Chloroplast Genome Engineering: A Plausible Approach to Combat Chili Thrips and Other Agronomic Insect Pests of Crops
Abstract
:1. Introduction
1.1. Feeding Mechanisms and Response of Plants to Thrips Invasion
1.2. Plastid Inheritance and Advantages of Chloroplast Engineering
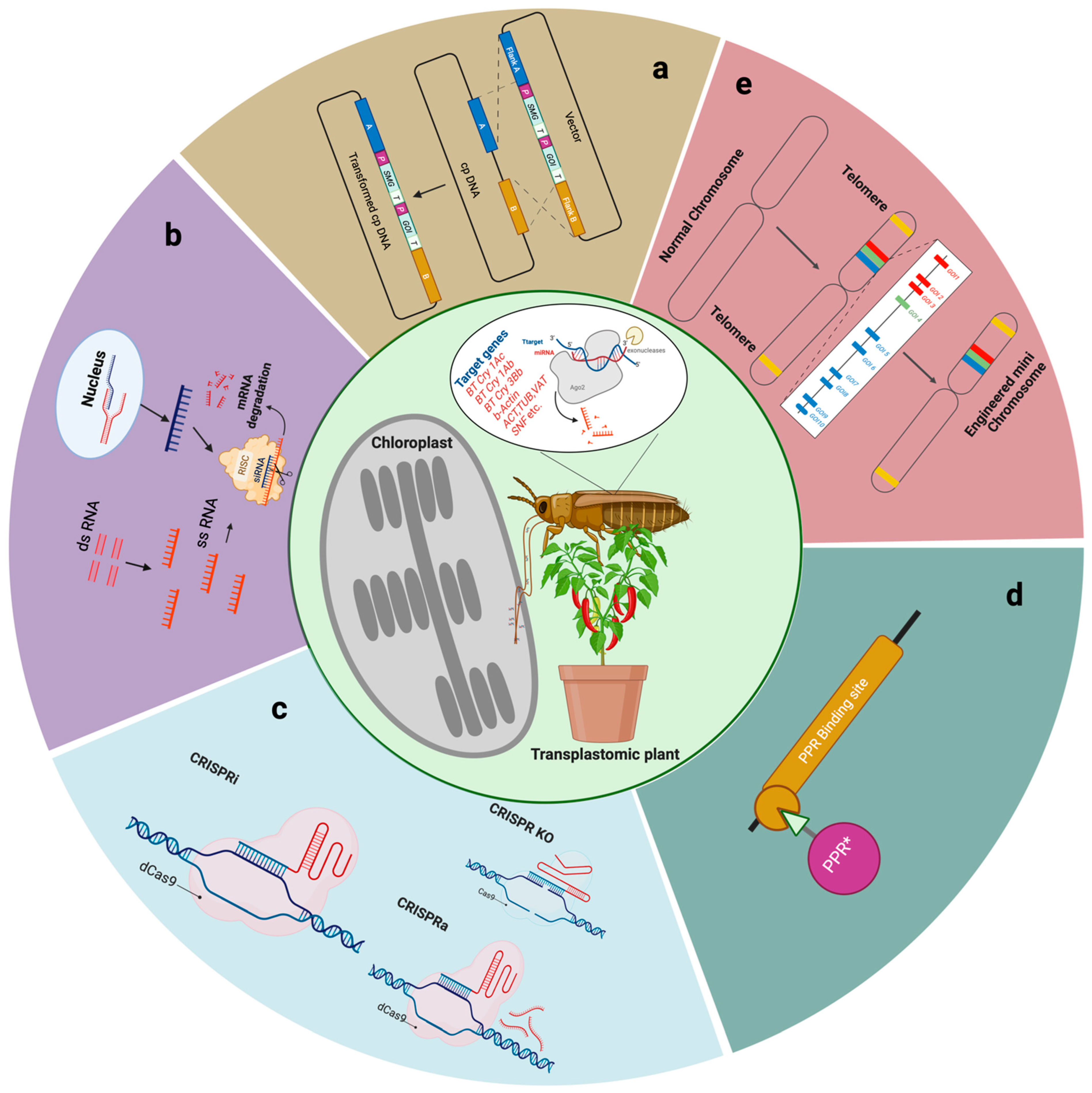
2. Biotechnological Approaches to Combat Invasive Pests
2.1. Deployment of Homology-Based Recombination in Chloroplasts
2.2. Plastid-Mediated RNAi Interference (PM-RNAi)
2.3. Engineering Chloroplasts Using CRISPR/Cas9 and Integrated Genome Editing Systems
2.4. RNA-Binding Proteins Regulate Gene Expression in Chloroplasts
2.5. Replicating Plant Mini Chromosomes for Chloroplast Genome Engineering
3. Conclusions and Perspectives
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Li, S.; Kim, D.S.; Zhang, J. Plastid-mediated RNA interference: A potential strategy for efficient pest control. Plant Cell Environ. 2023, 46, 2595–2605. [Google Scholar] [CrossRef] [PubMed]
- Mateos Fernández, R.; Petek, M.; Gerasymenko, I.; Juteršek, M.; Baebler, Š.; Kallam, K.; Moreno Giménez, E.; Gondolf, J.; Nordmann, A.; Gruden, K. Insect pest management in the age of synthetic biology. Plant Biotechnol. J. 2022, 20, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Kakkar, G.; McKenzie, C.L.; Seal, D.R.; Osborne, L.S. An overview of chilli thrips, Scirtothrips dorsalis (Thysanoptera: Thripidae) biology, distribution and management. In Weed and Pest Control: Conventional and New Challenges; Sonia, S., Marcelo, L., Eds.; InTech: Rijeka, Croatia, 2013; pp. 53–77. [Google Scholar]
- Tompe, A.; Hole, U.; Kulkarni, S.; Chaudhari, C.; Chavan, S. Studies on seasonal incidence of leaf eating caterpillar, Spodoptera litura (Fab.) infesting capsicum under polyhouse condition. J. Entomol. Zool. Stud. 2020, 8, 761–764. [Google Scholar]
- Pathak, V.M.; Verma, V.K.; Rawat, B.S.; Kaur, B.; Babu, N.; Sharma, A.; Dewali, S.; Yadav, M.; Kumari, R.; Singh, S. Current status of pesticide effects on environment, human health and it’s eco-friendly management as bioremediation: A comprehensive review. Front. Microbiol. 2022, 13, 962619. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.; Singha, A.; Haque, M.; Mondal, M.; Jiku, M.; Alam, M. Management of chilli insect pests by using trap crops. Thai J. Agric. Sci. 2021, 54, 212–221. [Google Scholar]
- Visschers, I.G.; Peters, J.L.; van de Vondervoort, J.A.; Hoogveld, R.H.; van Dam, N.M. Thrips resistance screening is coming of age: Leaf position and ontogeny are important determinants of leaf-based resistance in pepper. Front. Plant Sci. 2019, 10, 510. [Google Scholar] [CrossRef]
- Yang, C.-H.; Qiao, F.-J.; Lu, Z.; Li, C.-Y.; Liu, T.-X.; Gao, Y.-L.; Zhang, B. Interspecific competitions between Frankliniella intonsa and Frankliniella occidentalis on fresh lentil bean pods and pepper plants. Insects 2022, 14, 1. [Google Scholar] [CrossRef]
- Thorat, S.; Sisodiya, D.; Gangwar, R. Invasive Thrips, Thrips parvispinus (Karny) an Invasive Threat: A Review. Environ. Ecol. 2022, 40, 2170–2175. [Google Scholar]
- Mound, L.A.; Collins, D.W. A south east Asian pest species newly recorded from Europe: Thripsparvispinus (Thysanoptera: Thripidae), its confused identity and potential quarantine significance. Eur. J. Entomol 2000, 97, 197–200. [Google Scholar] [CrossRef]
- Gopal, G.V.; Lakshmi, K.V.; Babu, B.S.; Varma, P.K. Seasonal incidence of chilli thrips, Scirtothrips dorsalis hood in relation to weather parameters. J. Entomol. Zool. Stud. 2018, 6, 466–471. [Google Scholar]
