Auxin Transporter OsPIN1b, a Novel Regulator of Leaf Inclination in Rice (Oryza sativa L.)
Abstract
1. Introduction
2. Results
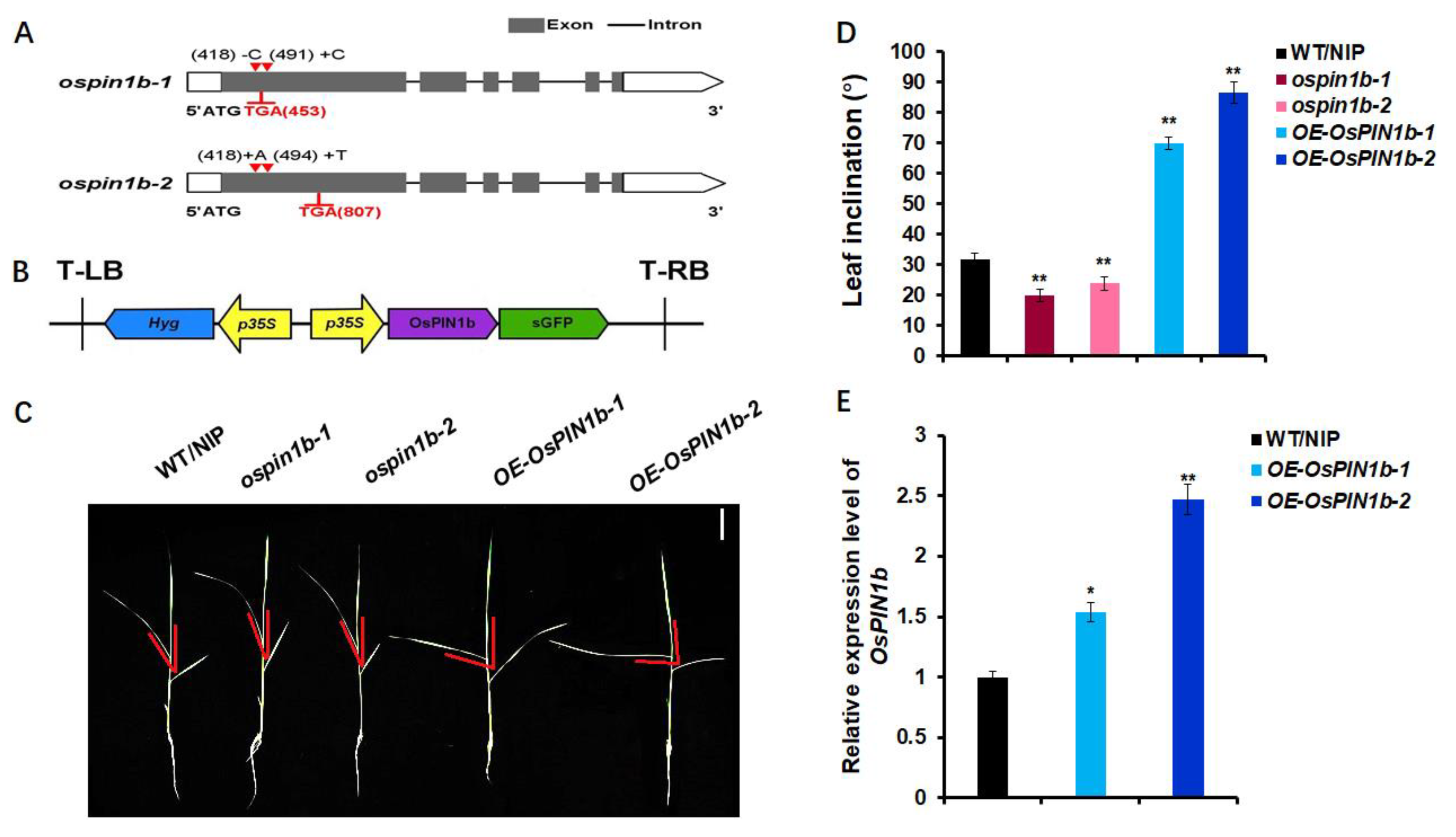
2.1. Identification of ospin1b Mutants and OsPIN1b Overexpression Lines
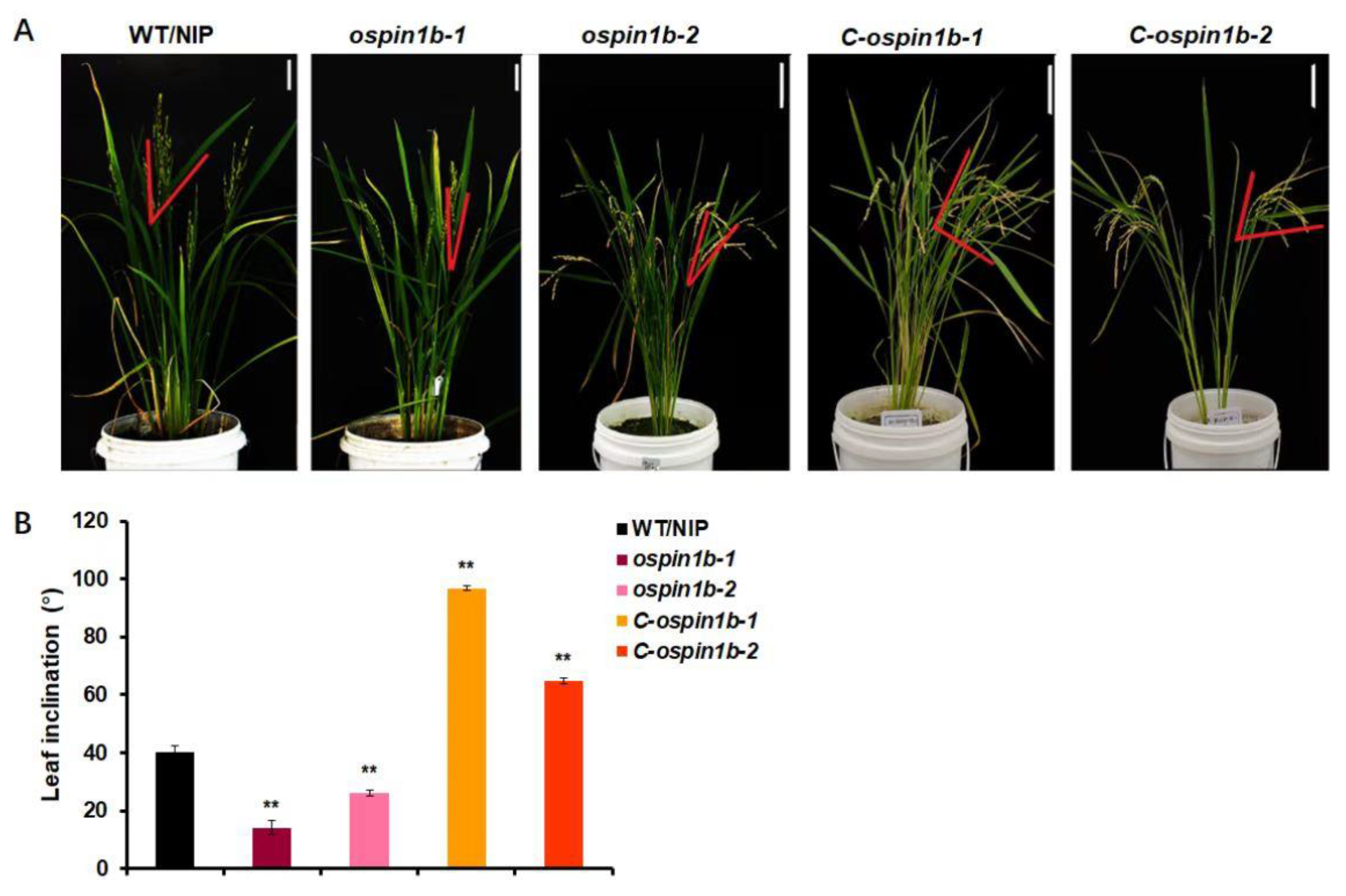
2.2. Characterization of Complementary ospin1b Lines
2.3. OsPIN1b Constitutively Expressed in Each Tissue Including Lamina Joint
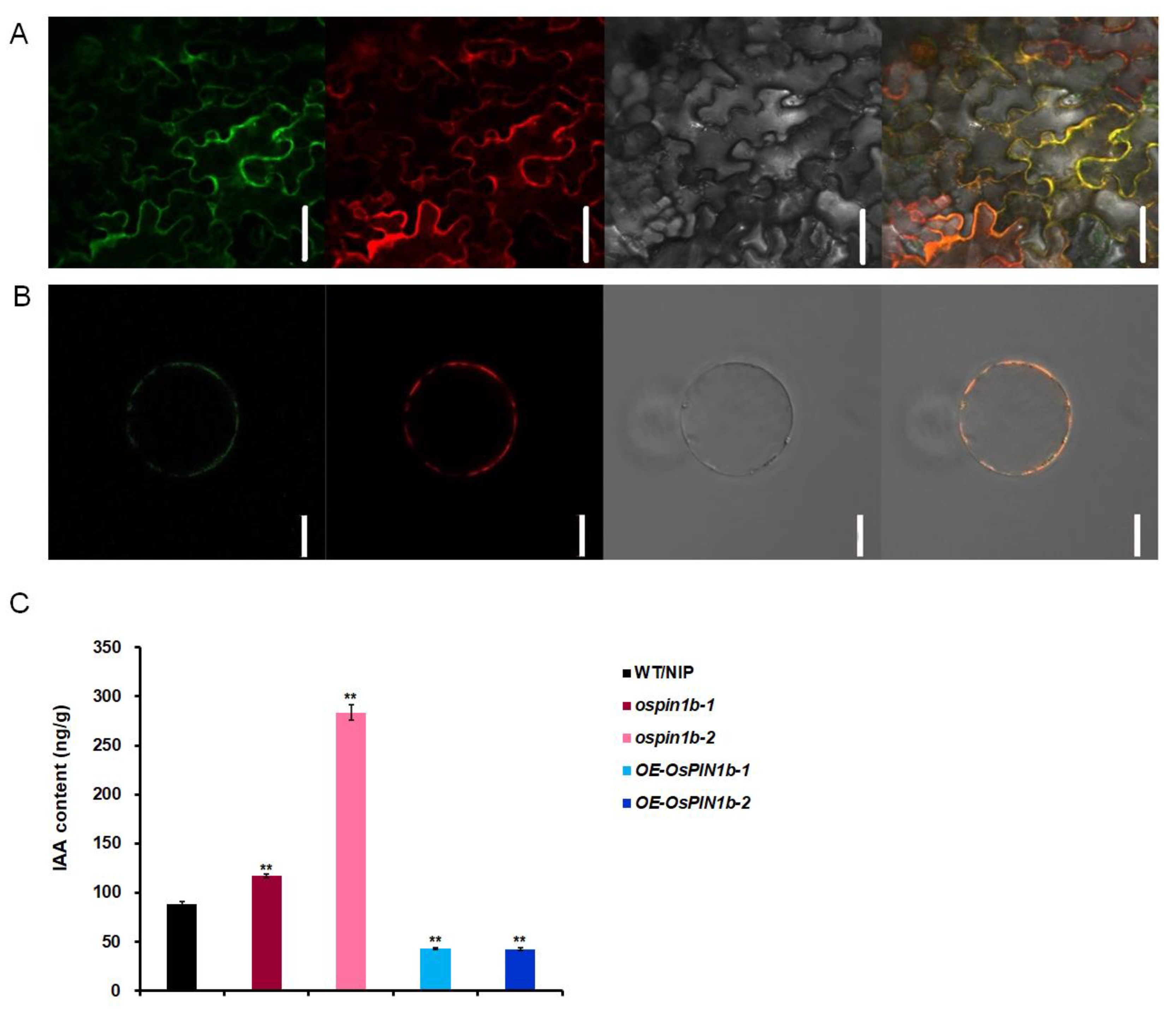
2.4. OsPIN1b Localized on Plasma Membrane and OsPIN1b Reduced the Free IAA Accumulation
2.5. OsPIN1b Enhances Adaxial Cell Division of Pulvinus to Increase Leaf Inclination
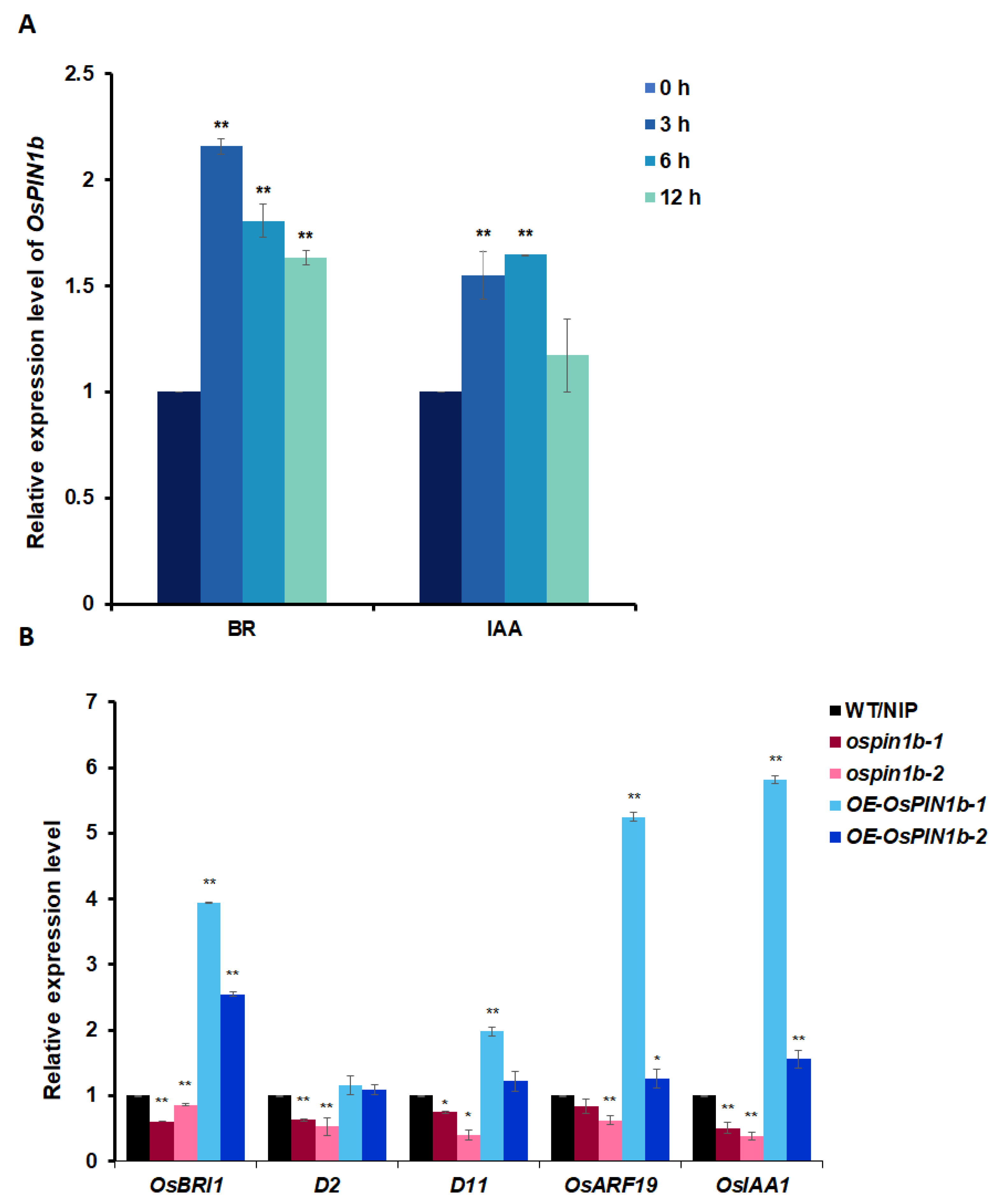
2.6. BR or Auxin Induces OsPIN1b Expression, and the Expression of the Genes Related Both Signaling Reduces in ospin1b Mutants
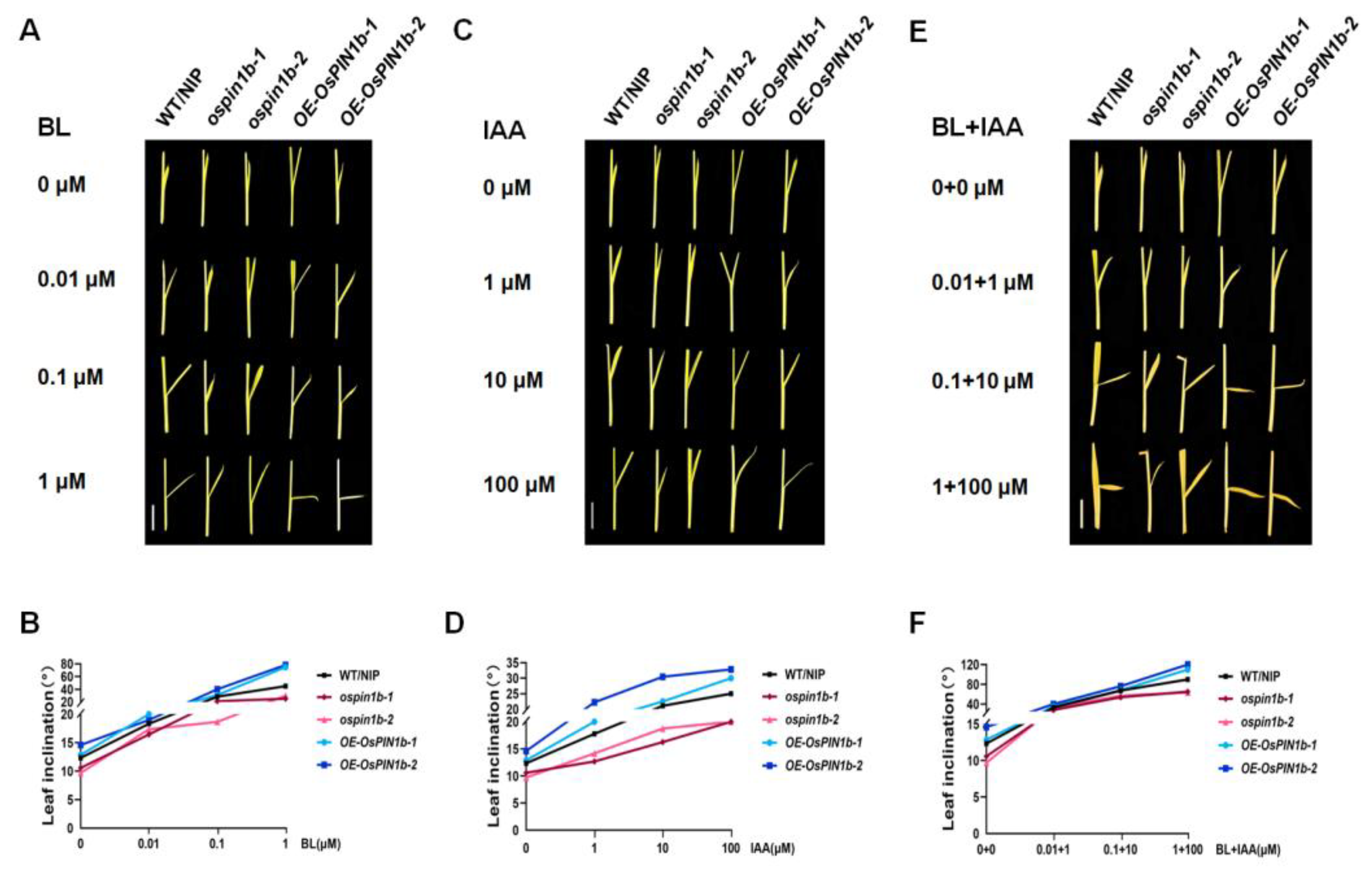
2.7. The Decreased Sensitivity to BR and IAA in ospin1b Mutants
3. Discussion
3.1. OsPIN1b Plays a Positive Role in Enlarging Leaf Inclination via Regulating Auxin Transport in Rice
3.2. OsPIN1b Participates in Adaxial Cell Enlargement of Lamina Joint through Complex Regulatory Mechanism of Auxin and BR
3.3. ospin1b Reduced the Plant Height and Leaf Inclination and Formed the Ideal Plant Architecture
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Construction and Identification of ospin1b Mutants
4.3. Construction and Transformation of Binary Vectors
4.4. Subcellular Localization of OsPIN1b
4.5. β- Glucuronidase (GUS) Staining
4.6. RNA Extraction, RT-PCR, and Real-Time Quantitative RT-PCR (qRT-PCR) Analyses
4.7. Exogenous Hormone Response Assay
4.8. Scanning Electron Microscopy (SEM)
4.9. Endogenous IAA Contents Analysis by HPLC
4.10. Accession Numbers
4.11. Primer Sequences
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sinclair, T.R.; Sheehy, J.E. Erect Leaves and Photosynthesis in Rice. Science 1999, 283, 1456–1457. [Google Scholar] [CrossRef]
- Sakamoto, T.; Morinaka, Y.; Ohnishi, T.; Sunohara, H.; Fujioka, S.; Ueguchi-Tanaka, M.; Mizutani, M.; Sakata, K.; Takatsuto, S.; Yoshida, S.; et al. Erect leaves caused by brassinosteroid deficiency increase biomass production and grain yield in rice. Nat. Biotechnol. 2006, 24, 105–109. [Google Scholar] [CrossRef] [PubMed]
