Transcriptional Regulation of Small Heat Shock Protein 17 (sHSP-17) by Triticum aestivum HSFA2h Transcription Factor Confers Tolerance in Arabidopsis under Heat Stress
Abstract
1. Introduction
2. Results
2.1. Identification of Transcripts Predicted to Be HS-Responsive Transcription Factor (HSF)
2.2. Cloning and In Silico Characterization of Putative HSFA2h
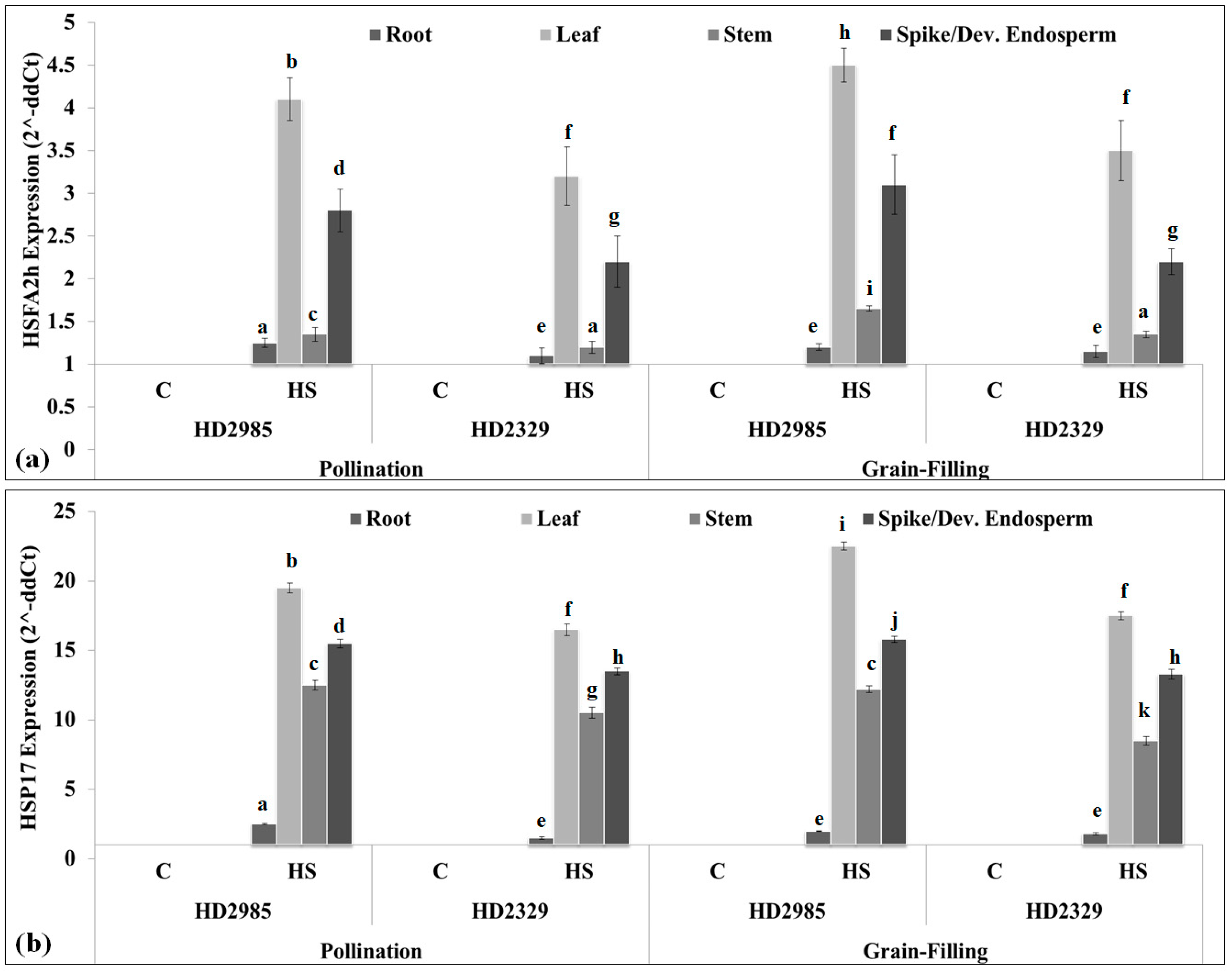
2.3. Expression Analysis of Cloned HSFA2h TF and Its Target Gene in Wheat under HS
2.4. Validation of Transgenic Arabidopsis Plants Overexpressing Wheat HSFA2h Transcription Factor
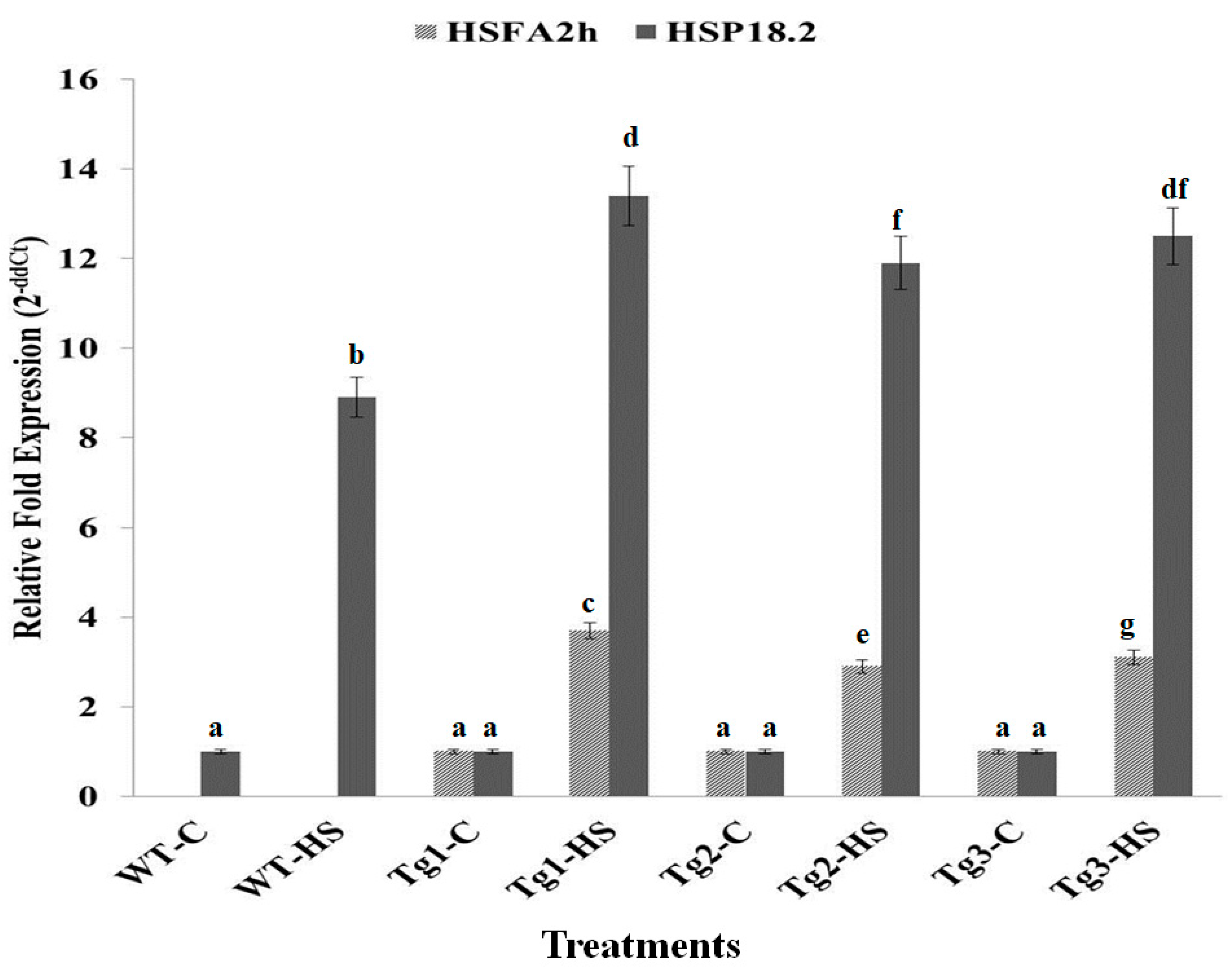
2.5. Expression Analysis of HSFA2h and Its Target Gene (HSP 18.2) in Transgenic Arabidopsis Exposed to HS at T3 Stage
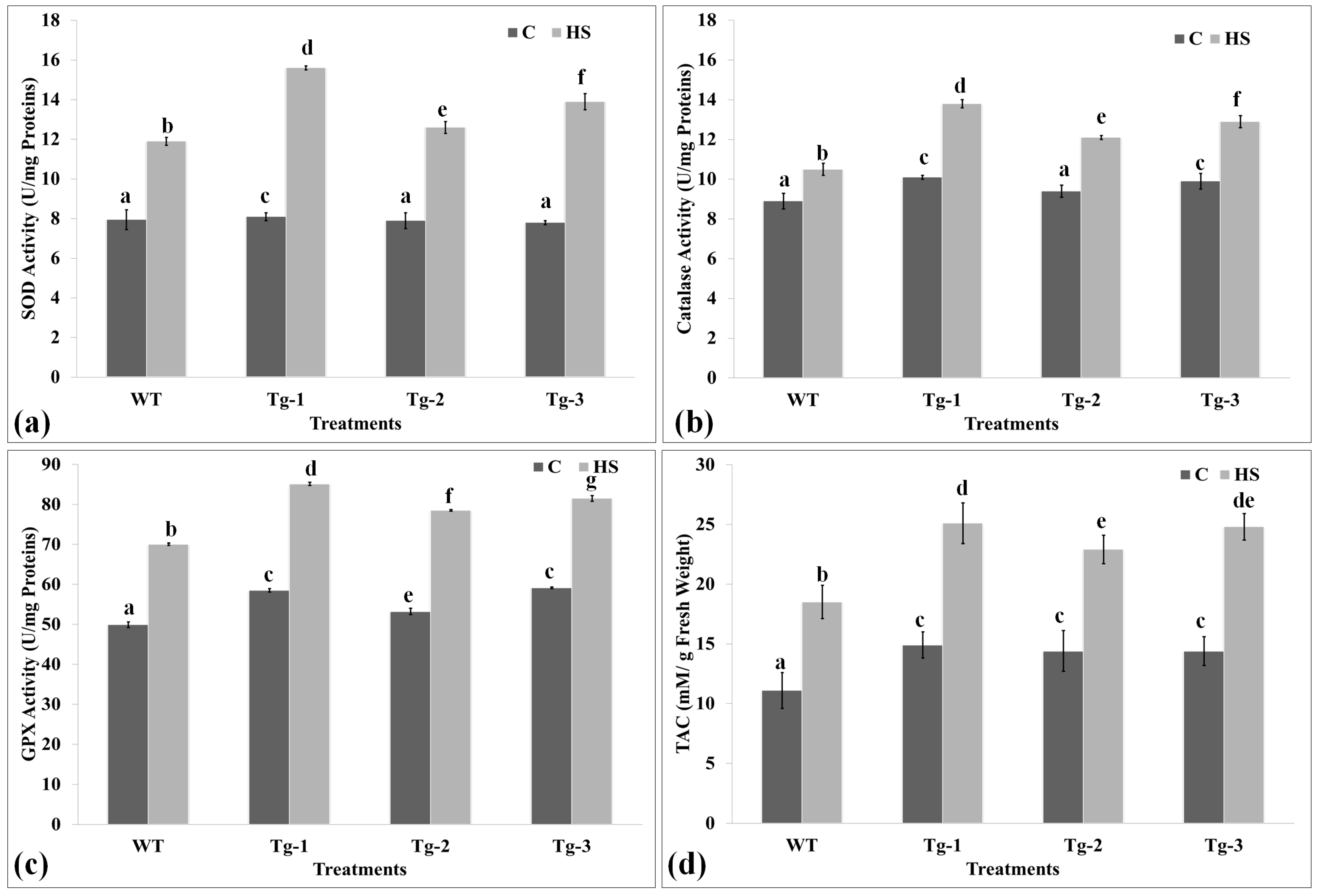
2.6. Biochemical Screening of Transgenic Arabidopsis under HS
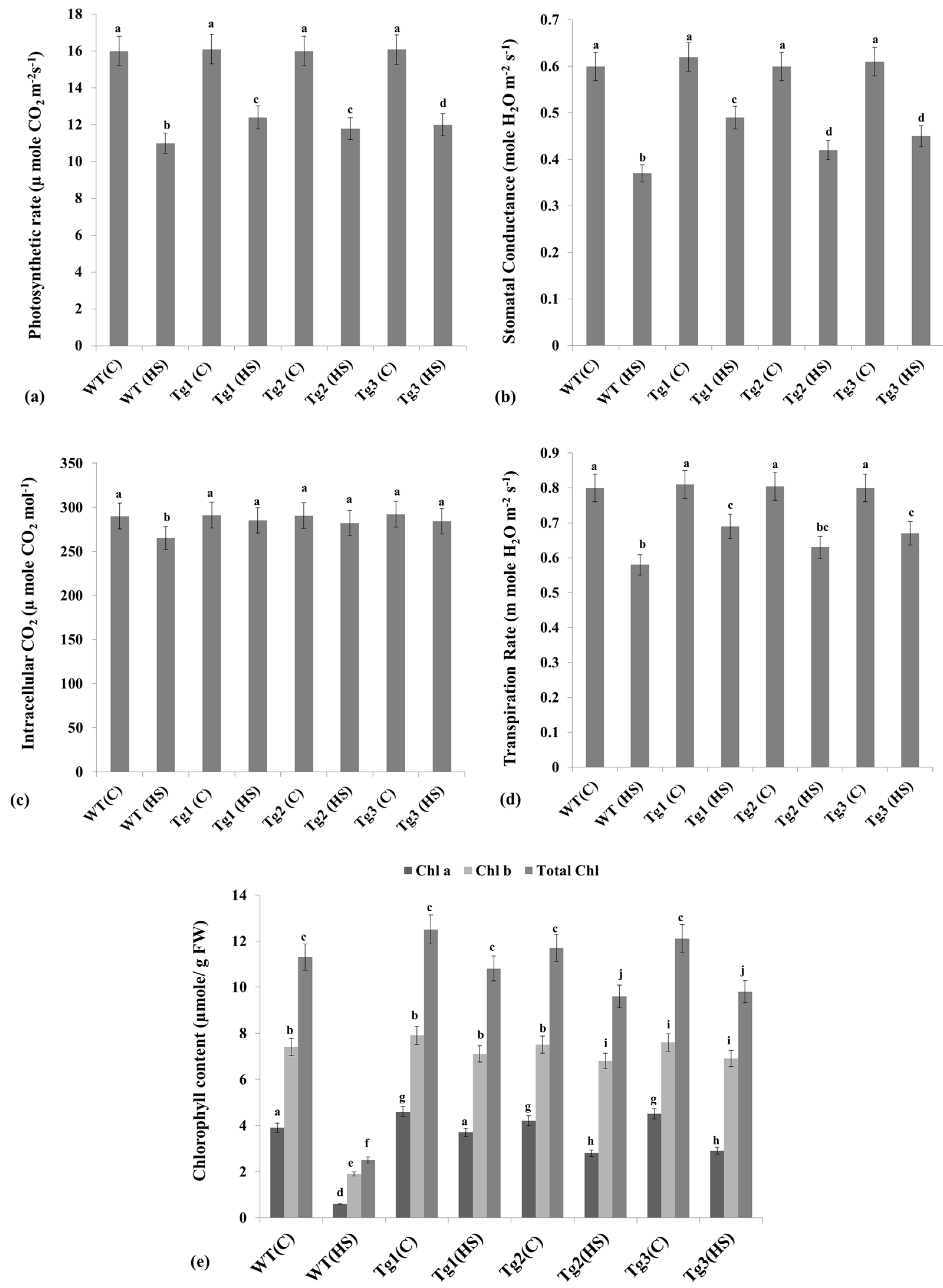
2.7. Alterations in the Photosynthesis-Associated Parameters under HS
3. Discussion
4. Materials and Methods
4.1. Plant Material and Stress Treatment
4.2. RNA-Seq for the Identification of HSF Transcripts
4.3. Molecular Cloning of Wheat HSF Gene
4.3.1. Transcript-Specific Oligo Designing
4.3.2. Isolation of Total RNA and RT-PCR Amplification of Gene
4.4. In Silico Characterization of the Cloned HSF Gene
4.5. Plasmid Construction of HSFA2h and Mobilization in Arabidopsis
4.6. Southern Blot Analysis to Confirm the Transgenic Plants
4.7. Northern Blot Analysis to Characterize the Expression of Wheat HSFA2h in Transgenic Arabidopsis
4.8. Validation of Transgenic Arabidopsis through Quantitative Real-Time PCR (qRT-PCR)
4.9. Biochemical Screening of HSFA2h Expressing Transgenic Arabidopsis for Thermotolerance
4.9.1. Estimation of Total Antioxidant Capacity (TAC)
4.9.2. Antioxidant Enzyme Assay in Transgenic Arabidopsis
4.10. Physiological Characterization of HSFA2h-Expressing Transgenic Arabidopsis for Thermotolerance
4.10.1. Infra-Red Gas Analyzer Analysis
4.10.2. Measurements of Chlorophyll Content