- Vanisree, K.; Upendhar, S.; Rajasekhar, P.; Ramachandra Rao, G. Effect of newer insecticides against chilli thrips, Scirtothrips dorsalis (Hood). J. Entomol. Zool. 2017, 5, 277–284. [Google Scholar]
- Khatun, M.F.; Jahan, M.; Das, K.R.; Lee, K.Y.; Kil, E.J. Population dynamics and biorational management of sucking insect vectors on chili (Capsicum annuum L.) in Bangladesh. Arch. Insect Biochem. Physiol. 2023, 112, e21980. [Google Scholar] [CrossRef] [PubMed]
- Kenna, D.; Graystock, P.; Gill, R.J. Toxic temperatures: Bee behaviours exhibit divergent pesticide toxicity relationships with warming. Glob. Chang. Biol. 2023, 29, 2981–2998. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, R.; Abney, M.R.; Lai, P.-C.; Culbreath, A.K.; Tallury, S.; Leal-Bertioli, S.C. Resistance to thrips in peanut and implications for management of thrips and thrips-transmitted orthotospoviruses in peanut. Front. Plant Sci. 2018, 9, 1604. [Google Scholar] [CrossRef]
- Reitz, S.R.; Gao, Y.; Kirk, W.D.; Hoddle, M.S.; Leiss, K.A.; Funderburk, J.E. Invasion biology, ecology, and management of western flower thrips. Annu. Rev. Entomol. 2020, 65, 17–37. [Google Scholar] [CrossRef]
- Scott-Brown, A.; Hodgetts, J.; Hall, J.; Simmonds, M.; Collins, D. Potential role of botanic garden collections in predicting hosts at risk globally from invasive pests: A case study using Scirtothrips dorsalis. J. Pest Sci. 2018, 91, 601–611. [Google Scholar] [CrossRef]
- Leiss, K.A.; Choi, Y.H.; Abdel-Farid, I.B.; Verpoorte, R.; Klinkhamer, P.G. NMR metabolomics of thrips (Frankliniella occidentalis) resistance in Senecio hybrids. J. Chem. Ecol. 2009, 35, 219–229. [Google Scholar] [CrossRef]
- Jones, D.R. Plant viruses transmitted by thrips. Eur. J. Plant Pathol. 2005, 113, 119–157. [Google Scholar] [CrossRef]
- Rotenberg, D.; Whitfield, A.E. Molecular interactions between tospoviruses and thrips vectors. Curr. Opin. Virol. 2018, 33, 191–197. [Google Scholar] [CrossRef]
- Abe, H.; Shimoda, T.; Ohnishi, J.; Kugimiya, S.; Narusaka, M.; Seo, S.; Narusaka, Y.; Tsuda, S.; Kobayashi, M. Jasmonate-dependent plant defense restricts thrips performance and preference. BMC Plant Biol. 2009, 9, 97. [Google Scholar] [CrossRef]
- Steenbergen, M.; Abd-el-Haliem, A.; Bleeker, P.; Dicke, M.; Escobar-Bravo, R.; Cheng, G.; Haring, M.A.; Kant, M.R.; Kappers, I.; Klinkhamer, P.G. Thrips advisor: Exploiting thrips-induced defences to combat pests on crops. J. Exp. Bot. 2018, 69, 1837–1848. [Google Scholar] [CrossRef] [PubMed]
- Kaur, B.; Kuraparthy, V.; Bacheler, J.; Fang, H.; Bowman, D.T. Screening germplasm and quantification of components contributing to thrips resistance in cotton. J. Econ. Entomol. 2018, 111, 2426–2434. [Google Scholar] [CrossRef] [PubMed]
- Badenes-Pérez, F.R.; López-Pérez, J.A. Resistance and susceptibility to powdery mildew, root-knot nematode, and western flower thrips in two types of winter cress (Brassicaceae). Crop Prot. 2018, 110, 41–47. [Google Scholar] [CrossRef]
- Maharijaya, A.; Vosman, B.; Steenhuis-Broers, G.; Harpenas, A.; Purwito, A.; Visser, R.G.; Voorrips, R.E. Screening of pepper accessions for resistance against two thrips species (Frankliniella occidentalis and Thrips parvispinus). Euphytica 2011, 177, 401–410. [Google Scholar] [CrossRef]
- Maharijaya, A.; Vosman, B.; Pelgrom, K.; Wahyuni, Y.; de Vos, R.C.; Voorrips, R.E. Genetic variation in phytochemicals in leaves of pepper (Capsicum) in relation to thrips resistance. Arthropod-Plant Interact. 2019, 13, 1–9. [Google Scholar] [CrossRef]
- Macel, M.; Visschers, I.G.; Peters, J.L.; Kappers, I.F.; de Vos, R.C.; van Dam, N.M. Metabolomics of thrips resistance in pepper (Capsicum spp.) reveals monomer and dimer acyclic diterpene glycosides as potential chemical defenses. J. Chem. Ecol. 2019, 45, 490–501. [Google Scholar] [CrossRef]
