Association Mapping of Amylose Content in Maize RIL Population Using SSR and SNP Markers
Abstract
1. Introduction
2. Results
2.1. Phenotypic Analysis for Amylose Content in RIL Population
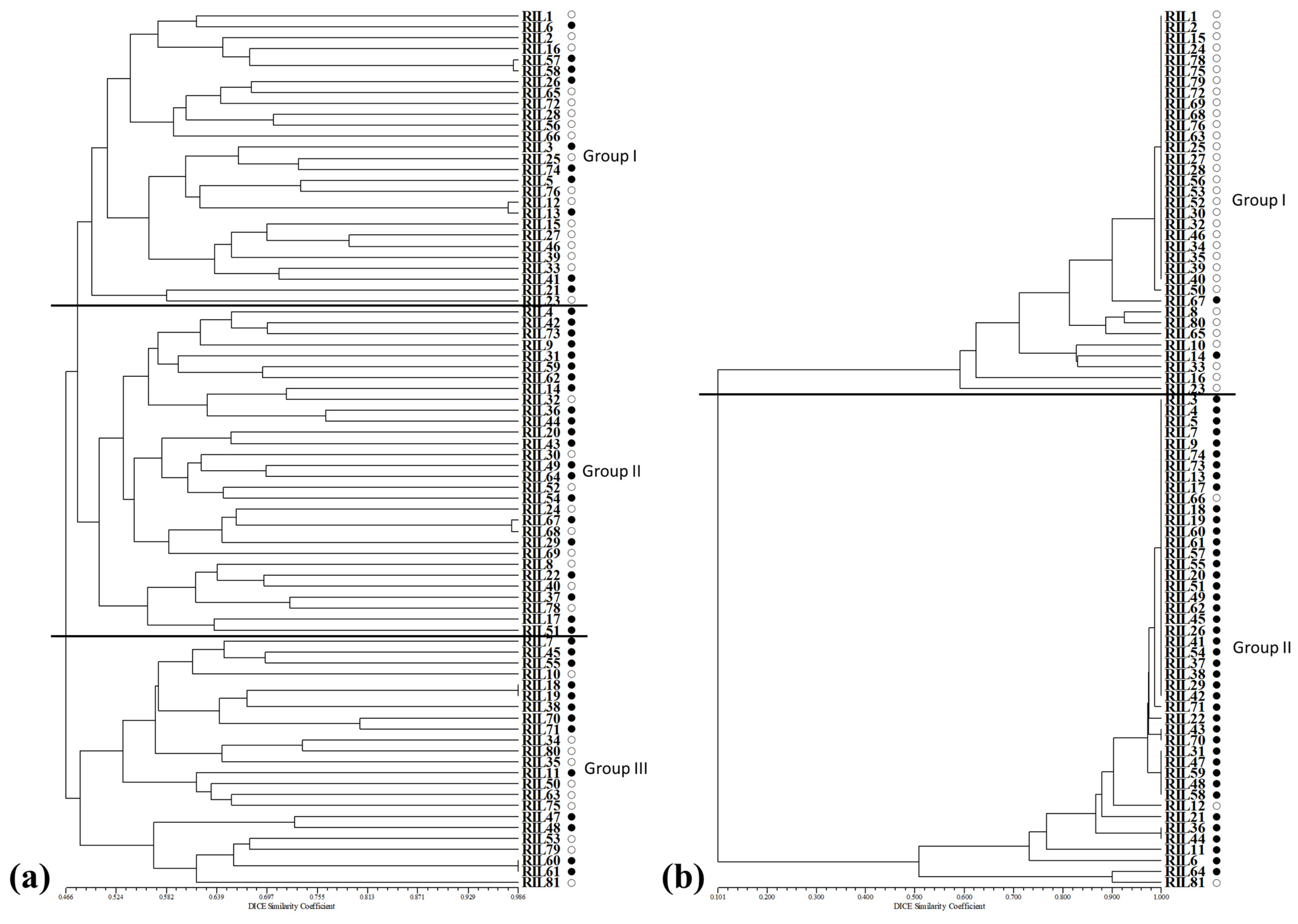
2.2. Cluster Analysis and Population Structure
2.3. Marker–Trait Association for Amylose Content
3. Discussion
4. Materials and Methods
4.1. Plant Material and Amylose Content Evaluation
4.2. SSR and SNP Genotype in RILs
4.3. Statistical Data Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nelson, O.; Pan, D. Starch synthesis in maize endosperms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1995, 46, 475–496. [Google Scholar] [CrossRef]
- Paraginskia, R.T.; Vanier, N.L.; Moomand, K.; de Oliveira, M.; Zavareze, E.R.; Silva, R.M.; Ferreira, C.D.; Elias, M.C. Characteristics of starch isolated from maize as a function of grain storage temperature. Carbohydr. Polym. 2014, 102, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Lin, F.; Zhou, L.; He, B.; Zhang, X.; Dai, H.; Qian, Y.; Ruan, L.; Zhao, H. QTL mapping for maize starch content and candidate gene prediction combined with co-expression network analysis. Theor. Appl. Genet. 2019, 132, 1931–1941. [Google Scholar] [CrossRef]
- Wang, T.; Wang, M.; Hu, S.; Xiao, Y.; Tong, H.; Pan, Q.; Xue, J.; Yan, J.; Li, J.; Yang, X. Genetic basis of maize kernel starch content revealed by high-density single nucleotide polymorphism markers in a recombinant inbred line population. BMC Plant Biol. 2015, 15, 288. [Google Scholar] [CrossRef]
- Li, C.; Huang, Y.; Huang, R.; Wu, Y.; Wang, W. The genetic architecture of amylose biosynthesis in maize kernel. Plant Biotechnol. J. 2018, 16, 688–695. [Google Scholar] [CrossRef] [PubMed]
- Whitt, S.R.; Wilson, L.M.; Tenaillon, M.I.; Gaut, B.S.; Buckler, E.S. Genetic diversity and selection in the maize starch pathway. Proc. Natl. Acad. Sci. USA 2002, 99, 12959–12962. [Google Scholar] [CrossRef] [PubMed]
- Mercier, C. The fine structure of corn starches of various amylose-percentage: Waxy, normal and amylomaize. Starch/Stärke 1973, 25, 78–83. [Google Scholar] [CrossRef]
- Devi, E.L.; Hossain, F.; Muthusamy, V.; Chhabra, R.; Zunjare, R.U.; Baveja, A.; Jaiswal, S.K.; Goswami, R.; Dosad, S. Microsatellite marker-based characterization of waxy maize inbreds for their utilization in hybrid breeding. 3 Biotech. 2017, 7, 316. [Google Scholar] [CrossRef] [PubMed]
- Edy; Numba, S.; Ibrahim, B. Increased amylopectin content potential in corn grains of quality protein maize (QPM). IOP Conf. Ser. Earth Environ. Sci. 2019, 334, 012011. [Google Scholar] [CrossRef]
- Jeon, J.S.; Ryoo, N.; Hahn, T.R.; Walia, H.; Nakamura, Y. Starch biosynthesis in cereal endosperm. Plant Physiol. Biochem. 2010, 48, 383–392. [Google Scholar] [CrossRef]
- Lu, D.; Lu, W. Effects of protein removal on the physico-chemical properties of waxy maize flours. Starch/Stärke 2012, 64, 874–881. [Google Scholar] [CrossRef]
- Zhang, W.; Yang, W.; Wang, M.; Wang, W.; Zeng, G.; Chen, Z.; Cai, Y. Increasing lysine content of waxy maize through introgression of opaque-2 and opaque-16 genes using molecular assisted and biochemical development. PLoS ONE 2013, 8, e56227. [Google Scholar] [CrossRef]
- Qi, X.; Dong, L.; Liu, C.; Mao, L.; Liu, F.; Zhang, X.; Cheng, B.; Xie, C. Systematic identification of endogenous RNA polymerase III promoters for efficient RNA guide-based genome editing technologies in maize. Crop J. 2018, 6, 314–320. [Google Scholar] [CrossRef]