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, P.; Song, H.; Li, C.; Li, P.; Li, A.; Guan, H.; Hou, L.; Wang, X. Genome-wide dissection of the heat shock transcription factor family genes in Arachis. Front. Plant Sci. 2017, 8, 106. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Liu, J.-H.; Ma, X.; Luo, D.-X.; Gong, Z.-H.; Lu, M.-H. The Plant Heat Stress Transcription Factors (HSFs): Structure, Regulation, and Function in Response to Abiotic Stresses. Front. Plant Sci. 2016, 7, 114. [Google Scholar] [CrossRef]
- Kumar, R.R.; Goswami, S.; Singh, K.; Dubey, K.; Singh, S.; Sharma, R.; Verma, N.; Kala, Y.K.; Rai, G.K.; Grover, M.; et al. Identification of Putative RuBisCo Activase (TaRca1)—The Catalytic Chaperone Regulating Carbon Assimilatory Pathway in Wheat (Triticum aestivum) under the Heat Stress. Front. Plant Sci. 2016, 7, 986. [Google Scholar] [CrossRef] [PubMed]
- Meena, S.; Samtani, H.; Khurana, P. Elucidating the functional role of heat stress transcription factor A6b (TaHsfA6b) in linking heat stress response and the unfolded protein response in wheat. Plant Mol. Biol. 2022, 108, 621–634. [Google Scholar] [CrossRef]
- Goswami, S.; Kumar, R.R.; Sharma, S.K.; Kala, Y.K.; Singh, K.; Gupta, R.; Dhavan, G.; Rai, G.K.; Singh, G.P.; Pathak, H.; et al. Calcium triggers protein kinases-induced signal transduction for augmenting the thermotolerance of developing wheat (Triticum aestivum) grain under the heat stress. J. Plant Biochem. Biotechnol. 2015, 24, 441–452. [Google Scholar] [CrossRef]
- Kumar, R.R.; Goswami, S.; Singh, K.; Dubey, K.; Rai, G.K.; Singh, B.; Singh, S.; Grover, M.; Mishra, D.; Kumar, S.; et al. Characterization of novel heat-responsive transcription factor (TaHSFA6e) gene involved in regulation of heat shock proteins (HSPs) —A key member of heat stress-tolerance network of wheat. J. Biotechnol. 2018, 279, 1–12. [Google Scholar] [CrossRef]
