Variation in Chloroplast Genome Size: Biological Phenomena and Technological Artifacts
Abstract
1. Introduction
2. Results
2.1. Data Acquisition
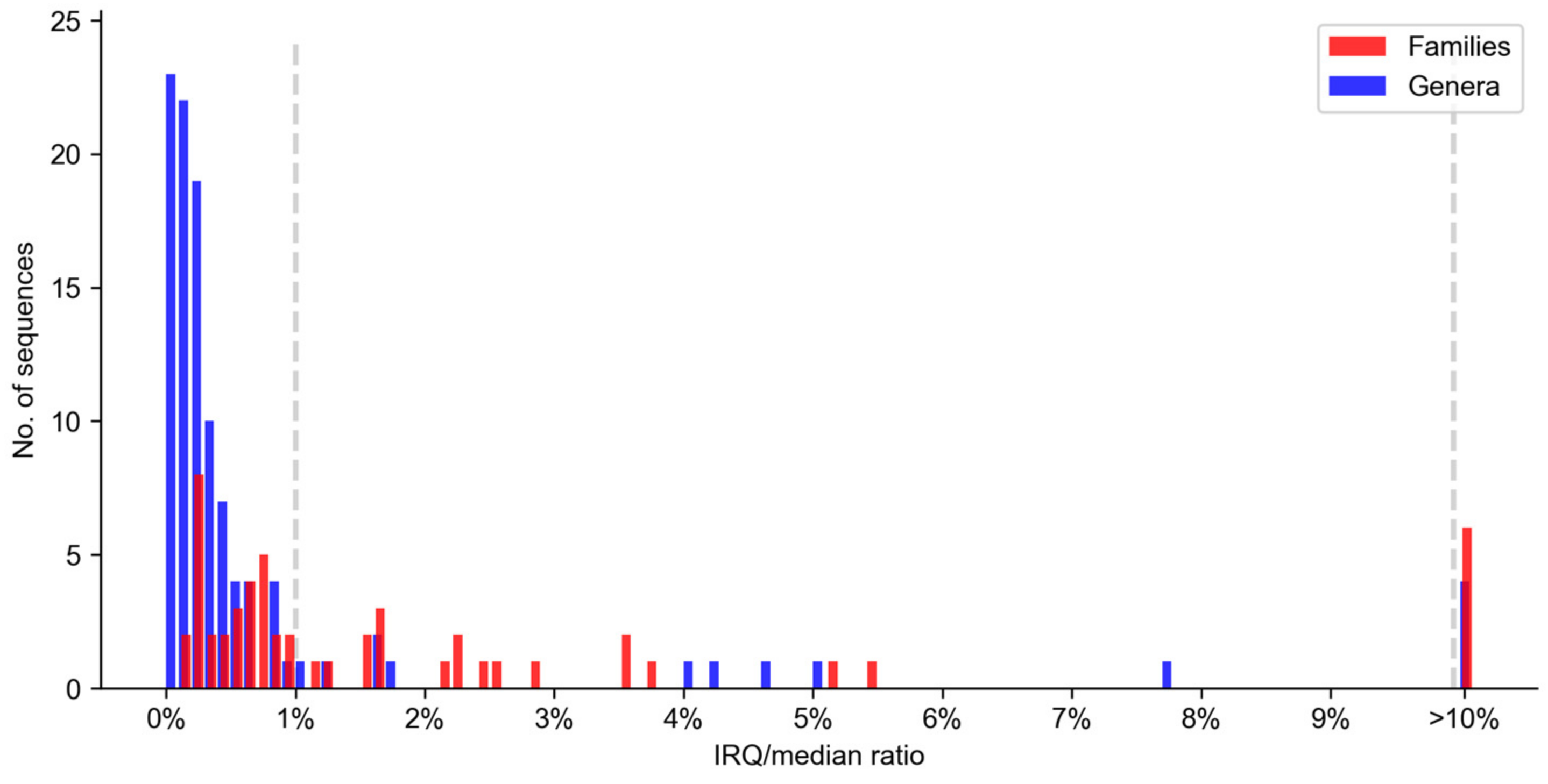
2.2. Distribution Assessment
2.3. Examination of Wide Distributions
2.3.1. Family Level
2.3.2. Genus Level
2.4. Outlier Detection at the Genus Level
3. Discussion
4. Materials and Methods
4.1. Data Acquisition
4.2. Bioinformatics Pipeline
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Jansen, R.K.; Ruhlman, T.A. Plastid Genomes of Seed Plants. Photosynthesis 2012, 35, 103–126. [Google Scholar]
- Deng, X.-W.; Wing, R.A.; Gruissem, W. The Chloroplast Genome Exists in Multimeric Forms. Proc. Natl. Acad. Sci. USA 1989, 86, 4156–4160. [Google Scholar] [CrossRef] [PubMed]
- Bendich, A.J.; Smith, S.B. Moving Pictures and Pulsed-Field Gel Electrophoresis Show Linear DNA Molecules from Chloroplasts and Mitochondria. Curr. Genet. 1990, 17, 421–425. [Google Scholar] [CrossRef]
- Bendich, A.J. Circular Chloroplast Chromosomes: The Grand Illusion. Plant Cell 2004, 16, 1661–1666. [Google Scholar] [CrossRef] [PubMed]
- Ohyama, K.; Fukuzawa, H.; Kohchi, T.; Shirai, H.; Sano, T.; Sano, S.; Umesono, K.; Shiki, Y.; Takeuchi, M.; Chang, Z.; et al. Chloroplast Gene Organization Deduced from Complete Sequence of Liverwort Marchantia Polymorpha Chloroplast DNA. Nature 1986, 322, 572–574. [Google Scholar] [CrossRef]
- Shinozaki, K.; Ohme, M.; Tanaka, M.; Wakasugi, T.; Hayashida, N.; Matsubayashi, T.; Zaita, N.; Chunwongse, J.; Obokata, J.; Yamaguchi-Shinozaki, K.; et al. The Complete Nucleotide Sequence of the Tobacco Chloroplast Genome: Its Gene Organization and Expression. EMBO J. 1986, 5, 2043–2049. [Google Scholar] [CrossRef]