- Varshney, R.K.; Bohra, A.; Yu, J.; Graner, A.; Zhang, Q.; Sorrells, M.E. Designing future crops: Genomics-assisted breeding comes of age. Trends Plant Sci. 2021, 26, 631–649. [Google Scholar] [CrossRef]
- Bock, R. Structure, function, and inheritance of plastid genomes. In Cell and Molecular Biology of Plastids; Springer: Berlin/Heidelberg, Germany, 2007; pp. 29–63. [Google Scholar]
- Azhagiri, A.K.; Maliga, P. Exceptional paternal inheritance of plastids in Arabidopsis suggests that low-frequency leakage of plastids via pollen may be universal in plants. Plant J. 2007, 52, 817–823. [Google Scholar] [CrossRef]
- Chung, K.P.; Gonzalez-Duran, E.; Ruf, S.; Endries, P.; Bock, R. Control of plastid inheritance by environmental and genetic factors. Nat. Plants 2023, 9, 68–80. [Google Scholar] [CrossRef]
- Daniell, H.; Kumar, S.; Dufourmantel, N. Breakthrough in chloroplast genetic engineering of agronomically important crops. Trends Biotechnol. 2005, 23, 238–245. [Google Scholar] [CrossRef]
- Svab, Z.; Hajdukiewicz, P.; Maliga, P. Stable transformation of plastids in higher plants. Proc. Natl. Acad. Sci. USA 1990, 87, 8526–8530. [Google Scholar] [CrossRef] [PubMed]
- Stewart Jr, C.N.; Halfhill, M.D.; Warwick, S.I. Transgene introgression from genetically modified crops to their wild relatives. Nat. Rev. Genet. 2003, 4, 806–817. [Google Scholar] [CrossRef] [PubMed]
- Chung, Y.H.; Church, D.; Koellhoffer, E.C.; Osota, E.; Shukla, S.; Rybicki, E.P.; Pokorski, J.K.; Steinmetz, N.F. Integrating plant molecular farming and materials research for next-generation vaccines. Nat. Rev. Mater. 2022, 7, 372–388. [Google Scholar] [CrossRef] [PubMed]
- Maliga, P. Engineering the plastid and mitochondrial genomes of flowering plants. Nat. Plants 2022, 8, 996–1006. [Google Scholar] [CrossRef]
- Hossain, M.J.; Bakhsh, A. Development and applications of transplastomic plants; a way towards eco-friendly agriculture. In Environment, Climate, Plant and Vegetation Growth; Springer: Cham, Switzerland, 2020; pp. 285–322. [Google Scholar]
- Horgan, F.G.; Garcia, C.P.F.; Haverkort, F.; de Jong, P.W.; Ferrater, J.B. Changes in insecticide resistance and host range performance of planthoppers artificially selected to feed on resistant rice. Crop Prot. 2020, 127, 104963. [Google Scholar] [CrossRef]
- Mansour, M.R.; Eryan, N.L. Effects of Certain Weather, Biotic Factors and Chemical Components on The population of Aphids in Egyptian Wheat Fields. Egypt. Acad. J. Biol. Sci. A Entomol. 2022, 15, 1–13. [Google Scholar]
- Joshi, M.J.; Prithiv Raj, V.; Solanki, C.B.; Birari Vaishali, V. Role of biotechnology in insect-pests management. Agric. Food E-Newsl. 2020, 2, 574–576. [Google Scholar]
- Kumari, P.; Jasrotia, P.; Kumar, D.; Kashyap, P.L.; Kumar, S.; Mishra, C.N.; Kumar, S.; Singh, G.P. Biotechnological Approaches for Host Plant Resistance to Insect Pests. Front. Genet. 2022, 13, 914029. [Google Scholar] [CrossRef]
- Yu, Q.; LaManna, L.M.; Kelly, M.E.; Lutz, K.A.; Maliga, P. New tools for engineering the Arabidopsis plastid genome. Plant Physiol. 2019, 181, 394–398. [Google Scholar] [CrossRef]
- Jin, S.; Kanagaraj, A.; Verma, D.; Lange, T.; Daniell, H. Release of hormones from conjugates: Chloroplast expression of β-glucosidase results in elevated phytohormone levels associated with significant increase in biomass and protection from aphids or whiteflies conferred by sucrose esters. Plant Physiol. 2011, 155, 222–235. [Google Scholar] [CrossRef]
