McAPRR2: The Key Regulator of Domesticated Pericarp Color in Bitter Gourd
Abstract
:1. Introduction
2. Results
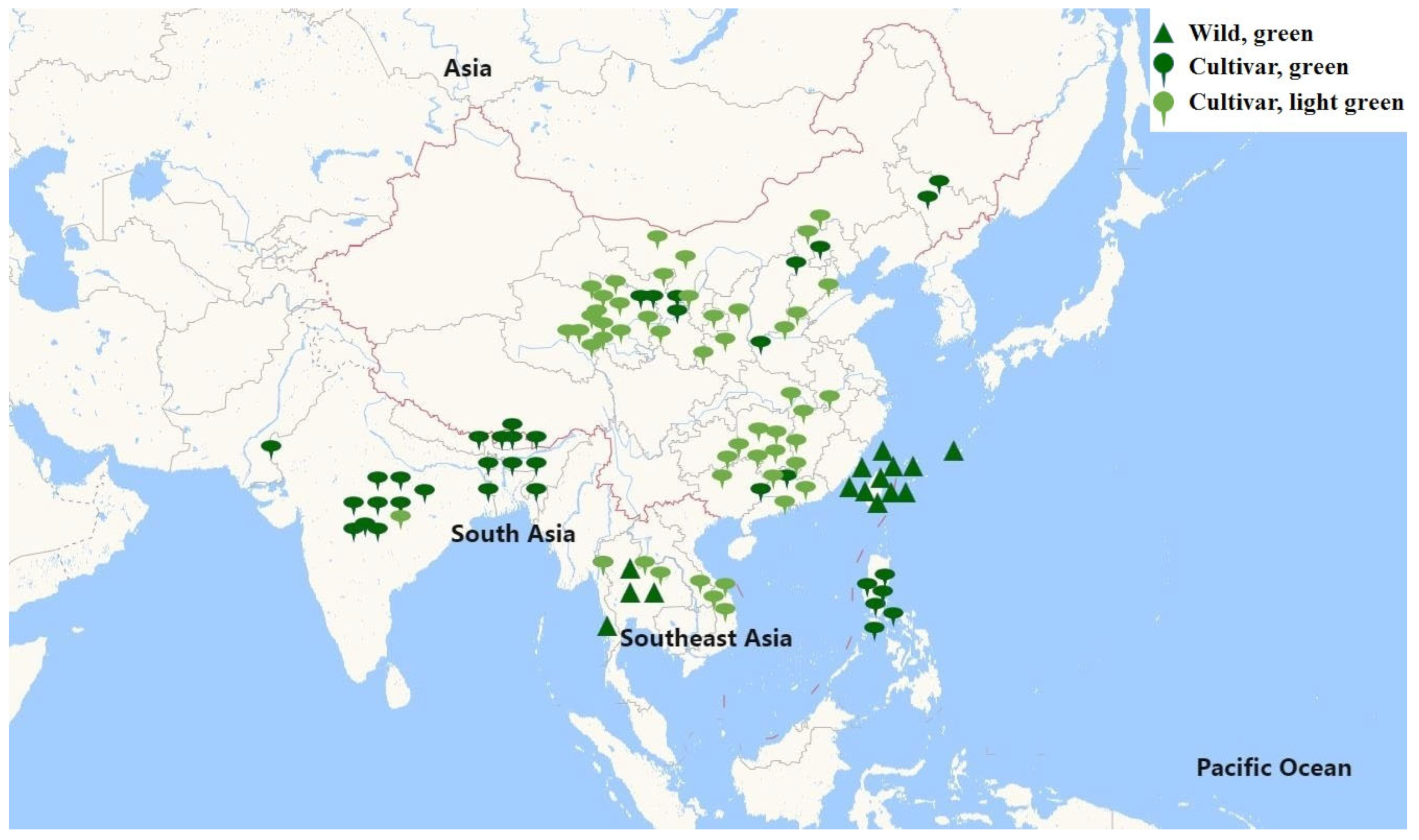
2.1. Genetic Characteristics of 106 Bitter Gourd Accessions
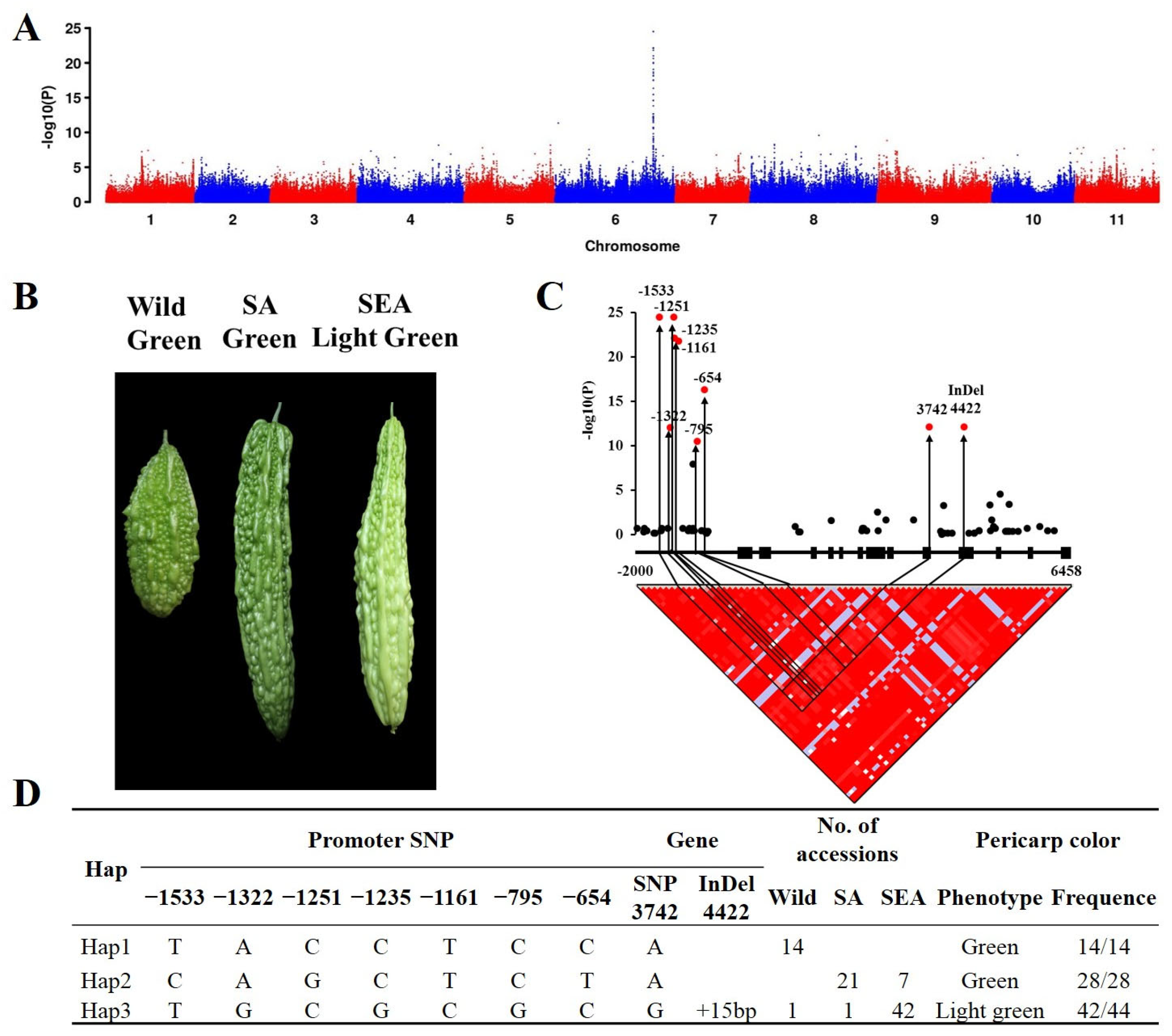
2.2. Genome-Wide Association of Pericarp Color in Bitter Gourd
2.3. The Haplotype of McAPRR2
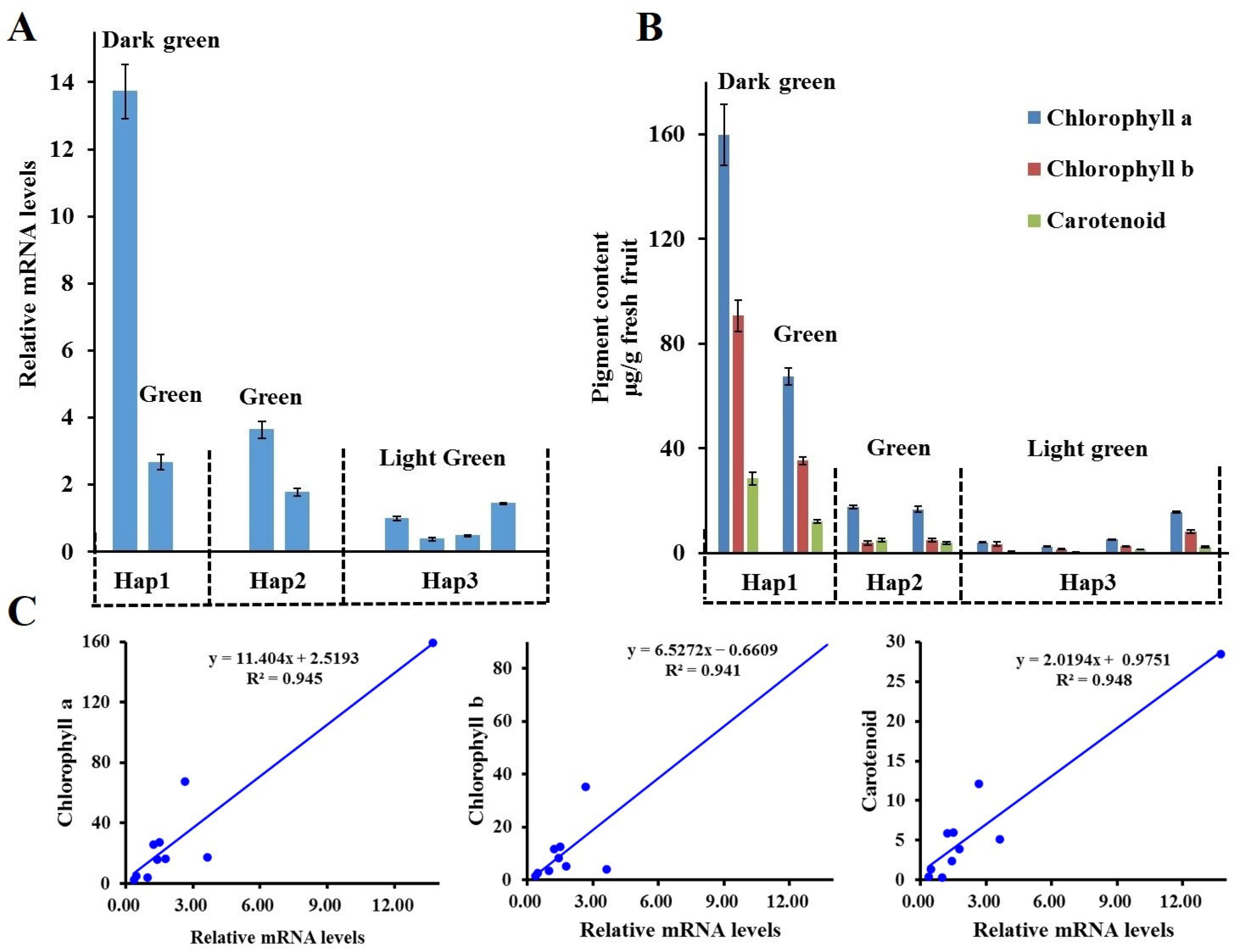
2.4. The Expression Pattern of McAPRR2 and Pigment Content in Pericarp Color
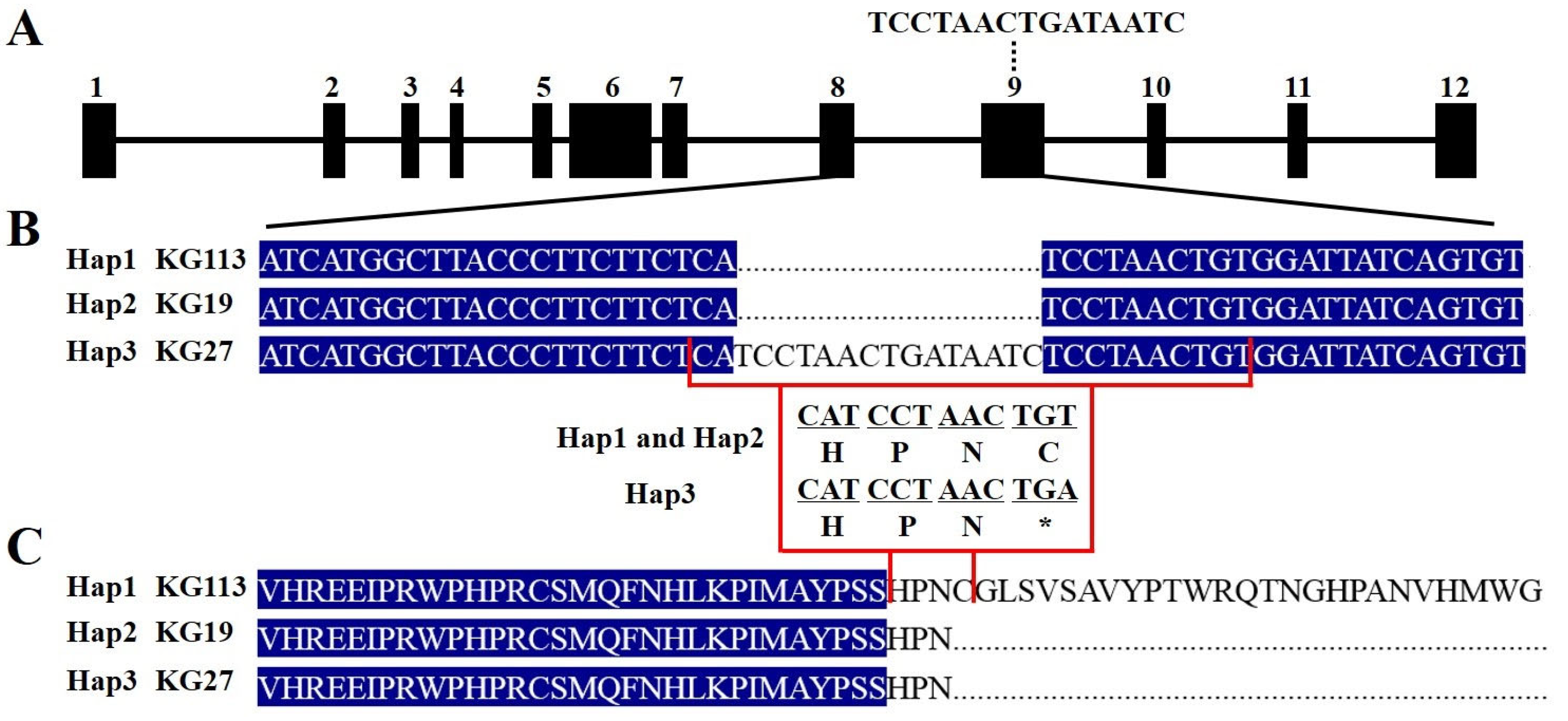
2.5. The 15 bp Insertion in McAPRR2 Alters Its Function
2.6. McAPRR2 Was under Domestication in Bitter Gourd
3. Discussion
4. Materials and Methods
4.1. Resequencing of 106 M. charantia Accessions
4.2. The SNP Calling and Filtering
4.3. Population Genetics Analyses
4.4. Genome-Wide Association (GWAS) of Pericarp Color
4.5. The Genetic Analysis of McAPRR2 Gene
4.6. RT-qPCR Analysis of McAPRR2
4.7. Analysis and Quantification of Chlorophyll and Carotenoid Contents
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Farooqi, A.A.; Khalid, S.; Tahir, F.; Sabitaliyevich, U.Y.; Yaylim, I.; Attar, R.; Xu, B. Bitter gourd (Momordica charantia) as a rich source of bioactive components to combat cancer naturally: Are we on the right track to fully unlock its potential as inhibitor of deregulated signaling pathways. Food Chem. Toxicol. 2018, 119, 98–105. [Google Scholar] [CrossRef]