- Shure, M.; Wessler, S.; Fedoroff, N. Molecular identification and isolation of the Waxy locus in maize. Cell 1983, 35, 225–233. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Wang, M.; Wang, W.; Yang, W. Marker-assisted selection for pyramiding the waxy and opaque-16 genes in maize using crosses and backcross schemes. Mol. Breed. 2013, 31, 767–775. [Google Scholar] [CrossRef]
- Dong, L.; Qi, X.; Zhu, J.; Liu, C.; Zhang, X.; Cheng, B.; Mao, L.; Xie, C. Supersweet and waxy: Meeting the diverse demands for specialty maize by genome editing. Plant Biotechnol. J. 2019, 17, 1853–1855. [Google Scholar] [CrossRef]
- Huang, B.Q.; Tian, M.L.; Zhang, J.J.; Huang, Y.B. Waxy locus and its mutant types in maize Zea mays L. J. Integr. Agric. 2010, 9, 1–10. [Google Scholar] [CrossRef]
- Pasam, R.K.; Sharma, R.; Malosetti, M.; van Eeuwijk, F.A.; Haseneyer, G.; Kilian, B.; Graner, A. Genome-wide association studies for agronomical traits in a worldwide spring barley collection. BMC Plant Biol. 2012, 12, 16. [Google Scholar] [CrossRef]
- Duvick, D.N.; Smith, J.S.C.; Cooper, R.M. Long-term selection in a commercial hybrid maize breeding program. Plant Breed. Rev. 2004, 24, 109–151. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, Y.; Wang, Y.; Peng, B.; Liu, C.; Liu, Z.; Tan, W.; Wang, D.; Shi, Y.; Sun, B.; et al. Correlations and QTL detection in maize family per se and testcross progenies for plant height and ear height. Plant Breed. 2011, 130, 617–624. [Google Scholar] [CrossRef]
- Yu, J.; Arbelbide, M.; Bernardo, R. Power of in silico QTL mapping from phenotypic, pedigree, and marker data in a hybrid breeding program. Theor. Appl. Genet. 2005, 10, 1061–1067. [Google Scholar] [CrossRef]
- Skøt, L.; Humphreys, M.O.; Armstead, I.; Heywood, S.; Skøt, K.P.; Sanderson, R.; Thomas, I.D.; Chorlton, K.H.; Hamilton, N.R.S. An association mapping approach to identify flowering time genes in natural populations of Lolium perenne (L.). Mol. Breed. 2005, 15, 233–245. [Google Scholar] [CrossRef]
- Flint-Garcia, S.A.; Thuillet, A.C.; Yu, J.M.; Pressoir, G.; Romero, S.M.; Mitchell, S.E.; Doebley, J.; Kresovich, S.; Goodman, M.M.; Buckler, E.S. Maize association population: A high-resolution platform for quantitative trait locus dissection. Plant J. 2005, 44, 1054–1064. [Google Scholar] [CrossRef]
- Yu, J.; Buckler, E.S. Genetic association mapping and genome organization of maize. Curr. Opin. Biotechnol. 2006, 17, 155–160. [Google Scholar] [CrossRef]
- Dong, Y.B.; Zhang, Z.W.; Shi, Q.L.; Wang, Q.L.; Zhou, Q.; Li, Y.L. QTL identification and meta-analysis for kernel composition traits across three generations in popcorn. Euphytica 2015, 204, 649–660. [Google Scholar] [CrossRef]
- Goldman, I.L.; Rocheford, T.R.; Dudley, J.W. Quantitative trait loci influencing protein and starch concentration in the Illinois long term selection maize strains. Theor. Appl. Genet. 1993, 87, 217–224. [Google Scholar] [CrossRef]
- Guo, Y.Q.; Yang, X.H.; Chander, S.; Yan, J.B.; Zhang, J.; Song, T.M.; Li, J.S. Identification of unconditional and conditional QTL for oil, protein and starch content in maize. Crop J. 2013, 1, 34–42. [Google Scholar] [CrossRef]
- Liu, Y.Y.; Dong, Y.B.; Niu, S.Z.; Cui, D.C.; Wang, Y.Z.; Wei, M.G.; Li, X.H.; Fu, J.F.; Zhang, Z.W.; Chen, H.Q.; et al. QTL identification of kernel composition traits with popcorn using both F2:3 and BC2F2 populations developed from the same cross. J. Cereal Sci. 2008, 48, 625–631. [Google Scholar] [CrossRef]