- Gielly, L.; Taberlet, P. The Use of Chloroplast DNA to Resolve Plant Phylogenies: Noncoding versus RbcL Sequences. Mol. Biol. Evol. 1994, 11, 769–777. [Google Scholar] [CrossRef]
- Ruhlman, T.A.; Jansen, R.K. The Plastid Genomes of Flowering Plants. Methods Mol. Biol. 2014, 1132, 3–38. [Google Scholar] [CrossRef]
- Bellot, S.; Renner, S.S. The Plastomes of Two Species in the Endoparasite Genus Pilostyles (Apodanthaceae) Each Retain Just Five or Six Possibly Functional Genes. Genome Biol. Evol. 2015, 8, 189–201. [Google Scholar] [CrossRef]
- Roquet, C.; Coissac, É.; Cruaud, C.; Boleda, M.; Boyer, F.; Alberti, A.; Gielly, L.; Taberlet, P.; Thuiller, W.; Van Es, J.; et al. Understanding the Evolution of Holoparasitic Plants: The Complete Plastid Genome of the Holoparasite Cytinus hypocistis (Cytinaceae). Ann. Bot. 2016, 118, 885–896. [Google Scholar] [CrossRef]
- Wicke, S.; Schneeweiss, G.M.; de Pamphilis, C.W.; Müller, K.F.; Quandt, D. The Evolution of the Plastid Chromosome in Land Plants: Gene Content, Gene Order, Gene Function. Plant Mol. Biol. 2011, 76, 273–297. [Google Scholar] [CrossRef]
- Downie, S.R.; Palmer, J.D. Restriction Site Mapping of the Chloroplast DNA Inverted Repeat: A Molecular Phylogeny of the Asteridae. Ann. Mo. Bot. Gard. 1992, 79, 266. [Google Scholar] [CrossRef][Green Version]
- Goulding, S.E.; Olmstead, R.G.; Morden, C.W.; Wolfe, K.H. Ebb and Flow of the Chloroplast Inverted Repeat. Mol. Gen. Genet. 1996, 252, 195–206. [Google Scholar] [CrossRef]
- Plunkett, G.M.; Downie, S.R. Expansion and Contraction of the Chloroplast Inverted Repeat in Apiaceae subfamily Apioideae. Syst. Bot. 2000, 25, 648. [Google Scholar] [CrossRef]