- Dia, V.P.; Krishnan, H.B. BG-4, a novel anticancer peptide from bitter gourd (Momordica charantia), promotes apoptosis in human colon cancer cells. Sci. Rep. 2016, 6, 33532. [Google Scholar] [CrossRef] [PubMed]
- Akihisa, T.; Higo, N.; Tokuda, H.; Ukiya, M.; Akazawa, H.; Tochigi, Y.; Kimura, Y.; Suzuki, T.; Nishino, H. Cucurbitane-type triterpenoids from the fruits of Momordica charantia and their cancer chemopreventive effects. J. Nat. Prod. 2007, 70, 1233–1239. [Google Scholar] [CrossRef] [PubMed]
- Lee-Huang, S.; Huang, P.L.; Nara, P.L.; Chen, H.C.; Kung, H.F.; Huang, P.; Huang, H.I.; Huang, P.L. MAP 30: A new inhibitor of HIV-1 infection and replication. FEBS Lett. 1990, 272, 12–18. [Google Scholar] [CrossRef]
- Schaefer, H.; Renner, S.S. A three-genome phylogeny of Momordica (Cucurbitaceae) suggests seven returns from dioecy to monoecy and recent long-distance dispersal to Asia. Mol. Phylogenet. Evol. 2010, 54, 553–560. [Google Scholar] [CrossRef]
- Chomicki, G.; Schaefer, H.; Renner, S.S. Origin and domestication of Cucurbitaceae crops: Insights from phylogenies, genomics and archaeology. New Phytol. 2020, 226, 1240–1255. [Google Scholar] [CrossRef] [PubMed]
- Miniraj, N.; Prasanna, K.; Peter, K. Bitter gourd Momordica spp. In Genetic Improvement of Vegetable Plants; Kalloo, G., Bergh, B.O., Eds.; Pergamon Press: Oxford, UK, 1993. [Google Scholar]