- Duan, K.; Li, L.; Hu, P.; Xu, S.P.; Xu, Z.H.; Xue, H.W. A brassinolide-suppressed rice MADS-box transcription factor, OsMDP1, has a negative regulatory role in BR signaling. Plant J. 2006, 47, 519–531. [Google Scholar] [CrossRef] [PubMed]
- Yamamuro, C.; Ihara, Y.; Xiong, W.; Noguchi, T.; Fujioka, S.; Takatsuto, S.; Ashikari, M.; Matsuoka, K.M. Loss of Function of a Rice brassinosteroid insensitive1 Homolog Prevents Internode Elongation and Bending of the Lamina Joint. Plant Cell 2000, 12, 1591–1605. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.Y.; Bai, M.Y.; Wu, J.; Zhu, J.Y.; Wang, H.; Zhang, Z.; Wang, W.; Sun, Y.; Zhao, J.; Sun, X.; et al. Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant Cell 2009, 21, 3767–3780. [Google Scholar] [CrossRef]
- Sun, S.; Chen, D.; Li, X.; Qiao, S.; Shi, C.; Li, C.; Shen, H.; Wang, X. Brassinosteroid Signaling Regulates Leaf Erectness in Oryza sativa via the Control of a Specific U-Type Cyclin and Cell Proliferation. Dev. Cell 2015, 34, 220–228. [Google Scholar] [CrossRef]
- Hong, Z.; Ueguchi-Tanaka, M.; Shimizu-Sato, S.; Inukai, Y.; Fujioka, S.; Shimada, Y.; Takatsuto, S.; Agetsuma, M.; Yoshida, S.; Watanabe, Y.; et al. Loss-of-function of a rice brassinosteroid biosynthetic enzyme, C-6 oxidase, prevents the organized arrangement and polar elongation of cells in the leaves and stem. Plant J. 2002, 32, 495–508. [Google Scholar] [CrossRef]
- Hong, Z.; Ueguchi-Tanaka, M.; Umemura, K.; Uozu, S.; Fujioka, S.; Takatsuto, S.; Yoshida, S.; Ashikari, M.; Kitano, H.; Matsuoka, M. A rice brassinosteroid-deficient mutant, ebisu dwarf (d2), is caused by a loss of function of a new member of cytochrome P450. Plant Cell 2003, 15, 2900–2910. [Google Scholar] [CrossRef]
- Bai, M.Y.; Zhang, L.Y.; Gampala, S.S.; Zhu, S.W.; Song, W.Y.; Chong, K.; Wang, Z.Y. Functions of OsBZR1 and 14-3-3 proteins in brassinosteroid signaling in rice. Proc. Natl. Acad. Sci. USA 2007, 104, 13839–13844. [Google Scholar] [CrossRef]