- Séne, M.; Causse, M.; Damerval, C.; Thévenot, C.; Prioul, J.L. Quantitative trait loci affecting amylose, amylopectin and starch content in maize recombinant inbred lines. Plant Physiol. Biochem. 2000, 38, 459–472. [Google Scholar] [CrossRef]
- Wang, Y.Z.; Li, J.Z.; Li, Y.L.; Wei, M.G.; Li, X.H.; Fu, J.F. QTL detection for grain oil and starch content and their associations in two connected F2:3 populations in high-oil maize. Euphytica 2010, 174, 239–252. [Google Scholar] [CrossRef]
- Wassom, J.J.; Wong, J.C.; Martinez, E.; King, J.J.; DeBaene, J.; Hotchkiss, J.R.; Mikkilineni, V.; Bohn, M.O.; Rocheford, T.R. QTL associated with maize kernel oil, protein, and starch concentrations; kernel mass; and grain yield in Illinois high oil × B73 backcross-derived lines. Crop Sci. 2008, 48, 243–252. [Google Scholar] [CrossRef]
- Yang, G.H.; Dong, Y.B.; Li, Y.L.; Wang, Q.L.; Shi, Q.L.; Zhou, Q. Verification of QTL for grain starch content and its genetic correlation with oil content using two connected RIL populations in high-oil maize. PLoS ONE 2013, 8, e53770. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhang, J.; Lu, X.Q.; Song, X.F.; Yan, J.B.; Song, T.M.; Dai, J.R.; Rocheford, T.; Li, J.S. Mapping quantitative trait loci for oil, starch, and protein concentrations in grain with high-oil maize by SSR markers. Euphytica 2008, 162, 335–344. [Google Scholar] [CrossRef]
- Salvi, S.; Tuberosa, R. To clone or not to clone plant QTLs: Present and future challenges. Trends Plant Sci. 2005, 10, 297–304. [Google Scholar] [CrossRef]
- Mackay, T.F.; Stone, E.A.; Ayroles, J.F. The genetics of quantitative traits: Challenges and prospects. Nat. Rev. Genet. 2009, 10, 565–577. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Xue, Y.; Guo, Z.; Li, W.; Tang, J. Genome-wide association study identifies candidate genes for starch content regulation in maize kernels. Front. Plant Sci. 2016, 7, 1046. [Google Scholar] [CrossRef]
- Khazaei, H.; Podder, R.; Caron, C.T.; Kundu, S.S.; Diapari, M.; Vandenberg, A.; Bett, K.E. Marker-trait association analysis of iron and zinc concentration in lentil (Lens culinaris Medik.) seeds. Plant Genome 2017, 10. [Google Scholar] [CrossRef]
- Zhao, K.; Aranzana, M.J.; Kim, S.; Lister, C.; Shindo, C.; Tang, C.; Toomajian, C.; Zheng, H.; Dean, C.; Marjoram, P.; et al. An Arabidopsis example of association mapping in structured samples. PLoS Genet. 2007, 3, 71–82. [Google Scholar] [CrossRef]
- Price, A.L.; Patterson, N.J.; Plenge, R.M.; Weinblatt, M.E.; Shadick, N.A.; Reich, D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006, 38, 904–909. [Google Scholar] [CrossRef]
- Yu, J.; Pressoir, G.; Briggs, W.; Bi, I.; Yamasaki, M.; Doebley, J.; McMullen, M.; Gaut, B.; Nielsen, D.; Holland, J.; et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 2006, 38, 203–208. [Google Scholar] [CrossRef]
- Yang, X.; Gao, S.; Xu, S.; Zhang, Z.; Prasanna, B.M.; Li, L.; Li, J.; Yan, J. Characterization of a global germplasm collection and its potential utilization for analysis of complex quantitative traits in maize. Mol. Breed. 2011, 28, 511–526. [Google Scholar] [CrossRef]
- Kaler, A.S.; Purcell, L.C. Estimation of a significance threshold for genome-wide association studies. BMC Genom. 2019, 20, 618. [Google Scholar] [CrossRef] [PubMed]