- Walters, T.W.; Decker-Walters, D.S. Balsam-pear (Momordica charantia, Cucurbitaceae). Econ. Bot. 1988, 42, 286–288. [Google Scholar]
- Matsumura, H.; Hsiao, M.C.; Lin, Y.P.; Toyoda, A.; Taniai, N.; Tarora, K.; Urasaki, N.; Anand, S.S.; Dhillon, N.P.S.; Schafleitner, R.; et al. Long-read bitter gourd (Momordica charantia) genome and the genomic architecture of nonclassic domestication. Proc. Natl. Acad. Sci. USA 2020, 117, 14543–14551. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Yang, Y.; Luo, S.; Wang, L.; Huang, R.; Wen, Q.; Han, X.; Miao, N.; Cheng, J.; Liu, Z.; et al. Whole-genome sequencing provides insights into the genetic diversity and domestication of bitter gourd (Momordica spp.). Hortic. Res. 2020, 7, 85. [Google Scholar] [CrossRef] [PubMed]
- Renner, S.S. Bitter gourd from Africa expanded to Southeast Asia and was domesticated there: A new insight from parallel studies. Proc. Natl. Acad. Sci. USA 2020, 117, 24630–24631. [Google Scholar] [CrossRef]
- Jin, J.; Huang, W.; Gao, J.P.; Yang, J.; Shi, M.; Zhu, M.Z.; Luo, D.; Lin, H.X. Genetic control of rice plant architecture under domestication. Nat. Genet. 2008, 40, 1365–1369. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarti, M.; Zhang, N.; Sauvage, C.; Muños, S.; Blanca, J.; Cañizares, J.; Diez, M.J.; Schneider, R.; Mazourek, M.; McClead, J.; et al. A cytochrome P450 regulates a domestication trait in cultivated tomato. Proc. Natl. Acad. Sci. USA 2013, 110, 17125–17130. [Google Scholar] [CrossRef]