- Cheng, H.; Hao, M.; Wang, W.; Mei, D.; Rachel, W.; Jia, L.; Wang, H.; Sang, S.; Tang, M.; Zhou, R. Integrative RNA- and miRNA-Profile Analysis Reveals a Likely Role of BR and Auxin Signaling in Branch Angle Regulation of B. napus. Int. J. Mol. Sci. 2017, 18, 887. [Google Scholar] [CrossRef]
- Sakamoto, T.; Morinaka, Y.; Inukai, Y.; Kitano, H.; Fujioka, S. Auxin signal transcription factor regulates expression of the brassinosteroid receptor gene in rice. Plant J. 2013, 73, 676–688. [Google Scholar] [CrossRef] [PubMed]
- Wada, K.; Marumo, S.; Ikekawa, N.; Morisaki, M.; Mori, K. Brassinolide and Homobrassinolide Promotion of Lamina Inclination of Rice Seedlings. Plant Cell Physiol. 1981, 22, 323–325. [Google Scholar]
- Bian, H.; Xie, Y.; Guo, F.; Ning, H.; Zhu, M. Distinctive expression patterns and roles of the miRNA393/TIR1 homolog module in regulating flag leaf inclination and primary and crown root growth in rice (Oryza sativa). N. Phytol. 2012, 196, 149–161. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; You, J.; Xiong, L. Characterization of OsIAA1 gene, a member of rice Aux/IAA family involved in auxin and brassinosteroid hormone responses and plant morphogenesis. Plant Mol. Biol. 2009, 70, 297–309. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.W.; Li, C.H.; Cao, J.; Zhang, Y.C.; Zhang, S.Q.; Xia, Y.F.; Sun, D.Y.; Sun, Y. Altered Architecture and Enhanced Drought Tolerance in Rice via the Down-Regulation of Indole-3-Acetic Acid by TLD1/OsGH3.13 Activation. Plant Physiol. 2009, 151, 1889–1901. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.Q.; Xiang, J.J.; Xue, H.W. Studies on the Rice LEAF INCLINATION1 (LC1), an IAA–amido Synthetase, Reveal the Effects of Auxin in Leaf Inclination Control. Mol. Plant 2013, 6, 174–187. [Google Scholar] [CrossRef]
- Liu, X.; Yang, C.Y.; Miao, R.; Zhou, C.L.; Cao, P.H.; Lan, J.; Zhu, X.J.; Mou, C.L.; Huang, Y.S.; Liu, S.J. DS1/OsEMF1 interacts with OsARF11 to control rice architecture by regulation of brassinosteroid signaling. Rice 2018, 11, 46. [Google Scholar] [CrossRef]
- Huang, G.; Hu, H.; van de Meene, A.; Zhang, J.; Dong, L.; Zheng, S.; Zhang, F.; Betts, N.S.; Liang, W.; Bennett, M.J.; et al. AUXIN RESPONSE FACTORS 6 and 17 control the flag leaf angle in rice by regulating secondary cell wall biosynthesis of lamina joints. Plant Cell 2021, 33, 3120–3133. [Google Scholar] [CrossRef]