- Sa, K.J.; Park, J.Y.; Woo, S.Y.; Ramekar, R.V.; Jang, C.S.; Lee, J.K. Mapping of QTL traits in corn using a RIL population derived from a cross of dent corn x waxy corn. Genes Genom. 2015, 37, 1–14. [Google Scholar] [CrossRef]
- Sa, K.J.; Choi, I.Y.; Park, J.Y.; Choi, J.K.; Ryu, S.H.; Lee, J.K. Mapping of QTL for agronomic traits using high-density SNPs with an RIL population in maize. Genes Genom. 2021, 43, 1403–1411. [Google Scholar] [CrossRef]
- Theune, M.L.; Bloss, U.; Brand, L.H.; Ladwig, F.; Wanke, D. Phylogenetic analyses and GAGA-motif binding studies of BBR/BPC proteins lend to clues in GAGA-motif recognition and a regulatory role in brassinosteroid signaling. Front. Plant Sci. 2019, 10, 466. [Google Scholar] [CrossRef]
- Butardo, V.M., Jr.; Anacleto, R.; Parween, S.; Samson, I.; de Guzman, K.; Alhambra, C.M.; Misra, G.; Sreenivasulu, N. Systems genetics identifies a novel regulatory domain of amylose synthesis. Plant Physiol. 2017, 173, 887–906. [Google Scholar] [CrossRef]
- Glowinski, A.; Flint-Garcia, S. Germplasm Resources for Mapping Quantitative Traits in Maize. In The Maize Genome. Compendium of Plant Genomes; Bennetzen, J., Flint-Garcia, S., Hirsch, C., Tuberosa, R., Eds.; Springer: Cham, Switzerland, 2018. [Google Scholar] [CrossRef]
- Zhang, H.; Jin, T.; Huang, Y.; Chen, J.; Zhu, L.; Zhao, Y.; Guo, J. Identification of quantitative trait loci underlying the protein, oil and starch contents of maize in multiple environments. Euphytica 2015, 205, 169–183. [Google Scholar] [CrossRef]
- Cook, J.P.; McMullen, M.D.; Holland, J.B.; Tian, F.; Bradbury, P.; Ross-Ibarra, J.; Buckler, E.S.; Flint-Garcia, S.A. Genetic architecture of maize kernel composition in the nested association mapping and inbred association panels. Plant Physiol. 2012, 158, 824–834. [Google Scholar] [CrossRef]
- Hu, S.; Wang, M.; Zhang, X.; Chen, W.; Song, X.; Fu, X.; Fang, H.; Xu, J.; Xiao, Y.; Li, Y.; et al. Genetic basis of kernel starch content decoded in a maize multi-parent population. Plant Biotechnol. J. 2021, 19, 2192–2205. [Google Scholar] [CrossRef]
- Sa, K.J.; Park, J.Y.; Park, K.C.; Lee, J.K. Analysis of genetic mapping in a waxy/dent maize RIL population using SSR and SNP markers. Genes Genom. 2012, 34, 157–164. [Google Scholar] [CrossRef]
- Ganal, M.W.; Durstewitz, G.; Polley, A.; Bérard, A.; Buckler, E.S.; Charcosset, A.; Clarke, J.D.; Graner, E.M.; Hansen, M.; Joets, J.; et al. A large maize (Zea mays L.) SNP genotyping array: Development and germplasm genotyping, and genetic mapping to compare with the B73 reference genome. PLoS ONE 2011, 6, e28334. [Google Scholar] [CrossRef] [PubMed]
- Dice, L.R. Measures of the amount of ecologic association between species. Ecology 1945, 26, 297–302. [Google Scholar] [CrossRef]
- Rohlf, F.J. NTSYS-pc: Numerical Taxonomy and Multivariate Analysis System; Version: 2.02; Exeter Software: Setauket, NY, USA, 1998. [Google Scholar]
- Pritchard, J.K.; Wen, W. Documentation for STRUCTURE Software: Version 2; University of Chicago: Chicago, IL, USA, 2003; Available online: http://pritc.h.bsd.uchicago.edu/structure.html (accessed on 1 April 2022).
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
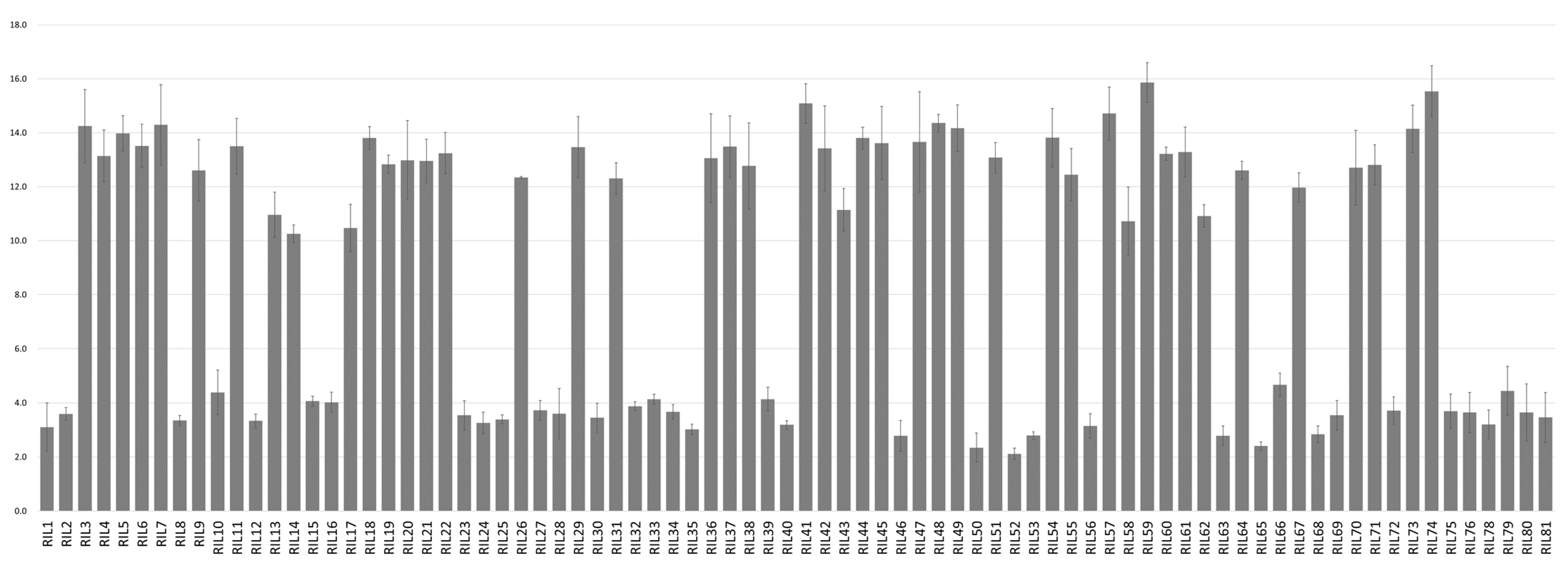

| Female (%) | Male (%) | All (n = 80) (%) | Low Group (n = 36) (%) | High Group (n = 44) (%) | |
|---|---|---|---|---|---|
| Mean | 14.4 | 2.9 | 8.8 | 3.4 | 13.1 |
| SD | 0.6 | 0.6 | 0.7 | 0.5 | 0.9 |
| Min | 2.1 | 2.1 | 10.3 | ||
| Max | 15.9 | 4.7 | 15.9 | ||
| t-test (p) | −45.403 (0.001) | ||||