- Guisinger, M.M.; Kuehl, J.V.; Boore, J.L.; Jansen, R.K. Extreme Reconfiguration of Plastid Genomes in the Angiosperm Family Geraniaceae: Rearrangements, Repeats, and Codon Usage. Mol. Biol. Evol. 2011, 28, 583–600. [Google Scholar] [CrossRef]
- Xiao-Ming, Z.; Junrui, W.; Li, F.; Sha, L.; Hongbo, P.; Lan, Q.; Jing, L.; Yan, S.; Weihua, Q.; Lifang, Z.; et al. Inferring the Evolutionary Mechanism of the Chloroplast Genome Size by Comparing Whole-Chloroplast Genome Sequences in Seed Plants. Sci. Rep. 2017, 7, 1555. [Google Scholar] [CrossRef]
- Bendich, A.J. Why Do Chloroplasts and Mitochondria Contain so Many Copies of Their Genome? BioEssays 1987, 6, 279–282. [Google Scholar] [CrossRef]
- Heinhorst, S.; Cannon, G.C. DNA Replication in Chloroplasts. J. Cell Sci. 1993, 104, 1–9. [Google Scholar] [CrossRef]
- Marks, R.A.; Hotaling, S.; Frandsen, P.B.; VanBuren, R. Representation and Participation across 20 Years of Plant Genome Sequencing. Nat. Plants 2021, 7, 1571–1578. [Google Scholar] [CrossRef]
- Jin, J.J.; Yu, W.B.; Yang, J.B.; Song, Y.; Depamphilis, C.W.; Yi, T.S.; Li, D.Z. Get Organelle: A Fast and Versatile Toolkit for Accurate de Novo Assembly of Organelle Genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef]
- McKain, M.R.; Wilson, M. Fast-Plast: Rapid de Novo Assembly and Finishing for Whole Chloroplast Genomes. Available online: https://github.com/mrmckain/Fast-Plast (accessed on 22 October 2022).
- Dierckxsens, N.; Mardulyn, P.; Smits, G. NOVOPlasty: De Novo Assembly of Organelle Genomes from Whole Genome Data. Nucleic Acids Res. 2017, 45, e18. [Google Scholar] [CrossRef]
- Zhong, X. Assembly, Annotation and Analysis of Chloroplast Genomes. 2020. Available online: https://research-repository.uwa.edu.au/en/publications/assembly-annotation-and-analysis-of-chloroplast-genomes (accessed on 22 October 2022).