- Kovach, M.J.; Sweeney, M.T.; McCouch, S.R. New insights into the history of rice domestication. Trends Genet. 2007, 23, 578–587. [Google Scholar] [CrossRef]
- Sweeney, M.; McCouch, S. The complex history of the domestication of rice. Ann. Bot. 2007, 100, 951–957. [Google Scholar] [CrossRef]
- Meyer, R.S.; Purugganan, M.D. Evolution of crop species: Genetics of domestication and diversification. Nat. Rev. Genet. 2013, 14, 840–852. [Google Scholar] [CrossRef] [PubMed]
- Olsen, K.M.; Purugganan, M.D. Molecular evidence on the origin and evolution of glutinous rice. Genetics 2002, 162, 941–950. [Google Scholar] [CrossRef] [PubMed]
- Dhillon, N.P.S.; Sanguansil, S.; Schafleitner, R.; Wang, Y.-W.; McCreight, J.D. Diversity Among a Wide Asian Collection of Bitter Gourd Landraces and their Genetic Relationships with Commercial Hybrid Cultivars. J. Am. Soc. Hortic. Sci. 2016, 141, 475–484. [Google Scholar] [CrossRef]
- Lun, Y.; Wang, X.; Zhang, C.; Yang, L.; Gao, D.; Chen, H.; Huang, S. A CsYcf54 variant conferring light green coloration in cucumber. Euphytica 2016, 208, 509–517. [Google Scholar] [CrossRef]
- Liu, H.; Jiao, J.; Liang, X.; Liu, J.; Meng, H.; Chen, S.; Li, Y.; Cheng, Z. Map-based cloning, identification and characterization of the w gene controlling white immature fruit color in cucumber (Cucumis sativus L.). Theor. Appl. Genet. 2016, 129, 1247–1256. [Google Scholar] [CrossRef] [PubMed]