| Marker Type | Marker | Chr | Position and Bin Value | p-Value | R2 (%) | Gene ID | Gene Name | Gene Description | FDR at 0.05 | FDR at 0.01 | −log10(p) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| SSR | umc1634 | 9 | 9.03 | 2.64E − 11 | 0.870 | GRMZM2G121333 | - | alpha/beta-Hydrolases superfamily protein | 3.34E − 06 | 6.68E − 07 | 10.58 ** |
| SNP | PZE-109022525 | 9 | 23,019,885 | 3.40E − 10 | 0.665 | GRMZM2G147319 | - | Zinc finger (C3HC4-type RING finger) family protein | 6.68E − 06 | 1.34E − 06 | 9.47 ** |
| SSR | umc2213 | 9 | 9.02 | 3.66E − 10 | 0.698 | GRMZM5G830776 | - | SNARE-interacting protein KEULE | 1.00E − 05 | 2.00E − 06 | 9.44 ** |
| SNP | SYN10618 | 9 | 22,668,399 | 1.01E − 09 | 0.620 | GRMZM2G092296 | rps22a | ribosomal protein S22 homolog | 1.34E − 05 | 2.67E − 06 | 9.00 ** |
| SNP | SYN34180 | 9 | 23,749,286 | 1.62E − 09 | 0.601 | GRMZM2G118355 | - | Histone H3 | 1.67E − 05 | 3.34E − 06 | 8.79 ** |
| SNP | SYN34181 | 9 | 23,765,962 | 1.62E − 09 | 0.601 | GRMZM2G118690 | bbr4 | BBR/BPC-transcription factor 4 | 3.01E − 05 | 6.01E − 06 | 8.79 ** |
| SNP | SYN34183 | 9 | 23,749,944 | 1.62E − 09 | 0.601 | GRMZM2G118355 | - | Histone H3 | 2.00E − 05 | 4.01E − 06 | 8.79 ** |
| SNP | SYN34184 | 9 | 23,749,967 | 1.62E − 09 | 0.601 | GRMZM2G118355 | - | Histone H3 | 2.34E − 05 | 4.68E − 06 | 8.79 ** |
| SNP | SYN34187 | 9 | 23,750,028 | 1.62E − 09 | 0.601 | GRMZM2G118355 | - | Histone H3 | 2.67E − 05 | 5.34E − 06 | 8.79 ** |
| SNP | PUT-163a-18172151-1376 | 9 | 22,689,623 | 3.50E − 09 | 0.570 | - | - | - | 3.67E − 05 | 7.35E − 06 | 8.46 ** |
| SNP | SYN10617 | 9 | 22,666,672 | 3.50E − 09 | 0.570 | GRMZM2G092296 | rps22a | ribosomal protein S22 homolog | 3.34E − 05 | 6.68E − 06 | 8.46 ** |
| SNP | PZE-109023492 | 9 | 23,538,268 | 5.80E − 09 | 0.550 | GRMZM2G396553 | adc1 | arginine decarboxylase1 | 4.01E − 05 | 8.02E − 06 | 8.24 ** |
| SNP | PZE-109023685 | 9 | 23,765,279 | 5.80E − 09 | 0.550 | GRMZM2G118687 | TIDP9202 | Peptidyl-prolyl cis-trans isomerase (PPIase) | 4.34E − 05 | 8.69E − 06 | 8.24 ** |
| SNP | PZE-109024175 | 9 | 24,345,287 | 7.42E − 09 | 0.540 | GRMZM2G370155 | - | - | 4.68E − 05 | 9.35E − 06 | 8.13 ** |
| SNP | PZE-109023327 | 9 | 23,467,856 | 8.13E − 09 | 0.537 | GRMZM2G121333 | umc1634 | alpha/beta-Hydrolases superfamily protein | 5.01E − 05 | 1.00E − 05 | 8.09 ** |
| SNP | PZE-109023851 | 9 | 24,080,158 | 2.52E − 08 | 0.493 | GRMZM2G082855 | er3 | erecta-like3 | 5.34E − 05 | 1.07E − 05 | 7.60 ** |
| SNP | PZE-109024053 | 9 | 24,147,325 | 2.52E − 08 | 0.493 | - | - | - | 5.68E − 05 | 1.14E − 05 | 7.60 ** |
| SNP | wx1 | 9 | 9.03 | 4.30E − 08 | 0.474 | GRMZM2G024993 | waxy1 | NDP-glucose-starch glucosyltransferase, starch granule-bound | 6.01E − 05 | 1.20E − 05 | 7.37 ** |
| SNP | PZE-109022101 | 9 | 22,340,830 | 5.91E − 08 | 0.462 | - | - | - | 6.35E − 05 | 1.27E − 05 | 7.23 ** |
| SSR | umc1893 | 9 | 9.02 | 7.93E − 08 | 0.473 | - | - | - | 6.68E − 05 | 1.34E − 05 | 7.10 ** |
| SNP | PZE-109021982 | 9 | 22,174,224 | 1.14E − 07 | 0.509 | - | - | - | 7.01E − 05 | 1.40E − 05 | 6.94 ** |
| SNP | SYN31842 | 9 | 22,336,574 | 1.14E − 07 | 0.509 | GRMZM2G107609 | - | Glycine-rich protein | 7.35E − 05 | 1.47E − 05 | 6.94 ** |
| SNP | PZE-109021558 | 9 | 21,857,603 | 1.22E − 07 | 0.436 | - | - | - | 7.68E − 05 | 1.54E − 05 | 6.91 ** |
| SNP | PZE-109021565 | 9 | 21,884,153 | 3.57E − 07 | 0.398 | - | - | - | 8.02E − 05 | 1.60E − 05 | 6.45 ** |