- Zheng, S.; Poczai, P.; Hyvönen, J.; Tang, J.; Amiryousefi, A. Chloroplot: An Online Program for the Versatile Plotting of Organelle Genomes. Front. Genet. 2020, 11, 576124. [Google Scholar] [CrossRef]
- Tillich, M.; Lehwark, P.; Pellizzer, T.; Ulbricht-Jones, E.S.; Fischer, A.; Bock, R.; Greiner, S. GeSeq—Versatile and Accurate Annotation of Organelle Genomes. Nucleic Acids Res. 2017, 45, W6–W11. [Google Scholar] [CrossRef] [PubMed]
- Qu, X.J.; Moore, M.J.; Li, D.Z.; Yi, T.S. PGA: A Software Package for Rapid, Accurate, and Flexible Batch Annotation of Plastomes. Plant Methods 2019, 15, 50. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.I.; Cronk, Q.C.B. Plann: A Command-Line Application for Annotating Plastome Sequences. Appl. Plant Sci. 2015, 3, 1500026. [Google Scholar] [CrossRef]
- Turudić, A.; Liber, Z.; Grdiša, M.; Jakše, J.; Varga, F.; Šatović, Z. Chloroplast Genome Annotation Tools: Prolegomena to the Identification of Inverted Repeats. Int. J. Mol. Sci 2022, 2022, 10804. [Google Scholar] [CrossRef] [PubMed]
- Fajardo, D.; Senalik, D.; Ames, M.; Zhu, H.; Steffan, S.A.; Harbut, R.; Polashock, J.; Vorsa, N.; Gillespie, E.; Kron, K.; et al. Complete Plastid Genome Sequence of Vaccinium Macrocarpon: Structure, Gene Content, and Rearrangements Revealed by next Generation Sequencing. Tree Genet. Genomes 2013, 9, 489–498. [Google Scholar] [CrossRef]
- Martínez-Alberola, F.; Del Campo, E.M.; Lázaro-Gimeno, D.; Mezquita-Claramonte, S.; Molins, A.; Mateu-Andrés, I.; Pedrola-Monfort, J.; Casano, L.M.; Barreno, E. Balanced Gene Losses, Duplications and Intensive Rearrangements Led to an Unusual Regularly Sized Genome in Arbutus Unedo Chloroplasts. PLoS ONE 2013, 8, e79685. [Google Scholar] [CrossRef]
- Wojciechowski, M.F.; Sanderson, M.J.; Hu, J.M. Evidence on the Monophyly of Astragalus (Fabaceae) and Its Major Subgroups Based on Nuclear Ribosomal DNA ITS and Chloroplast DNA TrnL Intron Data. Syst. Bot. 1999, 24, 409. [Google Scholar] [CrossRef]
- Wojciechowski, M.F.; Sanderson, M.J.; Steele, K.P.; Liston, A. Molecular Phylogeny of the “Temperate Herbaceous Tribes” of Papilionoid Legumes: A Supertree Approach. Adv. Legume Syst. 2000, 9, 277–298. [Google Scholar]
- Blazier, C.C.; Guisinger, M.M.; Jansen, R.K. Recent Loss of Plastid-Encoded Ndh Genes within Erodium (Geraniaceae). Plant Mol. Biol. 2011, 76, 263–272. [Google Scholar] [CrossRef]