- Oren, E.; Tzuri, G.; Vexler, L.; Dafna, A.; Meir, A.; Faigenboim, A.; Kenigswald, M.; Portnoy, V.; Schaffer, A.A.; Levi, A.; et al. The multi-allelic APRR2 gene is associated with fruit pigment accumulation in melon and watermelon. J. Exp. Bot. 2019, 70, 3781–3794. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Zhao, S.; Dou, J.; Ali, A.; Gebremeskel, H.; Gao, L.; He, N.; Lu, X.; Liu, W. Genetic mapping and development of molecular markers for a candidate gene locus controlling rind color in watermelon. Theor. Appl. Genet. 2019, 132, 2741–2753. [Google Scholar] [CrossRef]
- Zhao, G.; Lian, Q.; Zhang, Z.; Fu, Q.; He, Y.; Ma, S.; Ruggieri, V.; Monforte, A.J.; Wang, P.; Julca, I.; et al. A comprehensive genome variation map of melon identifies multiple domestication events and loci influencing agronomic traits. Nat. Genet. 2019, 51, 1607–1615. [Google Scholar] [CrossRef]
- Ma, L.; Liu, Z.; Cheng, Z.; Gou, J.; Chen, J.; Yu, W.; Wang, P. Identification and Application of BhAPRR2 Controlling Peel Colour in Wax Gourd (Benincasa hispida). Front. Plant Sci. 2021, 12, 716772. [Google Scholar] [CrossRef] [PubMed]
- Zhan, J.; Zhong, J.; Cheng, J.; Wang, Y.; Hu, K. Map-based cloning of the APRR2 gene controlling green stigma in bitter gourd (Momordica charantia). Front. Plant Sci. 2023, 14, 1128926. [Google Scholar] [CrossRef]