- Nover, L.; Bharti, K.; Döring, P.; Mishra, S.K.; Ganguli, A.; Scharf, K.-D. Arabidopsis and the heat stress transcription factor world: How many heat stress transcription factors do we need? Cell Stress Chaperones 2001, 6, 177. [Google Scholar] [CrossRef]
- Scharf, K.D.; Berberich, T.; Ebersberger, I.; Nover, L. The plant heat stress transcription factor (Hsf) family: Structure, function and evolution. Biochim. Biophys. Acta-Gene Regul. Mech. 2012, 1819, 104–119. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.P.; Fan, X.; You, M.H.; Wang, S.S.; Zong, R.X. Changes in sugar, pyruvic acid content and nitrate reductase activity of Elymus sibiricus reproductive branches during seed development. Acta Prataculturae Sin. 2016, 25, 69–77. [Google Scholar] [CrossRef][Green Version]
- Hu, X.-J.; Chen, D.; Lynne Mclntyre, C.; Fernanda Dreccer, M.; Zhang, Z.-B.; Drenth, J.; Kalaipandian, S.; Chang, H.; Xue, G.-P. Heat shock factor C2a serves as a proactive mechanism for heat protection in developing grains in wheat via an ABA-mediated regulatory pathway. Plant Cell Environ. 2017, 41, 79–98. [Google Scholar] [CrossRef]
- Xue, G.P.; Sadat, S.; Drenth, J.; McIntyre, C.L. The heat shock factor family from Triticum aestivum in response to heat and other major abiotic stresses and their role in regulation of heat shock protein genes. J. Exp. Bot. 2014, 65, 539–557. [Google Scholar] [CrossRef] [PubMed]
- Hahn, A.; Bublak, D.; Schleiff, E.; Scharf, K.-D. Crosstalk between Hsp90 and Hsp70 chaperones and heat stress transcription factors in tomato. Plant Cell 2011, 23, 741–755. [Google Scholar] [CrossRef]
- Lohmann, C.; Eggers-Schumacher, G.; Wunderlich, M.; Schöffl, F. Two different heat shock transcription factors regulate immediate early expression of stress genes in Arabidopsis. Mol. Genet. Genom. 2004, 271, 11–21. [Google Scholar] [CrossRef]