- Stefanović, S.; Olmstead, R.G. Testing the Phylogenetic Position of a Parasitic Plant (Cuscuta, Convolvulaceae, Asteridae): Bayesian Inference and the Parametric Bootstrap on Data Drawn from Three Genomes. Syst. Biol. 2004, 53, 384–399. [Google Scholar] [CrossRef]
- Park, I.; Song, J.H.; Yang, S.; Kim, W.J.; Choi, G.; Moon, B.C. Cuscuta Species Identification Based on the Morphology of Reproductive Organs and Complete Chloroplast Genome Sequences. Int. J. Mol. Sci. 2019, 20, 2726. [Google Scholar] [CrossRef]
- Li, X.; Zhang, T.C.; Qiao, Q.; Ren, Z.; Zhao, J.; Yonezawa, T.; Hasegawa, M.; Crabbe, M.J.C.; Li, J.; Zhong, Y. Complete Chloroplast Genome Sequence of Holoparasite Cistanche deserticola (Orobanchaceae) Reveals Gene Loss and Horizontal Gene Transfer from Its Host Haloxylon ammodendron (Chenopodiaceae). PLoS ONE 2013, 8, e58747. [Google Scholar] [CrossRef]
- Rabah, S.O.; Shrestha, B.; Hajrah, N.H.; Sabir, M.J.; Alharby, H.F.; Sabir, M.J.; Alhebshi, A.M.; Sabir, J.S.M.; Gilbert, L.E.; Ruhlman, T.A.; et al. Passiflora Plastome Sequencing Reveals Widespread Genomic Rearrangements. J. Syst. Evol. 2019, 57, 1–14. [Google Scholar] [CrossRef]
- Cauz-Santos, L.A.; da Costa, Z.P.; Callot, C.; Cauet, S.; Zucchi, M.I.; Bergès, H.; van den Berg, C.; Vieira, M.L.C. A Repertory of Rearrangements and the Loss of an Inverted Repeat Region in Passiflora Chloroplast Genomes. Genome Biol. Evol. 2020, 12, 1841–1857. [Google Scholar] [CrossRef]
- Chumley, T.W.; Palmer, J.D.; Mower, J.P.; Fourcade, H.M.; Calie, P.J.; Boore, J.L.; Jansen, R.K. The Complete Chloroplast Genome Sequence of Pelargonium × Hortorum: Organization and Evolution of the Largest and Most Highly Rearranged Chloroplast Genome of Land Plants. Mol. Biol. Evol. 2006, 23, 2175–2190. [Google Scholar] [CrossRef]
- Röschenbleck, J.; Wicke, S.; Weinl, S.; Kudla, J.; Müller, K.F. Genus-Wide Screening Reveals Four Distinct Types of Structural Plastid Genome Organization in Pelargonium (Geraniaceae). Genome Biol. Evol. 2017, 9, 64–76. [Google Scholar] [CrossRef][Green Version]
- Weng, M.L.; Ruhlman, T.A.; Jansen, R.K. Expansion of Inverted Repeat Does not Decrease Substitution Rates in Pelargonium Plastid Genomes. New Phytol. 2017, 214, 842–851. [Google Scholar] [CrossRef]
- Favre, A.; Pringle, J.S.; Heckenhauer, J.; Kozuharova, E.; Gao, Q.; Lemmon, E.M.; Lemmon, A.R.; Sun, H.; Tkach, N.; Gebauer, S.; et al. Phylogenetic Relationships and Sectional Delineation within Gentiana (Gentianaceae). Taxon 2020, 69, 1221–1238. [Google Scholar] [CrossRef]