- Pan, Y.; Bradley, G.; Pyke, K.; Ball, G.; Lu, C.; Fray, R.; Marshall, A.; Jayasuta, S.; Baxter, C.; van Wijk, R.; et al. Network inference analysis identifies an APRR2-like gene linked to pigment accumulation in tomato and pepper fruits. Plant Physiol. 2013, 161, 1476–1485. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Zhao, S.; Sun, H.; Wang, X.; Wu, S.; Lin, T.; Ren, Y.; Gao, L.; Deng, Y.; Zhang, J.; et al. Resequencing of 414 cultivated and wild watermelon accessions identifies selection for fruit quality traits. Nat. Genet. 2019, 51, 1616–1623. [Google Scholar] [CrossRef]
- Li, C.; Huang, Y.; Huang, R.; Wu, Y.; Wang, W. The genetic architecture of amylose biosynthesis in maize kernel. Plant Biotechnol. J. 2018, 16, 688–695. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Wei, X.; Sang, T.; Zhao, Q.; Feng, Q.; Zhao, Y.; Li, C.; Zhu, C.; Lu, T.; Zhang, Z.; et al. Genome-wide association studies of 14 agronomic traits in rice landraces. Nat. Genet. 2010, 42, 961–967. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Duan, S.; Xia, Q.; Liang, Z.; Dong, X.; Margaryan, K.; Musayev, M.; Goryslavets, S.; Zdunić, G.; Bert, P.F.; et al. Dual domestications and origin of traits in grapevine evolution. Science 2023, 379, 892–901. [Google Scholar] [CrossRef] [PubMed]