- Dufourmantel, N.; Dubald, M.; Matringe, M.; Canard, H.; Garcon, F.; Job, C.; Kay, E.; Wisniewski, J.P.; Ferullo, J.M.; Pelissier, B. Generation and characterization of soybean and marker-free tobacco plastid transformants over-expressing a bacterial 4-hydroxyphenylpyruvate dioxygenase which provides strong herbicide tolerance. Plant Biotechnol. J. 2007, 5, 118–133. [Google Scholar] [CrossRef] [PubMed]
- Kota, M.; Daniell, H.; Varma, S.; Garczynski, S.F.; Gould, F.; Moar, W.J. Overexpression of the Bacillus thuringiensis (Bt) Cry2Aa2 protein in chloroplasts confers resistance to plants against susceptible and Bt-resistant insects. Proc. Natl. Acad. Sci. USA 1999, 96, 1840–1845. [Google Scholar] [CrossRef]
- Chakrabarti, S.K.; Lutz, K.A.; Lertwiriyawong, B.; Svab, Z.; Maliga, P. Expression of the cry9Aa2 Bt gene in tobacco chloroplasts confers resistance to potato tuber moth. Transgenic Res. 2006, 15, 481–488. [Google Scholar] [CrossRef] [PubMed]
- Dauvillee, D.; Hilbig, L.; Preiss, S.; Johanningmeier, U. Minimal extent of sequence homology required for homologous recombination at the psb A locus in Chlamydomonas reinhardtii chloroplasts using PCR-generated DNA fragments. Photosynth. Res. 2004, 79, 219–224. [Google Scholar] [CrossRef] [PubMed]
- Scotti, N.; Cardi, T. Transgene-induced pleiotropic effects in transplastomic plants. Biotechnol. Lett. 2014, 36, 229–239. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.-p.; Luo, T.; Fu, H.-w.; Wang, L.; Tan, Y.-y.; Huang, J.-z.; Wang, Q.; Ye, G.-y.; Gatehouse, A.M.; Lou, Y.-g. Resistance of rice to insect pests mediated by suppression of serotonin biosynthesis. Nat. Plants 2018, 4, 338–344. [Google Scholar] [CrossRef]
- Mühlbauer, S.K.; Koop, H.U. External control of transgene expression in tobacco plastids using the bacterial lac repressor. Plant J. 2005, 43, 941–946. [Google Scholar] [CrossRef]
- Lössl, A.; Bohmert, K.; Harloff, H.; Eibl, C.; Mühlbauer, S.; Koop, H.-U. Inducible trans-activation of plastid transgenes: Expression of the R. eutropha phb operon in transplastomic tobacco. Plant Cell Physiol. 2005, 46, 1462–1471. [Google Scholar] [CrossRef]
- Emadpour, M.; Karcher, D.; Bock, R. Boosting riboswitch efficiency by RNA amplification. Nucleic Acids Res. 2015, 43, e66. [Google Scholar] [CrossRef]
- Baum, J.A.; Bogaert, T.; Clinton, W.; Heck, G.R.; Feldmann, P.; Ilagan, O.; Johnson, S.; Plaetinck, G.; Munyikwa, T.; Pleau, M. Control of coleopteran insect pests through RNA interference. Nat. Biotechnol. 2007, 25, 1322–1326. [Google Scholar] [CrossRef]
- Mao, Y.-B.; Cai, W.-J.; Wang, J.-W.; Hong, G.-J.; Tao, X.-Y.; Wang, L.-J.; Huang, Y.-P.; Chen, X.-Y. Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat. Biotechnol. 2007, 25, 1307–1313. [Google Scholar] [CrossRef] [PubMed]
- Zhu, K.Y.; Palli, S.R. Mechanisms, applications, and challenges of insect RNA interference. Annu. Rev. Entomol. 2020, 65, 293–311. [Google Scholar] [CrossRef] [PubMed]
- Borges, F.; Martienssen, R.A. The expanding world of small RNAs in plants. Nat. Rev. Mol. Cell Biol. 2015, 16, 727–741. [Google Scholar] [CrossRef]
- Wu, M.; Dong, Y.; Zhang, Q.; Li, S.; Chang, L.; Loiacono, F.V.; Ruf, S.; Zhang, J.; Bock, R. Efficient control of western flower thrips by plastid-mediated RNA interference. Proc. Natl. Acad. Sci. USA 2022, 119, e2120081119. [Google Scholar] [CrossRef]
- Dong, Y.; Wu, M.; Zhang, Q.; Fu, J.; Loiacono, F.V.; Yang, Y.; Wang, Z.; Li, S.; Chang, L.; Bock, R. Control of a sap-sucking insect pest by plastid-mediated RNA interference. Mol. Plant 2022, 15, 1176–1191. [Google Scholar] [CrossRef] [PubMed]