- Busch, W.; Wunderlich, M.; Schöffl, F. Identification of novel heat shock factor-dependent genes and biochemical pathways in Arabidopsis thaliana. Plant J. 2005, 41, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Charng, Y.-Y.; Liu, H.-C.; Liu, N.-Y.; Chi, W.-T.; Wang, C.-N.; Chang, S.-H.; Wang, T.-T. A heat-inducible transcription factor, HsfA2, is required for extension of acquired thermotolerance in Arabidopsis. Plant Physiol. 2007, 143, 251–262. [Google Scholar] [CrossRef] [PubMed]
- Schramm, F.; Ganguli, A.; Kiehlmann, E.; Englich, G.; Walch, D.; Von Koskull-Döring, P. The heat stress transcription factor HsfA2 serves as a regulatory amplifier of a subset of genes in the heat stress response in Arabidopsis. Plant Mol. Biol. 2006, 60, 759–772. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Salamó, I.; Papdi, C.; Rigó, G.; Zsigmond, L.; Vilela, B.; Lumbreras, V.; Nagy, I.; Horváth, B.; Domoki, M.; Darula, Z.; et al. The heat shock factor A4A confers salt tolerance and is regulated by oxidative stress and the mitogen-activated protein kinases MPK3 and MPK6. Plant Physiol. 2014, 165, 319–334. [Google Scholar] [CrossRef]
- Baniwal, S.K.; Bharti, K.; Chan, K.Y.; Fauth, M.; Ganguli, A.; Kotak, S.; Mishra, S.K.; Nover, L.; Port, M.; Scharf, K.; et al. Heat stress response in plants: A complex game with chaperones and more than twenty heat stress transcription factors. J. Biosci. 2004, 29, 471–487. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Wu, J.; Ji, Q.; Wang, C.; Luo, L.; Yuan, Y.; Wang, Y.; Wang, J. Genome-wide analysis of heat shock transcription factor families in rice and Arabidopsis. J. Genet. Genom. 2008, 35, 105–118. [Google Scholar] [CrossRef]
- Mittal, S.; Mallikarjuna, M.G.; Rao, A.R.; Jain, P.A.; Dash, P.K.; Thirunavukkarasu, N. Comparative analysis of CDPK family in maize, Arabidopsis, rice, and sorghum revealed potential targets for drought tolerance improvement. Front. Chem. 2017, 5, 115. [Google Scholar] [CrossRef] [PubMed]
- Nishizawa, A.; Yabuta, Y.; Yoshida, E.; Maruta, T.; Yoshimura, K.; Shigeoka, S. Arabidopsis heat shock transcription factor A2 as a key regulator in response to several types of environmental stress. Plant J. 2006, 48, 535–547. [Google Scholar] [CrossRef] [PubMed]