- Yoshikawa, T.; Ito, M.; Sumikura, T.; Nakayama, A.; Itoh, J.C. The rice FISH BONE gene encodes a tryptophan aminotransferase, which affects pleiotropic auxin-related processes. Plant J. Cell Mol. Biol. 2014, 78, 927–936. [Google Scholar] [CrossRef]
- Adamowski, M.; Friml, J. PIN-Dependent Auxin Transport: Action, Regulation, and Evolution. Plant Cell 2015, 27, 20–32. [Google Scholar] [CrossRef]
- Geisler, M.; Blakeslee, J.J.; Bouchard, R.; Lee, O.R.; Vincenzetti, V.; Bandyopadhyay, A.; Titapiwatanakun, B.; Peer, W.A.; Bailly, A.; Richards, E.L.; et al. Cellular efflux of auxin catalyzed by the Arabidopsis MDR/PGP transporter AtPGP1. Plant J. 2005, 44, 179–194. [Google Scholar] [CrossRef] [PubMed]
- Geisler, M.; Murphy, A.S. The ABC of auxin transport: The role of p-glycoproteins in plant development. FEBS Lett. 2006, 580, 1094–1102. [Google Scholar] [CrossRef] [PubMed]
- Swarup, R.; Péret, B. AUX/LAX family of auxin influx carriers—An overview. Front. Plant Sci. 2012, 3, 225. [Google Scholar] [CrossRef]
- Wang, M.; Qiao, J.Y.; Yu, C.L.; Chen, H.; Sun, C.D.; Huang, L.Z.; Li, C.Y.; Geisler, M.; Qian, Q.; Jiang, D.A.; et al. The auxin influx carrier, OsAUX3, regulates rice root development and responses to aluminium stress. Plant Cell Environ. 2019, 42, 1125–1138. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.L.; Sun, C.D.; Shen, C.; Wang, S.; Liu, F.; Liu, Y.; Chen, Y.L.; Li, C.; Qian, Q.; Aryal, B. The auxin transporter, OsAUX1, is involved in primary root and root hair elongation and in Cd stress responses in rice (Oryza sativa L.). Plant J. 2015, 83, 818–830. [Google Scholar] [CrossRef] [PubMed]
- Wiśniewska, J.; Xu, J.; Seifertová, D.; Brewer, P.B.; Růžička, K.; Blilou, I.; Rouquié, D.; Benková, E.; Scheres, B.; Friml, J. Polar PIN Localization Directs Auxin Flow in Plants. Science 2006, 312, 883. [Google Scholar] [CrossRef] [PubMed]
- Blilou, I.; Xu, J.; Wildwater, M.; Willemsen, V.; Paponov, I.; Friml, J.; Heidstra, R.; Aida, M.; Palme, K.; Scheres, B. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 2005, 433, 39–44. [Google Scholar] [CrossRef]