- McBride, K.E.; Svab, Z.; Schaaf, D.J.; Hogan, P.S.; Stalker, D.M.; Maliga, P. Amplification of a chimeric Bacillus gene in chloroplasts leads to an extraordinary level of an insecticidal protein in tobacco. Bio/Technol. 1995, 13, 362–365. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Zhang, Y.; Li, S.; Chang, L.; Wu, Y.; Zhang, J. Plastid-expressed Bacillus thuringiensis (Bt) cry3Bb confers high mortality to a leaf eating beetle in poplar. Plant Cell Rep. 2020, 39, 317–323. [Google Scholar] [CrossRef]
- Zhang, J.; Khan, S.A.; Hasse, C.; Ruf, S.; Heckel, D.G.; Bock, R. Full crop protection from an insect pest by expression of long double-stranded RNAs in plastids. Science 2015, 347, 991–994. [Google Scholar] [CrossRef]
- Priti; Mukherjee, S.K.; Ghosh, A. Silencing of Thrips palmi UHRF1BP1 and PFAS Using Antisense Oligos Induces Mortality and Reduces Tospovirus Titer in Its Vector. Pathogens 2022, 11, 1319. [Google Scholar] [CrossRef]
- Kaplanoglu, E.; Kolotilin, I.; Menassa, R.; Donly, C. Plastid transformation of micro-tom tomato with a Hemipteran double-stranded RNA results in RNA interference in multiple insect species. Int. J. Mol. Sci. 2022, 23, 3918. [Google Scholar] [CrossRef]
- Di Lelio, I.; Varricchio, P.; Di Prisco, G.; Marinelli, A.; Lasco, V.; Caccia, S.; Casartelli, M.; Giordana, B.; Rao, R.; Gigliotti, S. Functional analysis of an immune gene of Spodoptera littoralis by RNAi. J. Insect Physiol. 2014, 64, 90–97. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Guo, Z.; Yang, Z.; Han, H.; Wang, S.; Xu, H.; Yang, X.; Yang, F.; Wu, Q.; Xie, W. Whitefly hijacks a plant detoxification gene that neutralizes plant toxins. Cell 2021, 184, 1693–1705.e1617. [Google Scholar] [CrossRef] [PubMed]
- Dufourmantel, N.; Tissot, G.; Goutorbe, F.; Garcon, F.; Muhr, C.; Jansens, S.; Pelissier, B.; Peltier, G.; Dubald, M. Generation and analysis of soybean plastid transformants expressing Bacillus thuringiensis Cry1Ab protoxin. Plant Mol. Biol. 2005, 58, 659–668. [Google Scholar] [CrossRef] [PubMed]
- Cheng-Wei, L.; Chin-Chung, L.; Jinn-Chin, Y.; JW, C.J.; Menq-Jiau, T. Expression of a Bacillus thuringiensis toxin (cry1Ab) gene in cabbage (Brassica oleracea L. var. capitata L.) chloroplasts confers high insecticidal efficacy against Plutella xylostella. Theor. Appl. Genet. 2008, 117, 75–88. [Google Scholar]
- Wu, Y.; Xu, L.; Chang, L.; Ma, M.; You, L.; Jiang, C.; Li, S.; Zhang, J. Bacillus thuringiensis cry1C expression from the plastid genome of poplar leads to high mortality of leaf-eating caterpillars. Tree Physiol. 2019, 39, 1525–1532. [Google Scholar] [CrossRef] [PubMed]
- Whitten, M.M.; Facey, P.D.; Del Sol, R.; Fernández-Martínez, L.T.; Evans, M.C.; Mitchell, J.J.; Bodger, O.G.; Dyson, P.J. Symbiont-mediated RNA interference in insects. Proc. R. Soc. B Biol. Sci. 2016, 283, 20160042. [Google Scholar] [CrossRef]
- Singh, S.; Gupta, M.; Pandher, S.; Kaur, G.; Goel, N.; Rathore, P.; Palli, S.R. RNA sequencing, selection of reference genes and demonstration of feeding RNAi in Thrips tabaci (Lind.)(Thysanoptera: Thripidae). BMC Mol. Biol. 2019, 20, 6. [Google Scholar] [CrossRef]
- Andongma, A.A.; Greig, C.; Dyson, P.J.; Flynn, N.; Whitten, M.M. Optimization of dietary RNA interference delivery to western flower thrips Frankliniella occidentalis and onion thrips Thrips tabaci. Arch. Insect Biochem. Physiol. 2020, 103, e21645. [Google Scholar] [CrossRef]
- Liu, S.; Jaouannet, M.; Dempsey, D.M.A.; Imani, J.; Coustau, C.; Kogel, K.-H. RNA-based technologies for insect control in plant production. Biotechnol. Adv. 2020, 39, 107463. [Google Scholar] [CrossRef]
- Hang, D.Q.; Zhang, Z.Y.; Unver, T.; Zhang, B.H. CRISPR/Cas: A powerful tool for gene function study and crop improvement. J. Adv. Res. 2021, 29, 207–221. [Google Scholar] [CrossRef]