- Yokotani, N.; Ichikawa, T.; Kondou, Y.; Matsui, M.; Hirochika, H.; Iwabuchi, M.; Oda, K. Tolerance to various environmental stresses conferred by the salt-responsive rice gene ONAC063 in transgenic Arabidopsis. Planta 2009, 229, 1065–1075. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.C.; Liao, H.T.; Charng, Y.Y. The role of class A1 heat shock factors (HSFA1s) in response to heat and other stresses in Arabidopsis. Plant Cell Environ. 2011, 34, 738–751. [Google Scholar] [CrossRef]
- Kotak, S.; Larkindale, J.; Lee, U.; von Koskull-Döring, P.; Vierling, E.; Scharf, K.D. Complexity of the heat stress response in plants. Curr. Opin. Plant Biol. 2007, 10, 310–316. [Google Scholar] [CrossRef] [PubMed]
- Panchuk, I.I.; Volkov, R.A.; Schöffl, F. Heat stress-and heat shock transcription factor-dependent expression and activity of ascorbate peroxidase in Arabidopsis. Plant Physiol. 2002, 129, 838–853. [Google Scholar] [CrossRef] [PubMed]
- Evrard, A.; Kumar, M.; Lecourieux, D.; Lucks, J.; von Koskull-Döring, P.; Hirt, H. Regulation of the heat stress response in Arabidopsis by MPK6-targeted phosphorylation of the heat stress factor HsfA2. PeerJ 2013, 1, e59. [Google Scholar] [CrossRef] [PubMed]
- Che, P.; Gingerich, D.J.; Lall, S.; Howell, S.H. Global and hormone-induced gene expression changes during shoot development in Arabidopsis. Plant Cell 2002, 14, 2771–2785. [Google Scholar] [CrossRef]
- Chauhan, H.; Khurana, N.; Agarwal, P.; Khurana, J.P.; Khurana, P. A seed preferential heat shock transcription factor from wheat provides abiotic stress tolerance and yield enhancement in transgenic Arabidopsis under heat stress environment. PLoS ONE 2013, 8, e79577. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.L.; Yuan, S.N.; Zhang, H.N.; Zhang, Y.Y.; Zhang, Y.J.; Wang, G.Y.; Li, Y.Q.; Li, G.L. Heat-response patterns of the heat shock transcription factor family in advanced development stages of wheat (Triticum aestivum L.) and thermotolerance-regulation by TaHsfA2-10. BMC Plant Biol. 2020, 20, 1–18. [Google Scholar] [CrossRef]