| SNP | PZE-109021581 | 9 | 21,886,534 | 3.57E − 07 | 0.398 | GRMZM2G447455 | - | Armadillo repeat-containing kinesin-like protein 3 | 8.35E − 05 | 1.67E − 05 | 6.45 ** |
| SNP | PZE-109022064 | 9 | 22,338,567 | 4.16E − 07 | 0.459 | GRMZM2G107665 | - | Aminomethyltransferase | 8.69E − 05 | 1.74E − 05 | 6.38 ** |
| SNP | PZE-109021851 | 9 | 22,066,639 | 4.32E − 07 | 0.458 | GRMZM2G148106 | AY105451 | ATP-dependent Clp protease proteolytic subunit | 9.35E − 05 | 1.87E − 05 | 6.36 ** |
| SNP | SYN9832 | 9 | 22,066,507 | 4.32E − 07 | 0.458 | GRMZM2G148106 | AY105451 | ATP-dependent Clp protease proteolytic subunit | 9.02E − 05 | 1.80E − 05 | 6.36 ** |
| SNP | SYN11958 | 9 | 25,829,587 | 7.56E − 07 | 0.372 | - | - | - | 9.69E − 05 | 1.94E − 05 | 6.12 * |
| SNP | SYN11959 | 9 | 25,830,547 | 7.56E − 07 | 0.372 | GRMZM2G042080 | sod11 | superoxide dismutase11 | 1.07E − 04 | 2.14E − 05 | 6.12 * |
| SNP | SYN11960 | 9 | 25,829,682 | 7.56E − 07 | 0.372 | GRMZM2G042080 | sod11 | superoxide dismutase11 | 1.00E − 04 | 2.00E − 05 | 6.12 * |
| SNP | SYN11961 | 9 | 25,829,976 | 7.56E − 07 | 0.372 | GRMZM2G042080 | sod11 | superoxide dismutase11 | 1.04E − 04 | 2.07E − 05 | 6.12 * |
| SNP | PZE-109025646 | 9 | 25,833,120 | 2.49E − 06 | 0.332 | GRMZM2G344634 | acb2 | Acyl-CoA-binding protein2 | 1.10E − 04 | 2.20E − 05 | 5.60 * |
| SNP | PZE-109026003 | 9 | 26,499,170 | 2.49E − 06 | 0.332 | GRMZM2G393146 | - | Putative acyl-activating enzyme 19 | 1.14E − 04 | 2.27E − 05 | 5.60 * |
| SNP | PZE-109025124 | 9 | 25,102,451 | 5.68E − 06 | 0.305 | GRMZM2G335052 | si660005h06c | Putative receptor-like protein kinase family protein | 1.17E − 04 | 2.34E − 05 | NS |
| SNP | SYN34135 | 9 | 26,592,678 | 6.17E − 06 | 0.302 | GRMZM2G136838 | krp2 | kinesin-related protein2 | 1.20E − 04 | 2.41E − 05 | NS |
| SNP | PZE-109020811 | 9 | 20,840,270 | 1.41E − 05 | 0.276 | GRMZM2G134260 | hb77 | Homeobox-transcription factor 77 | 1.24E − 04 | 2.47E − 05 | NS |
| SNP | PZE-109025060 | 9 | 25,097,187 | 1.71E − 05 | 0.282 | - | - | - | 1.27E − 04 | 2.54E − 05 | NS |
| SSR | umc2338 | 9 | 9.03 | 2.75E − 05 | 0.255 | GRMZM2G153792 | polm3 | polymerase II transcription-mediator3 | 1.30E − 04 | NS | NS |
| SNP | PZA00583.4 | 9 | 20,609,016 | 3.62E − 05 | 0.246 | GRMZM2G089992 | bub3 | Budding inhibited by benzimidazoles homolog3 | 1.40E − 04 | NS | NS |
| SNP | PZE-109020028 | 9 | 20,403,620 | 3.62E − 05 | 0.246 | GRMZM2G000645 | - | Coatomer subunit beta’-3 | 1.37E − 04 | NS | NS |
| SNP | SYN9817 | 9 | 20,346,749 | 3.62E − 05 | 0.246 | GRMZM2G444801 | sfp5 | sulfate transporter5 | 1.34E − 04 | NS | NS |
| SNP | PZE-109021152 | 9 | 21,138,321 | 6.06E − 05 | 0.288 | - | - | - | 1.44E − 04 | NS | NS |
| SNP | PZE-109044991 | 9 | 77,785,458 | 6.44E − 05 | 0.229 | - | - | - | 1.47E − 04 | NS | NS |
| SSR | umc1170 | 9 | 9.02 | 7.77E − 05 | 0.223 | - | - | - | 1.50E − 04 | NS | NS |
| SNP | PZE-109021389 | 9 | 21,823,525 | 7.80E − 05 | 0.275 | GRMZM2G144421 | saur72 | small auxin up RNA72 | 1.54E − 04 | NS | NS |
| SSR | bnlg244 | 9 | 9.02 | 1.03E − 04 | 0.240 | - | - | - | 1.57E − 04 | NS | NS |
| SSR | phi065 | 9 | 9.03 | 1.14E − 04 | 0.263 | GRMZM2G083841 | pep1 | phosphoenolpyruvate carboxylase1 | 1.60E − 04 | NS | NS |
| SSR | umc2087 | 9 | 9.03 | 1.18E − 04 | 0.214 | GRMZM2G422069 | platz16 | PLATZ-transcription factor 16 | 1.64E − 04 | NS | NS |
| SNP | PZE-109020401 | 9 | 20,619,848 | 1.19E − 04 | 0.261 | - | - | - | 1.67E − 04 | NS | NS |