- Stetter, M.G.; Vidal-Villarejo, M.; Schmid, K.J. Parallel Seed Color Adaptation during Multiple Domestication Attempts of an Ancient New World Grain. Mol. Biol. Evol. 2020, 37, 1407–1419. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 2015, 4, 7. [Google Scholar] [CrossRef]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef] [PubMed]
- Barrett, J.C.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 2005, 21, 263–265. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, S.; Yang, J.; Fu, Y.; Zhang, X.; Zhang, J.; Zhao, H.; Hu, Q.; Liu, P.; He, W.; Han, X.; et al. McAPRR2: The Key Regulator of Domesticated Pericarp Color in Bitter Gourd. Plants 2023, 12, 3585. https://doi.org/10.3390/plants12203585
Tian S, Yang J, Fu Y, Zhang X, Zhang J, Zhao H, Hu Q, Liu P, He W, Han X, et al. McAPRR2: The Key Regulator of Domesticated Pericarp Color in Bitter Gourd. Plants. 2023; 12(20):3585. https://doi.org/10.3390/plants12203585
Chicago/Turabian StyleTian, Shouwei, Jingjing Yang, Yiqian Fu, Xiaofei Zhang, Jian Zhang, Hong Zhao, Qi Hu, Pangyuan Liu, Weiming He, Xiangyang Han, and et al. 2023. "McAPRR2: The Key Regulator of Domesticated Pericarp Color in Bitter Gourd" Plants 12, no. 20: 3585. https://doi.org/10.3390/plants12203585
APA StyleTian, S., Yang, J., Fu, Y., Zhang, X., Zhang, J., Zhao, H., Hu, Q., Liu, P., He, W., Han, X., & Wen, C. (2023). McAPRR2: The Key Regulator of Domesticated Pericarp Color in Bitter Gourd. Plants, 12(20), 3585. https://doi.org/10.3390/plants12203585