- Saidi, M.N.; Mahjoubi, H.; Yacoubi, I. Transcriptome meta-analysis of abiotic stresses-responsive genes and identification of candidate transcription factors for broad stress tolerance in wheat. Protoplasma 2023, 260, 707–721. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Li, M.; Zhang, H.; Zhao, B.; Liu, Z.; Duan, S.; Meng, X.; Li, G.; Guo, X. Alternative Splicing of TaHsfA2-7 Is Involved in the Improvement of Thermotolerance in Wheat. Int. J. Mol. Sci. 2023, 24, 1014. [Google Scholar] [CrossRef] [PubMed]
- Wen, J.; Qin, Z.; Sun, L.; Zhang, Y.; Wang, D.; Peng, H.; Yao, Y.; Hu, Z.; Ni, Z.; Sun, Q.; et al. Alternative splicing of TaHSFA6e modulates heat shock protein–mediated translational regulation in response to heat stress in wheat. New Phytol. 2023, 239, 2235–2247. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.T.; Guo, X.N.; Zhu, K.X. Effect of phosphate salts on the shelf-life and quality characteristics of semi-dried noodles. Food Chem. 2022, 384, 132481. [Google Scholar] [CrossRef]
- Waters, E.R. The evolution, function, structure, and expression of the plant sHSPs. J. Exp. Bot. 2013, 64, 391–403. [Google Scholar] [CrossRef]
- Heckathorn, S.; North, G.; Wang, D.; Zhu, C. Editorial: Climate Change and Plant Nutrient Relations. Front. Plant Sci. 2020, 11, 869. [Google Scholar] [CrossRef] [PubMed]
- Wan, X.L.; Yang, J.; Li, X.B.; Zhou, Q.; Guo, C.; Bao, M.Z.; Zhang, J.W. Over-expression of PmHSP17.9 in transgenic Arabidopsis thaliana confers thermotolerance. Plant Mol. Biol. Rep. 2016, 34, 899–908. [Google Scholar] [CrossRef]
- Large, E.C. Growth stages in cereals illustration of the feekes scale. Plant Pathol. 1954, 3, 128–129. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Green, M.R.; Sambrook, J. Molecular Cloning. A Laboratory Manual, 4th ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2012. [Google Scholar]