- Kang, B.-C.; Bae, S.-J.; Lee, S.; Lee, J.S.; Kim, A.; Lee, H.; Baek, G.; Seo, H.; Kim, J.; Kim, J.-S. Chloroplast and mitochondrial DNA editing in plants. Nat. Plants 2021, 7, 899–905. [Google Scholar] [CrossRef] [PubMed]
- Daniell, H.; Jin, S.; Zhu, X.G.; Gitzendanner, M.A.; Soltis, D.E.; Soltis, P.S. Green giant—A tiny chloroplast genome with mighty power to produce high-value proteins: History and phylogeny. Plant Biotechnol. J. 2021, 19, 430–447. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Brueggeman, A.J.; Horken, K.M.; Plucinak, T.M.; Weeks, D.P. Successful transient expression of Cas9 and single guide RNA genes in Chlamydomonas reinhardtii. Eukaryot. Cell 2014, 13, 1465–1469. [Google Scholar] [CrossRef] [PubMed]
- Yoo, B.-C.; Yadav, N.S.; Orozco Jr, E.M.; Sakai, H. Cas9/gRNA-mediated genome editing of yeast mitochondria and Chlamydomonas chloroplasts. PeerJ 2020, 8, e8362. [Google Scholar] [CrossRef]
- Li, C.; Brant, E.; Budak, H.; Zhang, B. CRISPR/Cas: A Nobel Prize award-winning precise genome editing technology for gene therapy and crop improvement. J. Zhejiang Univ. Sci. B 2021, 22, 253. [Google Scholar] [CrossRef]
- Khan, M.S.; Maliga, P. Fluorescent antibiotic resistance marker for tracking plastid transformation in higher plants. Nat. Biotechnol. 1999, 17, 910–915. [Google Scholar] [CrossRef]
- Gammage, P.A.; Moraes, C.T.; Minczuk, M. Mitochondrial genome engineering: The revolution may not be CRISPR-Ized. Trends Genet. 2018, 34, 101–110. [Google Scholar] [CrossRef]
- Nakazato, I.; Okuno, M.; Yamamoto, H.; Tamura, Y.; Itoh, T.; Shikanai, T.; Takanashi, H.; Tsutsumi, N.; Arimura, S.-I. Targeted base editing in the plastid genome of Arabidopsis thaliana. Nat. Plants 2021, 7, 906–913. [Google Scholar] [CrossRef]
- Mok, Y.G.; Hong, S.; Bae, S.-J.; Cho, S.-I.; Kim, J.-S. Targeted A-to-G base editing of chloroplast DNA in plants. Nat. Plants 2022, 8, 1378–1384. [Google Scholar] [CrossRef]
- Glisovic, T.; Bachorik, J.L.; Yong, J.; Dreyfuss, G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008, 582, 1977–1986. [Google Scholar] [CrossRef]
- Barkan, A.; Small, I. Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant Biol. 2014, 65, 415–442. [Google Scholar] [CrossRef] [PubMed]
- Gully, B.S.; Cowieson, N.; Stanley, W.A.; Shearston, K.; Small, I.D.; Barkan, A.; Bond, C.S. The solution structure of the pentatricopeptide repeat protein PPR10 upon binding atpH RNA. Nucleic Acids Res. 2015, 43, 1918–1926. [Google Scholar] [CrossRef] [PubMed]
- Coquille, S.; Filipovska, A.; Chia, T.; Rajappa, L.; Lingford, J.P.; Razif, M.F.; Thore, S.; Rackham, O. An artificial PPR scaffold for programmable RNA recognition. Nat. Commun. 2014, 5, 5729. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.; Zhang, D.; Guan, Z.; Liu, Y.; Yang, Z.; Yang, Y.; Wang, X.; Wang, Q.; Zhang, Q.; Fan, S. Structural basis for specific single-stranded RNA recognition by designer pentatricopeptide repeat proteins. Nat. Commun. 2016, 7, 11285. [Google Scholar] [CrossRef]
- McDermott, J.J.; Watkins, K.P.; Williams-Carrier, R.; Barkan, A. Ribonucleoprotein capture by in vivo expression of a designer pentatricopeptide repeat protein in Arabidopsis. Plant Cell 2019, 31, 1723–1733. [Google Scholar] [CrossRef]
- Manavski, N.; Mathieu, S.; Rojas, M.; Méteignier, L.-V.; Brachmann, A.; Barkan, A.; Hammani, K. In vivo stabilization of endogenous chloroplast RNAs by customized artificial pentatricopeptide repeat proteins. Nucleic Acids Res. 2021, 49, 5985–5997. [Google Scholar] [CrossRef]