- Friml, J.; Vieten, A.; Sauer, M.; Weijers, D.; Schwarz, H.; Hamann, T.; Offringa, R.; Juergens, G. Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 2003, 426, 147–153. [Google Scholar] [CrossRef]
- Gälweiler, L.; Guan, C.; M-Ller, A.; Wisman, E.; Mendgen, K.; Yephremov, A.; Palme, K. Regulation of Polar Auxin Transport by AtPIN1 in Arabidopsis Vascular Tissue. Science 1998, 282, 2226–2230. [Google Scholar] [CrossRef]
- Okada, K.; Ueda, J.; Komaki, M.K.; Shimura, B.Y. Requirement of the Auxin Polar Transport System in Early Stages of Arabidopsis Floral Bud Formation. Plant Cell 1991, 3, 677–684. [Google Scholar] [CrossRef]
- Wang, J.R.; Hu, H.; Wang, G.H.; Jing, L.; Chen, J.Y.; Ping, W. Expression of PIN genes in rice (Oryza sativa L.): Tissue specificity and regulation by hormones. Mol. Plant 2009, 2, 823–831. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Tao, J.; Bi, Y.; Hou, M.; Lou, J.; Chen, X.; Zhang, X.; Le, L.; Xie, X.; Koichi, Y. OsPIN1b is Involved in Rice Seminal Root Elongation by Regulating Root Apical Meristem Activity in Response to Low Nitrogen and Phosphate. Sci. Rep. 2018, 8, 13014. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Zhu, L.; Shou, H.; Wu, P. A PIN1 Family Gene, OsPIN1, involved in Auxin-dependent Adventitious Root Emergence and Tillering in Rice. Plant Cell Physiol. 2005, 46, 1674–1681. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Fan, X.; Song, W.; Zhang, Y.; Xu, G. Over-expression of OsPIN2 leads to increased tiller numbers, angle and shorter plant height through suppression of OsLAZY1. Plant Biotechnol. J. 2012, 10, 139–149. [Google Scholar] [CrossRef] [PubMed]
- Lu, G.; Coneva, V.; Casaretto, J.A.; Ying, S.; Mahmood, K.; Liu, F.; Nambara, E.; Bi, Y.M.; Rothstein, S.J. OsPIN5b modulates rice (Oryza sativa) plant architecture and yield by changing auxin homeostasis, transport and distribution. Plant J. 2015, 83, 913–925. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Wen, J.; Zhao, W.; Wang, Q.; Huang, W. Rational Improvement of Rice Yield and Cold Tolerance by Editing the Three Genes OsPIN5b, GS3, and OsMYB30 With the CRISPR-Cas9 System. Front. Plant Sci. 2020, 10, 1663. [Google Scholar] [CrossRef] [PubMed]