- Procunier, J.D.; Xu, J.; Kasha, K.J. A rapid and reliable DNA extraction method for higher plants. Barley Genet. Newsl. 1990, 20, 74–75. [Google Scholar]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleid Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Benzie, I.F.F.; Strain, J.J. [2] Ferric reducing/antioxidant power assay: Direct measure of total antioxidant activity of biological fluids and modified version for simultaneous measurement of total antioxidant power and ascorbic acid concentration. Methods Enzymol. 1999, 299, 15–27. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Mika, A.; Lüthje, S. Properties of guaiacol peroxidase activities isolated from corn root plasma membranes. Plant Physiol. 2003, 132, 1489–1498. [Google Scholar] [CrossRef] [PubMed]
- Rucińska, R.; Waplak, S.; Gwóźdź, E.A. Free radical formation and activity of antioxidant enzymes in lupin roots exposed to lead. Plant Physiol. Biochem. 1999, 37, 187–194. [Google Scholar] [CrossRef]
- Giannopolitis, C.N.; Ries, S.K. Superoxide Dismutases: I. Occurrence in Higher Plants. Plant Physiol. 1977, 59, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Long, S.P.; Bernacchi, C.J. Gas exchange measurements, what can they tell us about the underlying limitations to photosynthesis? Procedures and sources of error. J. Exp. Bot. 2003, 54, 2393–2401. [Google Scholar] [CrossRef]
- Hiscox, J.D.; Israelstam, G.F. A method for the extraction of chlorophyll from leaf tissue without maceration. Can. J. Bot. 1979, 57, 1332–1334. [Google Scholar] [CrossRef]



| * Primer ID | Oligo Sequence (5′-3′) | Tm (°C) |
|---|---|---|
| Fp_HSFA2h | ATGGACCCGGTGCCGAGTCTG | 58 °C |
| Rp_HSFA2h | TAGGTTGAGAGGGTTGGGCTATTTG | 58 °C |
| FpHSFA2h-pRI101 | GGAATTCCATATGGACCCGGTGCCGAGTCTG | 62 °C |
| RpHSFA2h-pRI101 | GGGGTACCCTAGGTTGAGAGGGTTGGGCTATTTG | 63.2 °C |
| qFp_HSFA2h | ACAGAGCCACAGGATTTTGG | 58 °C |
| qRp_HSFA2h | TGAGAGGGTTGGGCTATTTG | 58 °C |