- Yu, W.; Yau, Y.Y.; Birchler, J.A. Plant artificial chromosome technology and its potential application in genetic engineering. Plant Biotechnol. J. 2016, 14, 1175–1182. [Google Scholar] [CrossRef]
- Houben, A.; Dawe, R.K.; Jiang, J.; Schubert, I. Engineered plant minichromosomes: A bottom-up success? Plant Cell 2008, 20, 8–10. [Google Scholar] [CrossRef]
- Birchler, J.A.; Graham, N.D.; Swyers, N.C.; Cody, J.P.; McCaw, M.E. Plant minichromosomes. Curr. Opin. Biotechnol. 2016, 37, 135–142. [Google Scholar] [CrossRef]
- Jakubiec, A.; Sarokina, A.; Choinard, S.; Vlad, F.; Malcuit, I.; Sorokin, A.P. Replicating minichromosomes as a new tool for plastid genome engineering. Nat. Plants 2021, 7, 932–941. [Google Scholar] [CrossRef]
- Yin, X.; Zhang, Y.; Chen, Y.; Wang, J.; Wang, R.R.-C.; Fan, C.; Hu, Z. Precise characterization and tracking of stably inherited artificial minichromosomes made by telomere-mediated chromosome truncation in Brassica napus. Front. Plant Sci. 2021, 12, 743792. [Google Scholar] [CrossRef] [PubMed]
- Kan, M.; Huang, T.; Zhao, P. Artificial chromosome technology and its potential application in plants. Front. Plant Sci. 2022, 13, 970943. [Google Scholar] [CrossRef] [PubMed]
- Kiani, S.; Mohamed, B.B.; Shehzad, K.; Jamal, A.; Shahid, M.N.; Shahid, A.A.; Husnain, T. Chloroplast-targeted expression of recombinant crystal-protein gene in cotton: An unconventional combat with resistant pests. J. Biotechnol. 2013, 166, 88–96. [Google Scholar] [CrossRef]
- Singh, R.K.; Sreenivasulu, N.; Prasad, M. Potential of underutilized crops to introduce the nutritional diversity and achieve zero hunger. Funct. Integr. Genom. 2022, 22, 1459–1465. [Google Scholar] [CrossRef]
- You, L.; Song, Q.; Wu, Y.; Li, S.; Jiang, C.; Chang, L.; Yang, X.; Zhang, J. Accumulation of glycine betaine in transplastomic potato plants expressing choline oxidase confers improved drought tolerance. Planta 2019, 249, 1963–1975. [Google Scholar] [CrossRef]
- Qaim, M. The economics of genetically modified crops. Annu. Rev. Resour. Econ. 2009, 1, 665–694. [Google Scholar] [CrossRef]
- Li, C.; Chu, W.; Gill, R.A.; Sang, S.; Shi, Y.; Hu, X.; Yang, Y.; Zaman, Q.U.; Zhang, B.H. Computational tools and resources for CRISPR/Cas genome editing. Genom. Proteom. Bioinform. 2023, 21, 108–126. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Kim, S.-T.; Ryu, J.; Kang, B.-C.; Kim, J.-S.; Kim, S.-G. CRISPR/Cpf1-mediated DNA-free plant genome editing. Nat. Commun. 2017, 8, 14406. [Google Scholar] [CrossRef]
- Kim, D.; Hager, M.; Brant, E. Efficient genome editing in wheat using Cas9 and Cpf1 (AsCpf1 and LbCpf1) nucleases. Funct. Integr. Genom. 2021, 21, 355–366. [Google Scholar] [CrossRef]
- Zhang, Y.; Tian, L.; Lu, C. Chloroplast gene expression: Recent advances and perspectives. Plant Commun. 2023, 4, 100611. [Google Scholar] [CrossRef]
- Maher, M.F.; Nasti, R.A.; Vollbrecht, M.; Starker, C.G.; Clark, M.D.; Voytas, D.F. Plant gene editing through de novo induction of meristems. Nat. Biotechnol. 2020, 38, 84–89. [Google Scholar] [CrossRef] [PubMed]
- Atkins, P.A.; Voytas, D.F. Overcoming bottlenecks in plant gene editing. Curr. Opin. Plant Biol. 2020, 54, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Bull, T.; Debernardi, J.; Reeves, M.; Hill, T.; Bertier, L.; Van Deynze, A.; Michelmore, R. GRF–GIF chimeric proteins enhance in vitro regeneration and Agrobacterium-mediated transformation efficiencies of lettuce (Lactuca spp.). Plant Cell Rep. 2023, 42, 629–643. [Google Scholar] [CrossRef]
- Liu, W.; Zhang, B.H. The landscape of genome sequencing and assembling in plants. Funct. Integr. Genom. 2022, 22, 1147–1152. [Google Scholar] [CrossRef] [PubMed]