- Spalik, K.; Reduron, J.P.; Downie, S.R. The Phylogenetic Position of Peucedanum sensu Lato and Allied Genera and Their Placement in Tribe Selineae (Apiaceae, Subfamily apioideae). Plant Syst. Evol. 2004, 243, 189–210. [Google Scholar] [CrossRef]
- Lohmann, L.G. Untangling the Phylogeny of Neotropical Lianas (Bignonieae, Bignoniaceae). Am. J. Bot. 2006, 93, 304–318. [Google Scholar] [CrossRef] [PubMed]
- Mast, A.R.; Kelso, S.; Richards, A.J.; Lang, D.J.; Feller, D.M.S.; Conti, E. Phylogenetic Relationships in Primula L. and Related Genera (Primulaceae) Based on Noncoding Chloroplast DNA. Int. J. Plant Sci. 2001, 162, 1381–1400. [Google Scholar] [CrossRef]
- Trift, I.; Källersjö, M.; Anderberg, A.A. The Monophyly of Primula (Primulaceae) Evaluated by Analysis of Sequences from the Chloroplast Gene RbcL. Syst. Bot. 2002, 27, 396–407. [Google Scholar]
- Schmidt-Lebuhn, A.N.; de Vos, J.M.; Keller, B.; Conti, E. Phylogenetic Analysis of Primula Section Primula Reveals Rampant Non-Monophyly among Morphologically Distinct Species. Mol. Phylogenet. Evol. 2012, 65, 23–34. [Google Scholar] [CrossRef]
- Steinmann, V.W. The Submersion of Pedilanthus into Euphorbia (Euphorbiaceae). Acta Bot. Mex. 2003, 65, 45–50. [Google Scholar] [CrossRef][Green Version]
- Yang, Y.; Riina, R.; Morawetz, J.J.; Haevermans, T.; Aubriot, X.; Berry, P.E. Molecular Phylogenetics and Classification of Euphorbia Subgenus Chamaesyce (Euphorbiaceae). Taxon 2012, 61, 764–789. [Google Scholar] [CrossRef]
- Lammers, T.G. Circumscription and Phylogeny of the Campanulales. Ann. Mo. Bot. Gard. 1992, 79, 388. [Google Scholar] [CrossRef]
- Lanfear, R.; Kokko, H.; Eyre-Walker, A. Population Size and the Rate of Evolution. Trends Ecol. Evol. 2014, 29, 33–41. [Google Scholar] [CrossRef]
- Mehl, T.; Gruenstaeudl, M. Airpg: Automatically Accessing the Inverted Repeats of Archived Plastid Genomes. BMC Bioinform. 2021, 22, 413. [Google Scholar] [CrossRef]
- Turudić, A.; Liber, Z.; Grdiša, M.; Jakše, J.; Varga, F.; Šatović, Z. Towards the Well-Tempered Chloroplast DNA Sequences. Plants 2021, 10, 1360. [Google Scholar] [CrossRef]
- David, C.H. Tukey Exploratory Data Analysis by John W. Tukey. Biometrics 1977, 33, 311–318. [Google Scholar] [CrossRef]