| SSR | umc1700 | 9 | 9.03 | 1.42E − 04 | 0.206 | GRMZM2G422069 | platz16 | PLATZ-transcription factor 16 | 1.70E − 04 | NS | NS |
| SNP | PZE-109021109 | 9 | 20,982,506 | 1.54E − 04 | 0.253 | - | - | - | 1.74E − 04 | NS | NS |
| SNP | SYN15749 | 9 | 20,988,151 | 1.54E − 04 | 0.253 | GRMZM2G105957 | - | - | 1.77E − 04 | NS | NS |
| Marker Type | Marker | Chr | Position and Bin Value | p-Value | R2 (%) | Gene ID | Gene Name | Gene Description | FDR at 0.05 | FDR at 0.01 | −log10(p) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| SSR | umc1634 | 9 | 9.03 | 1.74E − 10 | 0.619 | GRMZM2G121333 | - | alpha/beta-Hydrolases superfamily protein | 3.34E − 06 | 6.68E − 07 | 9.76 ** |
| SNP | PZE-109022525 | 9 | 23,019,885 | 1.73E − 09 | 0.48 | GRMZM2G147319 | - | Zinc finger (C3HC4-type RING finger) family protein | 6.68E − 06 | 1.34E − 06 | 8.76 ** |
| SSR | umc2213 | 9 | 9.02 | 3.18E − 09 | 0.481 | GRMZM5G830776 | - | SNARE-interacting protein KEULE | 1.00E − 05 | 2.00E − 06 | 8.50 ** |
| SNP | SYN10618 | 9 | 22,668,399 | 4.89E − 09 | 0.446 | GRMZM2G092296 | rps22a | ribosomal protein S22 homolog | 1.34E − 05 | 2.67E − 06 | 8.31 ** |
| SNP | SYN34180 | 9 | 23,749,286 | 1.29E − 08 | 0.416 | GRMZM2G118355 | - | Histone H3 | 1.67E − 05 | 3.34E − 06 | 7.89 ** |
| SNP | SYN34181 | 9 | 23,765,962 | 1.29E − 08 | 0.416 | GRMZM2G118690 | bbr4 | BBR/BPC-transcription factor 4 | 3.01E − 05 | 6.01E − 06 | 7.89 ** |
| SNP | SYN34183 | 9 | 23,749,944 | 1.29E − 08 | 0.416 | GRMZM2G118355 | - | Histone H3 | 2.00E − 05 | 4.01E − 06 | 7.89 ** |
| SNP | SYN34184 | 9 | 23,749,967 | 1.29E − 08 | 0.416 | GRMZM2G118355 | - | Histone H3 | 2.34E − 05 | 4.68E − 06 | 7.89 ** |
| SNP | SYN34187 | 9 | 23,750,028 | 1.29E − 08 | 0.416 | GRMZM2G118355 | - | Histone H3 | 2.67E − 05 | 5.34E − 06 | 7.89 ** |
| SNP | PUT-163a-18172151-1376 | 9 | 22,689,623 | 2.48E − 08 | 0.396 | - | - | - | 3.67E − 05 | 7.35E − 06 | 7.61 ** |
| SNP | SYN10617 | 9 | 22,666,672 | 2.48E − 08 | 0.396 | GRMZM2G092296 | rps22a | ribosomal protein S22 homolog | 3.34E − 05 | 6.68E − 06 | 7.61 ** |
| SNP | PZE-109024175 | 9 | 24,345,287 | 6.47E − 08 | 0.367 | GRMZM2G370155 | - | - | 4.01E − 05 | 8.02E − 06 | 7.19 ** |
| SNP | PZE-109023492 | 9 | 23,538,268 | 6.86E − 08 | 0.365 | GRMZM2G396553 | adc1 | arginine decarboxylase1 | 4.34E − 05 | 8.69E − 06 | 7.16 ** |
| SNP | PZE-109023685 | 9 | 23,765,279 | 6.86E − 08 | 0.365 | GRMZM2G118687 | TIDP9202 | Peptidyl-prolyl cis-trans isomerase (PPIase) | 4.68E − 05 | 9.35E − 06 | 7.16 ** |
| SNP | PZE-109023327 | 9 | 23,467,856 | 8.06E − 08 | 0.361 | GRMZM2G121333 | umc1634 | alpha/beta-Hydrolases superfamily protein | 5.01E − 05 | 1.00E − 05 | 7.09 ** |
| SNP | PZE-109023851 | 9 | 24,080,158 | 3.23E − 07 | 0.321 | GRMZM2G082855 | er3 | erecta-like3 | 5.34E − 05 | 1.07E − 05 | 6.49** |
| SNP | PZE-109024053 | 9 | 24,147,325 | 3.23E − 07 | 0.321 | - | - | - | 5.68E − 05 | 1.14E − 05 | 6.49 ** |
| SNP | PZE-109022101 | 9 | 22,340,830 | 3.59E − 07 | 0.318 | - | - | - | 6.01E − 05 | 1.20E − 05 | 6.45 ** |
| SNP | PZE-109021558 | 9 | 21,857,603 | 4.12E − 07 | 0.314 | - | - | - | 6.35E − 05 | 1.27E − 05 | 6.39 ** |
| SSR | umc1893 | 9 | 9.02 | 5.53E − 07 | 0.318 | - | - | - | 6.68E − 05 | 1.34E − 05 | 6.26 ** |
| SNP | wx1 | 9 | 9.03 | 5.73E − 07 | 0.305 | GRMZM2G024993 | waxy1 | NDP-glucose-starch glucosyltransferase, starch granule-bound | 7.01E − 05 | 1.40E − 05 | 6.24 ** |
| SNP | PZE-109021982 | 9 | 22,174,224 | 6.19E − 07 | 0.356 | - | - | - | 7.35E − 05 | 1.47E − 05 | 6.21 ** |