| Gene Name | Function | Insect Species/Order Investigated | Molecular Approach | Host | References |
|---|---|---|---|---|---|
| Thrips palmiUHRF1BP1 | Mortality and to impair virus transmission | Thrips palmi (Thysanoptera: Thripidae) | Silencing | Thrips palmi | [62] |
| Thrips palmi PFAS | |||||
| Western flower thrip Actin (ACT) | Mortality of western flower thrip | Western flower thrip (WFT, Frankliniella occidentalis) | Silencing | Nicotiana tabacum | [57] |
| Western flower thrip Tubulin (TUB) | |||||
| Western flower thrip Catalytic subunit B of vacuolar ATPase (VAT) | |||||
| Western flower thrip Endosomal sorting com-plex for transport III subunit SNF7 (SNF) | |||||
| Phenacoccussolenopsis v-ATPaseA | Efficient control of leaf-chewing and lacerate-and-flush feeding insects | Brown marmorated stink bug (BMSB) (Halyomorphahalys) Madeira mealybug (MMB) (Phenacoccusmadeirensis) Colorado potato beetle (CPB) (Leptinotarsa decemlineata) | Silencing | Solanum lycopersicum | [63] |
| MyzuspersicaeMpDhc64C, Helicoverpaarmigera NDUFVII | Efficient control of sap-sucking pests | Myzuspersicae, Acyrthosiphonpisum, Adelphocorissuturalis, Rhopalosiphumpadi, Nilaparvatalugens | Silencing | Nicotiana tabacum | [58] |
| Spodoptera littoralis 102 (Sl102) | 1.Reduced food consumption of larvae 2. Impairment of hemocyte-mediated encapsulation response 3. Enhanced biopesticide activity | Spodoptera littoralis | Silencing | Spodoptera littoralis | [64] |
| Phenolic glucoside malonyltransferase gene BtPMaT1 | Enhanced impeding the whiteflies’ capacity for detoxification | Whitefly (Beniciatabaci) | Silencing | Solanum lycopersicum | [65] |
| Beta-glucosidase (Bgl-1) | Elevated phytohormone levels and conferred resistance to whiteflies and aphids | Whitefly and aphids | Overexpression | Nicotiana tabacum | [43] |
| Cry1Ab | Resistance to Caterpillar | Caterpillar (Anticarsiagemmatalis | Overexpression | Glycine max | [66] |
| Cry9Aa2 | Resistance to potato tuber moth | Potato tuber moth (Phthorimaeaoperculella) | Overexpression | Nicotiana tabacum | [46] |
| cry2Aa2 | Resistance to moth | Heliothisvirescens, Helicoverpazea, Spodoptera exigua | Overexpression | Nicotiana tabacum | [45] |
| β-actin encoding gene of whitefly ACTB | Control of sap-sucking pest Whitefly | Bemisiatabaci | Silencing | Nicotiana tabacum | [58] |
| cry1Ab | Enhanced resistance to Plutella xylostella | Plutella xylostella | Overexpression | Brassica oleracea | [67] |
| cry1C | Improved toxicity to Hyphantria cunea and Lymantria dispar | H. cunea and L. dispar | Overexpression | Hybrid Popular (P. davidiana × Populus bolleana) | [68] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bulle, M.; Sheri, V.; Aileni, M.; Zhang, B. Chloroplast Genome Engineering: A Plausible Approach to Combat Chili Thrips and Other Agronomic Insect Pests of Crops. Plants 2023, 12, 3448. https://doi.org/10.3390/plants12193448
Bulle M, Sheri V, Aileni M, Zhang B. Chloroplast Genome Engineering: A Plausible Approach to Combat Chili Thrips and Other Agronomic Insect Pests of Crops. Plants. 2023; 12(19):3448. https://doi.org/10.3390/plants12193448
Chicago/Turabian StyleBulle, Mallesham, Vijay Sheri, Mahender Aileni, and Baohong Zhang. 2023. "Chloroplast Genome Engineering: A Plausible Approach to Combat Chili Thrips and Other Agronomic Insect Pests of Crops" Plants 12, no. 19: 3448. https://doi.org/10.3390/plants12193448
APA StyleBulle, M., Sheri, V., Aileni, M., & Zhang, B. (2023). Chloroplast Genome Engineering: A Plausible Approach to Combat Chili Thrips and Other Agronomic Insect Pests of Crops. Plants, 12(19), 3448. https://doi.org/10.3390/plants12193448