- Hou, M.; Luo, F.; Wu, D.; Zhang, X.; Zhang, Y. OsPIN9, an auxin efflux carrier, is required for the regulation of rice tiller bud outgrowth by ammonium. N. Phytol. 2020, 229, 935–949. [Google Scholar] [CrossRef] [PubMed]
- Miyashita, Y.; Takasugi, T.; Ito, Y. Identification and expression analysis of PIN genes in rice. Plant Sci. 2010, 178, 424–428. [Google Scholar] [CrossRef]
- Zhang, Q.; Li, J.; Zhang, W.; Yan, S.; Wang, R.; Zhao, J.; Li, Y.; Qi, Z.; Sun, Z.; Zhu, Z. The putative auxin efflux carrier OsPIN3t is involved in the drought stress response and drought tolerance. Plant J. 2012, 72, 805–816. [Google Scholar] [CrossRef] [PubMed]
- Paponov, I.A.; Teale, W.D.; Trebar, M.; Blilou, I.; Palme, K. The PIN auxin efflux facilitators: Evolutionary and functional perspectives. Trends Plant Sci. 2005, 10, 170–177. [Google Scholar] [CrossRef]
- Nelson, B.K.; Cai, X.; Nebenführ, A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 2007, 51, 1126–1136. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Wang, S.; Xu, Y.; Yu, C.; Shen, C.; Qian, Q.; Geisler, M.; Jiang, D.A.; Qi, Y. The auxin response factor, OsARF19, controls rice leaf angles through positively regulating OsGH3-5 and OsBRI1. Plant Cell Environ. 2015, 38, 638–654. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Chen, S. Brassinosteroid-induced rice lamina joint inclination and its relation to indole-3-acetic acid and ethylene. Plant Growth Regul. 1995, 16, 189–196. [Google Scholar] [CrossRef]
- Zhao, S.Q.; Hu, J.; Guo, L.B.; Qian, Q.; Xue, H.W. Rice leaf inclination2, a VIN3-like protein, regulates leaf angle through modulating cell division of the collar. Cell Res. 2010, 20, 935–947. [Google Scholar] [CrossRef]
- He, Z.; Wang, Z.Y.; Li, J.; Zhu, Q.; Lamb, C.; Ronald, P.; Chory, J. Perception of brassinosteroids by the extracellular domain of the receptor kinase BRI1. Science 2000, 288, 2360–2363. [Google Scholar] [CrossRef]
- Kinoshita, T.; Caño-Delgado, A.; Seto, H.; Hiranuma, S.; Fujioka, S.; Yoshida, S.; Chory, J. Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature 2005, 433, 167–171. [Google Scholar] [CrossRef]
