Molecular Mechanism of Response and Adaptation of Antioxidant Enzyme System to Salt Stress in Leaves of Gymnocarpos przewalskii
Abstract
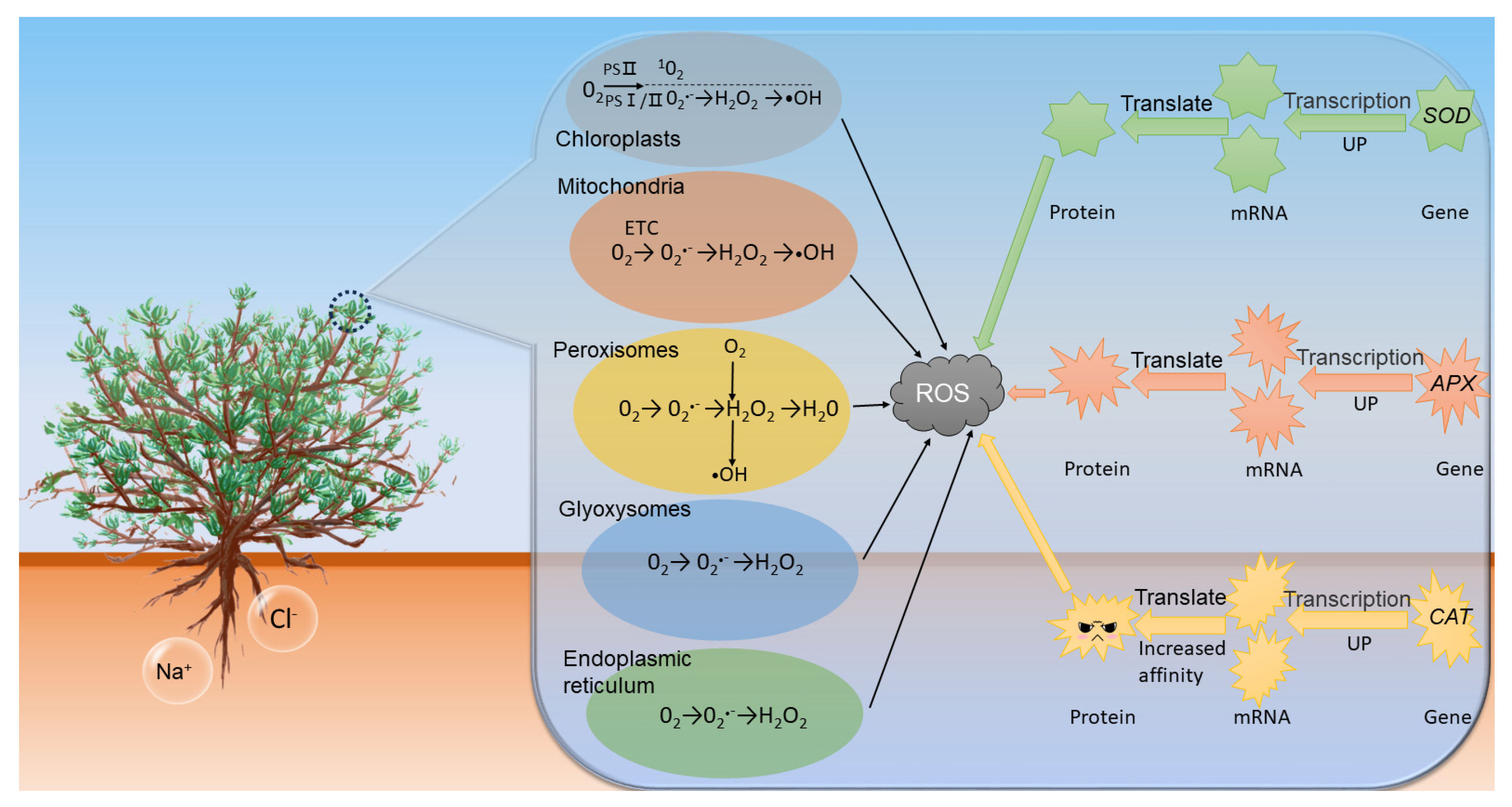
1. Introduction
2. Materials and Methods
2.1. Experimental Materials
2.2. Transcriptome Sequencing and Transcript Assembly
2.3. Enzyme Activity Determination
2.4. Gene Identification
2.5. Reconstruction of Phylogenetic Relationships and Analysis of Gene Structure, Conserved Motifs and Conserved Domains of Proteins