- Cock, P.J.A.; Antao, T.; Chang, J.T.; Chapman, B.A.; Cox, C.J.; Dalke, A.; Friedberg, I.; Hamelryck, T.; Kauff, F.; Wilczynski, B.; et al. Biopython: Freely Available Python Tools for Computational Molecular Biology and Bioinformatics. Bioinformatics 2009, 25, 1422–1423. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Serra, F.; Bork, P. ETE 3: Reconstruction, Analysis, and Visualization of Phylogenomic Data. Mol. Biol. Evol. 2016, 33, 1635–1638. [Google Scholar] [CrossRef]
- Hunter, J.D. Matplotlib: A 2D Graphics Environment. Comput. Sci. Eng. 2007, 9, 90–95. [Google Scholar] [CrossRef]


| Parameter | Family Level | Genus Level | ||||
|---|---|---|---|---|---|---|
| Asterids | Rosids | Total | Asterids | Rosids | Total | |
| Total number of taxa | 68 | 89 | 157 | 690 | 905 | 1595 |
| No. of taxa analyzed | 26 | 28 | 54 | 49 | 59 | 108 |
| No. sequences in analyses | 2337 | 2739 | 5076 | 1285 | 1410 | 2695 |
| Minimum no. of sequences | 21 | 21 | 21 | 10 | 10 | 10 |
| Balsaminaceae | Ulmaceae | Rhododendron | Glycine | |||
| Maximum no. of sequences | 543 | 480 | 543 | 178 | 78 | 178 |
| Asteraceae | Fabaceae | Solanum | Acer | |||
| Examination of wide distributions | ||||||
| No. and % of sequences IQR/median > 1% | 11 | 13 | 24 | 8 | 6 | 14 |
| (42.31%) | (46.43%) | (44.44%) | (16.33%) | (10.17%) | (12.96%) | |
| No. and % of sequences IQR/median > 10% | 3 | 3 | 6 | 1 | 3 | 4 |
| (11.54%) | (10.71%) | (11.11%) | (2.04%) | (5.08%) | (3.70%) | |
| Outlier detection | ||||||
| No. of distributions with outliers | 18 | 22 | 40 | 31 | 38 | 69 |
| (69.23%) | (78.57%) | (74.07%) | (63.27%) | (64.41%) | (63.89%) | |
| Total number of outliers | 188 | 129 | 317 | 103 | 97 | 200 |
| (8.04%) | (4.71%) | (6.25%) | (8.02%) | (6.88%) | (7.42%) | |
| Family | No. of Genera | No. of Sequences | IQR/Median Ratio | Genera Showing Wide Distributions |
|---|---|---|---|---|
| Ericaceae a | 9 | 22 | 73.54% | Rhododendron |
| Convolvulaceae a | 11 | 52 | 43.30% | Cuscuta |
| Orobanchaceae a | 22 | 49 | 39.07% | - |
| Fabaceae r | 223 | 480 | 18.53% | Medicago, Lathyrus |
| Geraniaceae r | 5 | 42 | 12.95% | Pelargonium, Erodium |
| Passifloraceae r | 4 | 53 | 10.60% | Passiflora |
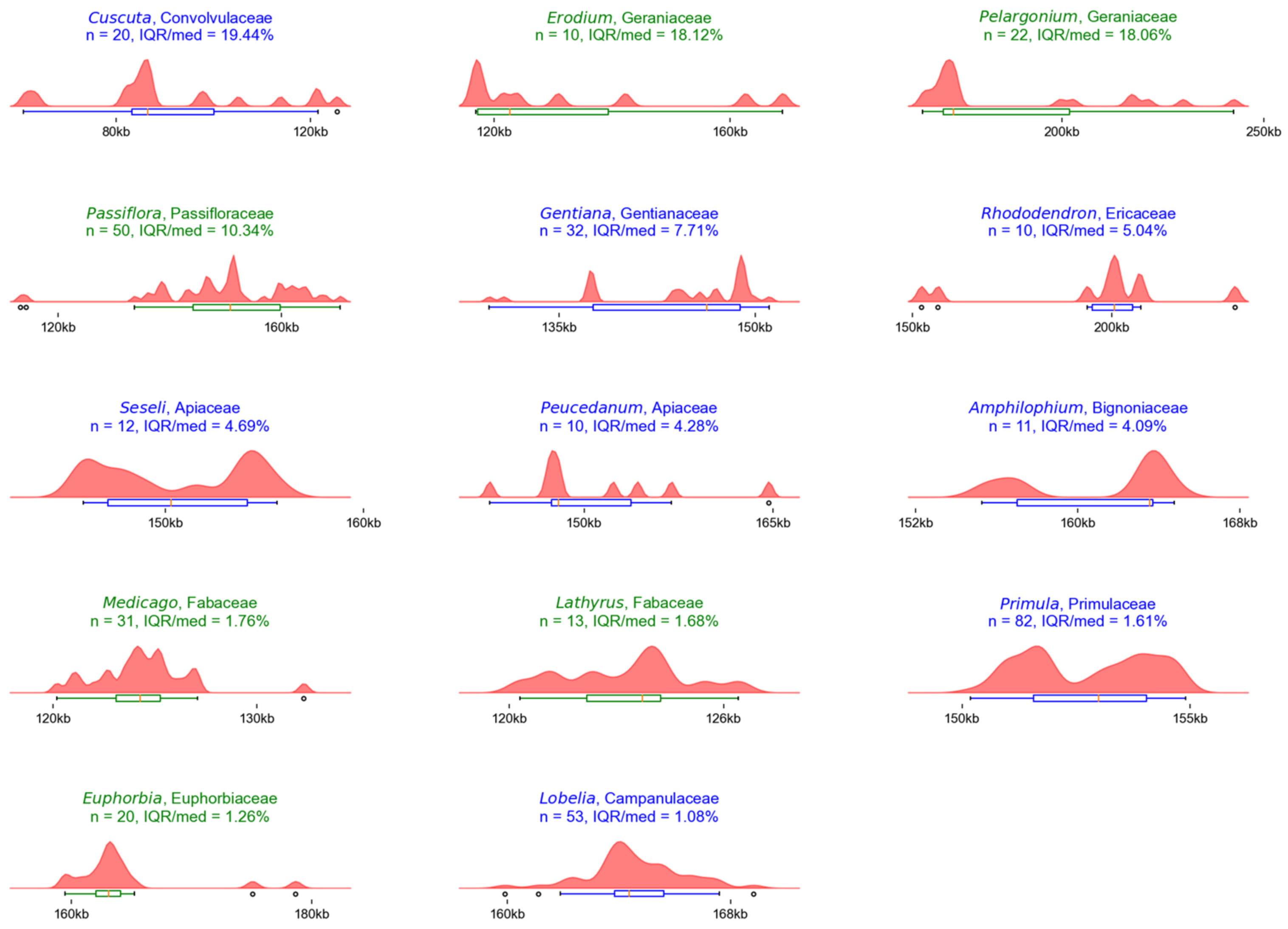
| Genus | Family | No. of Sequences | IQR/Median Ratio | Reported Factor |
|---|---|---|---|---|
| Cuscuta | Convolvulaceae a | 20 | 19.44% | Parasitic life form [34,35] |
| Erodium | Geraniaceae r | 10 | 18.12% | IR loss [15,33] |
| Pelargonium | Geraniaceae r | 22 | 18.06% | IR expansions [15,39,40,41] |