- Li, J.; Chory, J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 1997, 90, 929–938. [Google Scholar] [CrossRef]
- Wang, Z.Y.; Seto, H.; Fujioka, S.; Yoshida, S.; Chory, J. BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 2001, 410, 380–383. [Google Scholar] [CrossRef]
- Zhao, J.; Wu, C.; Yuan, S.; Yin, L.; Sun, W.; Zhao, Q.; Zhao, B.; Li, X. Kinase activity of OsBRI1 is essential for brassinosteroids to regulate rice growth and development. Plant Sci. 2013, 199–200, 113–120. [Google Scholar] [CrossRef]
- Tanabe, S.; Ashikari, M.; Fujioka, S.; Takatsuto, S.; Yoshida, S.; Yano, M.; Yoshimura, A.; Kitano, H.; Matsuoka, M.; Fujisawa, Y.; et al. A novel cytochrome P450 is implicated in brassinosteroid biosynthesis via the characterization of a rice dwarf mutant, dwarf11, with reduced seed length. Plant Cell 2005, 17, 776–790. [Google Scholar] [CrossRef]
- Yu, X.; Wang, H.; Leung, D.W.M.; He, Z.; Zhang, J.; Peng, X.; Liu, E. Overexpression of OsIAAGLU reveals a role for IAA–glucose conjugation in modulating rice plant architecture. Plant Cell Rep. 2019, 38, 731–739. [Google Scholar] [CrossRef] [PubMed]
- Gallavotti, A. The role of auxin in shaping shoot architecture. J. Exp. Bot. 2013, 64, 2593–2608. [Google Scholar] [CrossRef]
- McSteen, P. Auxin and Monocot Development. Cold Spring Harbor Perspect. Biol. 2010, 2, a001479. [Google Scholar] [CrossRef] [PubMed]
- Petrasek, J.; Friml, J. Auxin transport routes in plant development. Development 2009, 136, 2675–2688. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Gao, L.; Ke, M.; Gao, Z.; Tu, T.; Huang, L.; Chen, J.; Guan, Y.; Huang, X.; Chen, X. GmPIN1-mediated auxin asymmetry regulates leaf petiole angle and plant architecture in soybean. J. Integr. Plant Biol. 2022, 64, 1325–1338. [Google Scholar] [CrossRef] [PubMed]
- Qiao, J.; Zhang, Y.; Han, S.; Chang, S.; Gao, Z.; Qi, Y.; Qian, Q. OsARF4 regulates leaf inclination via auxin and brassinosteroid pathways in rice. Front. Plant Sci. 2022, 13, 979033. [Google Scholar] [CrossRef] [PubMed]
