Hussain, H.A.; Men, S.; Hussain, S.; Zhang, Q.; Ashraf, U.; Anjum, S.A.; Ali, I.; Wang, L. Maize tolerance against drought and chilling stresses varied with root morphology and antioxidative defense system. Plants 2020, 9, 720. [Google Scholar] [CrossRef] [PubMed]
- He, R.Y.; Zheng, J.J.; Chen, Y.; Pan, Z.Y.; Yang, T.; Zhou, Y.; Li, X.F.; Nan, X.; Li, Y.Z.; Cheng, M.J.; et al. QTL-seq and transcriptomic integrative analyses reveal two positively regulated genes that control the low-temperature germination ability of MTP-maize introgression lines. Theor. Appl. Genet. 2023, 136, 116. [Google Scholar] [CrossRef] [PubMed]
- Qin, F.; Sakuma, Y.; Li, J.; Liu, Q.; Li, Y.Q.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Cloning and functional analysis of a novel DREB1/CBF transcription factor involved in cold-responsive gene expression in Zea mays L. Plant Cell Physiol. 2004, 45, 1042–1052. [Google Scholar] [CrossRef] [PubMed]
- Zeng, R.; Li, Z.; Shi, Y.; Fu, D.; Yin, P.; Cheng, J.; Jiang, C.; Yang, S. Natural variation in a type-A response regulator confers maize chilling tolerance. Nat. Commun. 2021, 12, 4713. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Shi, Y.; Liu, J.; Li, Z.; Fu, D.; Wu, S.; Li, M.; Yang, Z.; Shi, Y.; Lai, J.; et al. Natural polymorphism of ZmICE1 contributes to amino acid metabolism that impacts cold tolerance in maize. Nat. Plants 2022, 8, 1176–1190. [Google Scholar] [CrossRef]
- Li, Z.; Fu, D.; Wang, X.; Zeng, R.; Zhang, X.; Tian, J.; Zhang, S.; Yang, X.; Tian, F.; Lai, J.; et al. The transcription factor bZIP68 negatively regulates cold tolerance in maize. Plant Cell 2022, 34, 2833–2851. [Google Scholar] [CrossRef]
- Han, L.; Zhong, W.; Qian, J.; Jin, M.; Tian, P.; Zhu, W.; Zhang, H.; Sun, Y.; Feng, J.W.; Liu, X.; et al. A multi-omics integrative network map of maize. Nat. Genet. 2023, 55, 144–153. [Google Scholar] [CrossRef]
- Xiong, J.; Cui, X.; Yuan, X.; Yu, X.; Sun, J.; Gong, Q. The Hippo/STE20 homolog SIK1 interacts with MOB1 to regulate cell proliferation and cell expansion in Arabidopsis. J. Exp. Bot. 2016, 67, 1461–1475. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Chiang, Y.H.; Toruño, T.Y.; Lee, D.; Ma, M.; Liang, X.; Lal, N.K.; Lemos, M.; Lu, Y.J.; Ma, S.; et al. The MAP4 kinase SIK1 ensures robust extracellular ROS burst and antibacterial immunity in plants. Cell Host Microbe 2018, 24, 379–391. [Google Scholar] [CrossRef]
- Zhang, P.; Yu, X.; Bai, J.; Gong, Q. The Arabidopsis STE20/Hippo kinase SIK1 regulates polarity independently of PIN proteins. Biochem. Bioph. Res. Co. 2021, 549, 21–26. [Google Scholar] [CrossRef]
- Klopffleisch, K.; Phan, N.; Augustin, K.; Bayne, R.S.; Booker, K.S.; Booker, J.R.; Carpita, N.C.; Carr, T.; Chen, J.G.; Cooke, T.R.; et al. Arabidopsis G-protein interactome reveals connections to cell wall carbohydrates and morphogenesis. Mol. Syst. Biol. 2011, 7, 532. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Yue, X.; Cui, X.; Song, L.; Cheng, Y. AtMOB1 genes regulate jasmonate accumulation and plant development. Plant Physiol. 2020, 182, 1481–1493. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.M.; Liang, Y.; Mei, J.; Liao, H.Z.; Wang, P.; Hu, K.; Chen, L.Q.; Zhang, X.Q.; Ye, D. The Arabidopsis AGC kinases NDR2/4/5 interact with MOB1A/1B and play important roles in pollen development and germination. Plant J. 2021, 105, 1035–1052. [Google Scholar] [CrossRef]
- Bögre, L.; Okrész, L.; Henriques, R.; Anthony, R.G. Growth signalling pathways in Arabidopsis and the AGC protein kinases. Trends Plant Sci. 2003, 8, 424–431. [Google Scholar] [CrossRef] [PubMed]
- Yoon, H.S.; Fujino, K.; Liu, S.; Takano, T.; Tsugama, D. NDR/LATS-family protein kinase genes are indispensable for embryogenesis in Arabidopsis. FEBS Open Bio. 2021, 11, 2600–2606. [Google Scholar] [CrossRef]
- Lu, X.; Liu, J.; Ren, W.; Yang, Q.; Chai, Z.; Chen, R.; Wang, L.; Zhao, J.; Lang, Z.; Wang, H.; et al. Gene-indexed mutations in maize. Mol. Plant 2018, 11, 496–504. [Google Scholar] [CrossRef] [PubMed]
- Xie, S.; Zhang, X.; Zhou, Z.; Li, X.; Huang, Y.; Zhang, J.; Weng, J. Identification of genes alternatively spliced in developing maize endosperm. Plant Biol. 2018, 20, 59–66. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, R.; Xie, S.; Zhang, J.; Liu, H.; Li, Y.; Hu, Y.; Huang, Y.; Liu, Y. Identification of Morphogenesis-Related NDR Kinase Signaling Network and Its Regulation on Cold Tolerance in Maize. Plants 2023, 12, 3639. https://doi.org/10.3390/plants12203639
Tian R, Xie S, Zhang J, Liu H, Li Y, Hu Y, Huang Y, Liu Y. Identification of Morphogenesis-Related NDR Kinase Signaling Network and Its Regulation on Cold Tolerance in Maize. Plants. 2023; 12(20):3639. https://doi.org/10.3390/plants12203639
Chicago/Turabian StyleTian, Ran, Sidi Xie, Junjie Zhang, Hanmei Liu, Yangping Li, Yufeng Hu, Yubi Huang, and Yinghong Liu. 2023. "Identification of Morphogenesis-Related NDR Kinase Signaling Network and Its Regulation on Cold Tolerance in Maize" Plants 12, no. 20: 3639. https://doi.org/10.3390/plants12203639
APA StyleTian, R., Xie, S., Zhang, J., Liu, H., Li, Y., Hu, Y., Huang, Y., & Liu, Y. (2023). Identification of Morphogenesis-Related NDR Kinase Signaling Network and Its Regulation on Cold Tolerance in Maize. Plants, 12(20), 3639. https://doi.org/10.3390/plants12203639