| Passiflora | Passifloraceae r | 50 | 10.34% | IR expansions and contractions [37,38] |
| Gentiana | Gentianaceae a | 32 | 7.71% | Polyphyly [42] |
| Rhododendron | Ericaceae a | 10 | 5.04% | IR loss [29,30] |
| Seseli | Apiaceae a | 12 | 4.96% | Polyphyly [43] |
| Peucedanum | Apiaceae a | 10 | 4.28% | Polyphyly [43] |
| Amphilophium | Bignoniaceae a | 11 | 4.09% | Polyphyly [44] |
| Medicago | Fabaceae r | 31 | 1.76% | IR loss [31,32] |
| Lathyrus | Fabaceae r | 13 | 1.68% | IR loss [31,32] |
| Primula | Primulaceae a | 82 | 1.61% | Polyphyly [45,46,47] |
| Euphorbia | Euphorbiaceae r | 20 | 1.26% | Polyphyly [48,49] |
| Lobelia | Campanulaceae a | 53 | 1.08% | Polyphyly [50] |
| Species | Genus Median | RefSeq Sequence | Alternative Sequence | ||||
|---|---|---|---|---|---|---|---|
| Accession | Length | Publication Date | Accession | Length | Publication Date | ||
| Angelica sinensis | 146,962 | NC_042826 | 142,485 | 25 June 2019 | MW820164 | 146,952 | 5 September 2021 |
| Ficus auriculata | 160,363 | NC_053837 | 162,558 | 26 March 2021 | MZ662866 | 160,361 | 31 August 2022 |
| Fragaria mandshurica | 155,621 | NC_018767 | 129,805 | 14 October 2012 | MW537846 | 155,640 | 30 March 2022 |
| Fragaria vesca | 155,621 | NC_018766 | 129,788 | 14 October 2012 | KC507757 | 155,620 | 26 July 2016 |
| Vitis romanetii | 160,971 | NC_056348 | 232,020 | 20 June 2021 | MW592524 | 160,976 | 16 March 2022 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Turudić, A.; Liber, Z.; Grdiša, M.; Jakše, J.; Varga, F.; Šatović, Z. Variation in Chloroplast Genome Size: Biological Phenomena and Technological Artifacts. Plants 2023, 12, 254. https://doi.org/10.3390/plants12020254
Turudić A, Liber Z, Grdiša M, Jakše J, Varga F, Šatović Z. Variation in Chloroplast Genome Size: Biological Phenomena and Technological Artifacts. Plants. 2023; 12(2):254. https://doi.org/10.3390/plants12020254
Chicago/Turabian StyleTurudić, Ante, Zlatko Liber, Martina Grdiša, Jernej Jakše, Filip Varga, and Zlatko Šatović. 2023. "Variation in Chloroplast Genome Size: Biological Phenomena and Technological Artifacts" Plants 12, no. 2: 254. https://doi.org/10.3390/plants12020254
APA StyleTurudić, A., Liber, Z., Grdiša, M., Jakše, J., Varga, F., & Šatović, Z. (2023). Variation in Chloroplast Genome Size: Biological Phenomena and Technological Artifacts. Plants, 12(2), 254. https://doi.org/10.3390/plants12020254