- Khush, G.S. Green revolution: Preparing for the 21st century. Genome 1999, 42, 646–655. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.K.; Zhang, S.N.; Sun, C.D.; Xu, Y.X.; Chen, Y.; Yu, C.L.; Qian, Q.; Jiang, D.A.; Qi, Y.H. Auxin response factor (OsARF12), a novel regulator for phosphate homeostasis in rice (Oryza sativa). N. Phytol. 2013, 201, 91–103. [Google Scholar] [CrossRef]
- Xie, K.; Minkenberg, B.; Yang, Y. Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc. Natl. Acad. Sci. USA 2015, 112, 3570–3575. [Google Scholar] [CrossRef]
- Hiei, Y.; Ohta, S.; Komari, T.; Kumashiro, T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 1994, 6, 271–282. [Google Scholar] [CrossRef]
- Qi, Y.; Wang, S.; Shen, C.; Zhang, S.; Chen, Y.; Xu, Y.; Liu, Y.; Wu, Y.; Jiang, D. OsARF12, a transcription activator on auxin response gene, regulates root elongation and affects iron accumulation in rice (Oryza sativa). N. Phytol. 2012, 193, 109–120. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; Zhou, H.Z.; Wu, Y.; Zhang, H.; Lin, J.; Jiang, X.; He, Q.; Zhu, J.; Li, Y.; Yu, H. OsSPL3, an SBP-Domain Protein, Regulates Crown Root Development in Rice. Plant Cell 2019, 31, 1257–1275. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Han, S.; Lin, Y.; Qiao, J.; Han, N.; Li, Y.; Feng, Y.; Li, D.; Qi, Y. Auxin Transporter OsPIN1b, a Novel Regulator of Leaf Inclination in Rice (Oryza sativa L.). Plants 2023, 12, 409. https://doi.org/10.3390/plants12020409
Zhang Y, Han S, Lin Y, Qiao J, Han N, Li Y, Feng Y, Li D, Qi Y. Auxin Transporter OsPIN1b, a Novel Regulator of Leaf Inclination in Rice (Oryza sativa L.). Plants. 2023; 12(2):409. https://doi.org/10.3390/plants12020409
Chicago/Turabian StyleZhang, Yanjun, Shaqila Han, Yuqing Lin, Jiyue Qiao, Naren Han, Yanyan Li, Yaning Feng, Dongming Li, and Yanhua Qi. 2023. "Auxin Transporter OsPIN1b, a Novel Regulator of Leaf Inclination in Rice (Oryza sativa L.)" Plants 12, no. 2: 409. https://doi.org/10.3390/plants12020409
APA StyleZhang, Y., Han, S., Lin, Y., Qiao, J., Han, N., Li, Y., Feng, Y., Li, D., & Qi, Y. (2023). Auxin Transporter OsPIN1b, a Novel Regulator of Leaf Inclination in Rice (Oryza sativa L.). Plants, 12(2), 409. https://doi.org/10.3390/plants12020409