| SNP | SYN31842 | 9 | 22,336,574 | 6.19E − 07 | 0.356 | GRMZM2G107609 | - | Glycine-rich protein | 7.68E − 05 | 1.54E − 05 | 6.21 ** |
| SNP | PZE-109021565 | 9 | 21,884,153 | 1.64E − 06 | 0.276 | - | - | - | 8.02E − 05 | 1.60E − 05 | 5.79 * |
| SNP | PZE-109021581 | 9 | 21,886,534 | 1.64E − 06 | 0.276 | GRMZM2G447455 | - | Armadillo repeat-containing kinesin-like protein 3 | 8.35E − 05 | 1.67E − 05 | 5.79 * |
| SNP | PZE-109021851 | 9 | 22,066,639 | 1.66E − 06 | 0.326 | GRMZM2G148106 | AY105451 | ATP-dependent Clp protease proteolytic subunit | 9.02E − 05 | 1.80E − 05 | 5.78 * |
| SNP | SYN9832 | 9 | 22,066,507 | 1.66E − 06 | 0.326 | GRMZM2G148106 | AY105451 | ATP-dependent Clp protease proteolytic subunit | 8.69E − 05 | 1.74E − 05 | 5.78 * |
| SNP | PZE-109022064 | 9 | 22,338,567 | 2.99E − 06 | 0.309 | GRMZM2G107665 | - | Aminomethyltransferase | 9.35E − 05 | 1.87E − 05 | 5.52 * |
| SNP | SYN11958 | 9 | 25,829,587 | 1.02E − 05 | 0.228 | - | - | - | 9.69E − 05 | 1.94E − 05 | NS |
| SNP | SYN11959 | 9 | 25,830,547 | 1.02E − 05 | 0.228 | GRMZM2G042080 | sod11 | superoxide dismutase11 | 1.07E − 04 | 2.14E − 05 | NS |
| SNP | SYN11960 | 9 | 25,829,682 | 1.02E − 05 | 0.228 | GRMZM2G042080 | sod11 | superoxide dismutase11 | 1.00E − 04 | 2.00E − 05 | NS |
| SNP | SYN11961 | 9 | 25,829,976 | 1.02E − 05 | 0.228 | GRMZM2G042080 | sod11 | superoxide dismutase11 | 1.04E − 04 | 2.07E − 05 | NS |
| SNP | PZE-109020811 | 9 | 20,840,270 | 2.59E − 05 | 0.205 | GRMZM2G134260 | hb77 | Homeobox-transcription factor 77 | 1.10E − 04 | NS | NS |
| SNP | PZE-109025646 | 9 | 25,833,120 | 5.64E − 05 | 0.186 | GRMZM2G344634 | acb2 | Acyl-CoA-binding protein2 | 1.14E − 04 | NS | NS |
| SNP | PZE-109026003 | 9 | 26,499,170 | 5.64E − 05 | 0.186 | GRMZM2G393146 | - | Putative acyl-activating enzyme 19 | 1.17E − 04 | NS | NS |
| SNP | PZE-109025124 | 9 | 25,102,451 | 9.59E − 05 | 0.173 | GRMZM2G335052 | si660005h06c | Putative receptor-like protein kinase family protein | 1.20E − 04 | NS | NS |
| SNP | PZA00583.4 | 9 | 20,609,016 | 9.82E − 05 | 0.172 | GRMZM2G089992 | bub3 | Budding inhibited by benzimidazoles homolog3 | 1.30E − 04 | NS | NS |
| SNP | PZE-109020028 | 9 | 20,403,620 | 9.82E − 05 | 0.172 | GRMZM2G000645 | - | Coatomer subunit beta’-3 | 1.27E − 04 | NS | NS |
| SNP | SYN9817 | 9 | 20,346,749 | 9.82E − 05 | 0.172 | GRMZM2G444801 | sfp5 | sulfate transporter5 | 1.24E − 04 | NS | NS |
| SNP | SYN34135 | 9 | 26,592,678 | 1.30E − 04 | 0.166 | GRMZM2G136838 | krp2 | kinesin-related protein2 | 1.34E − 04 | NS | NS |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sa, K.J.; Park, H.; Jang, S.J.; Lee, J.K. Association Mapping of Amylose Content in Maize RIL Population Using SSR and SNP Markers. Plants 2023, 12, 239. https://doi.org/10.3390/plants12020239
Sa KJ, Park H, Jang SJ, Lee JK. Association Mapping of Amylose Content in Maize RIL Population Using SSR and SNP Markers. Plants. 2023; 12(2):239. https://doi.org/10.3390/plants12020239
Chicago/Turabian StyleSa, Kyu Jin, Hyeon Park, So Jung Jang, and Ju Kyong Lee. 2023. "Association Mapping of Amylose Content in Maize RIL Population Using SSR and SNP Markers" Plants 12, no. 2: 239. https://doi.org/10.3390/plants12020239
APA StyleSa, K. J., Park, H., Jang, S. J., & Lee, J. K. (2023). Association Mapping of Amylose Content in Maize RIL Population Using SSR and SNP Markers. Plants, 12(2), 239. https://doi.org/10.3390/plants12020239