| qFp_HSP17 | GAGGGAGGAGAAGGAGGAC | 57.7 °C |
| qRp_HSP17 | TCGCTACTCTCTGCTTCGAT | 57.9 °C |
| qFp_HSP18.2 | CTGCAGATTAGCGGAGAGAG | 58 °C |
| qRp_HSP18.2 | ACAACCGTAAGCACACCATT | 58 °C |
| qFp_AT-actin-2 | AAGCTGGGGTTTTATGAATGG | 58 °C |
| qRp_AT-actin-2 | GGGACTAAAACGCAAAACGA | 58 °C |
| qFp_β-Actin-F | GCG GTCGAACAACTGGTATT | 63.7 °C |
| qFp_ β-Actin-F | GGT CCAAACGAAGGATAGCA | 63.8 °C |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumar, R.R.; Dubey, K.; Goswami, S.; Rai, G.K.; Rai, P.K.; Salgotra, R.K.; Bakshi, S.; Mishra, D.; Mishra, G.P.; Chinnusamy, V. Transcriptional Regulation of Small Heat Shock Protein 17 (sHSP-17) by Triticum aestivum HSFA2h Transcription Factor Confers Tolerance in Arabidopsis under Heat Stress. Plants 2023, 12, 3598. https://doi.org/10.3390/plants12203598
Kumar RR, Dubey K, Goswami S, Rai GK, Rai PK, Salgotra RK, Bakshi S, Mishra D, Mishra GP, Chinnusamy V. Transcriptional Regulation of Small Heat Shock Protein 17 (sHSP-17) by Triticum aestivum HSFA2h Transcription Factor Confers Tolerance in Arabidopsis under Heat Stress. Plants. 2023; 12(20):3598. https://doi.org/10.3390/plants12203598
Chicago/Turabian StyleKumar, Ranjeet R., Kavita Dubey, Suneha Goswami, Gyanendra K. Rai, Pradeep K. Rai, Romesh K. Salgotra, Suman Bakshi, Dwijesh Mishra, Gyan P. Mishra, and Viswanathan Chinnusamy. 2023. "Transcriptional Regulation of Small Heat Shock Protein 17 (sHSP-17) by Triticum aestivum HSFA2h Transcription Factor Confers Tolerance in Arabidopsis under Heat Stress" Plants 12, no. 20: 3598. https://doi.org/10.3390/plants12203598
APA StyleKumar, R. R., Dubey, K., Goswami, S., Rai, G. K., Rai, P. K., Salgotra, R. K., Bakshi, S., Mishra, D., Mishra, G. P., & Chinnusamy, V. (2023). Transcriptional Regulation of Small Heat Shock Protein 17 (sHSP-17) by Triticum aestivum HSFA2h Transcription Factor Confers Tolerance in Arabidopsis under Heat Stress. Plants, 12(20), 3598. https://doi.org/10.3390/plants12203598