2.6. Functional Annotation of Genes and Prediction of Protein Interactions
2.7. Gene Expression and qRT-PCR Analysis
2.8. Physical and Chemical Properties and Molecular Evolution Analysis of Protein
2.9. Protein Structural Modeling and Comparison
3. Results
3.1. Gene Identification Results
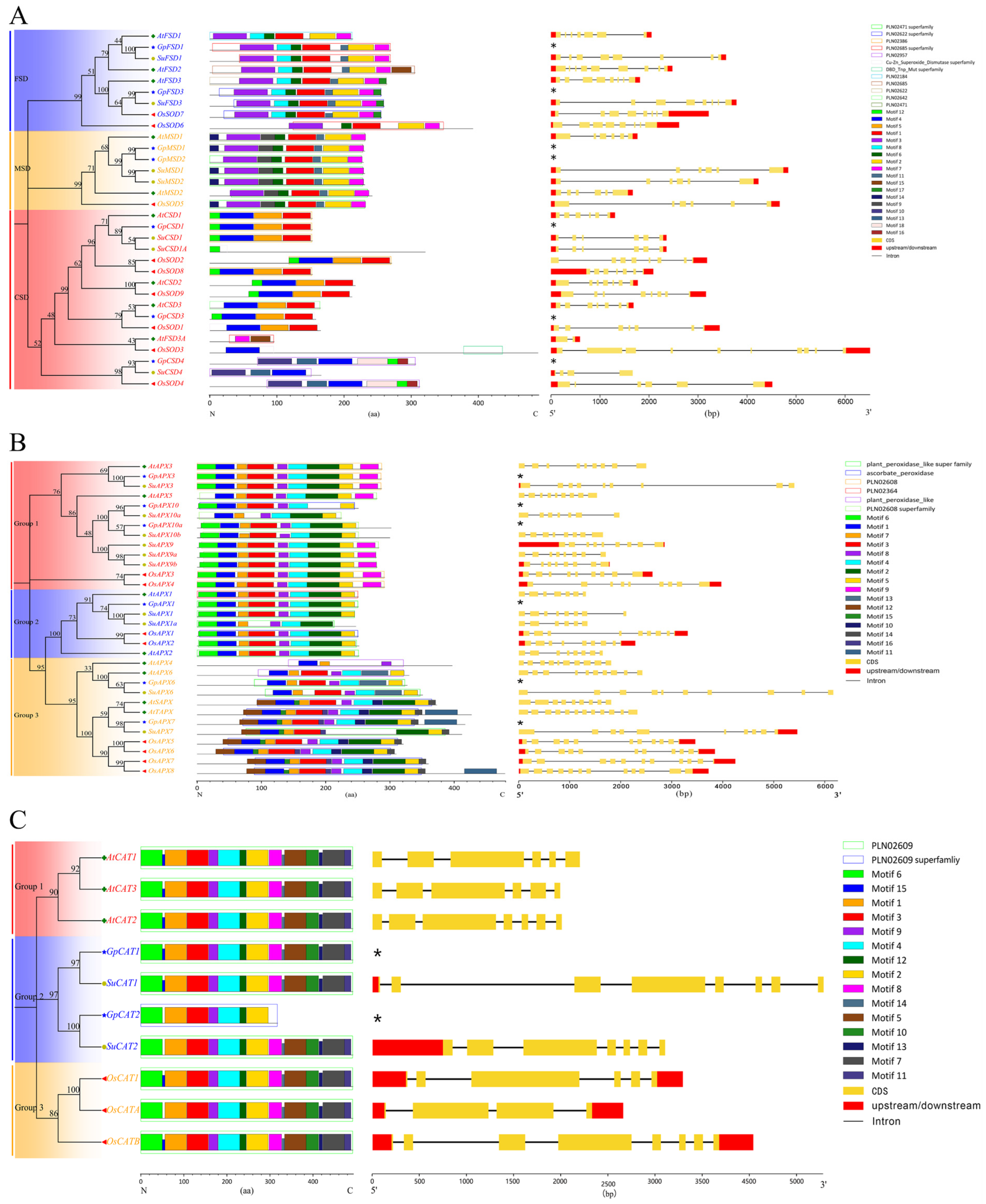
3.2. Genetic Relationship, Gene Structure, Motifs and Domains
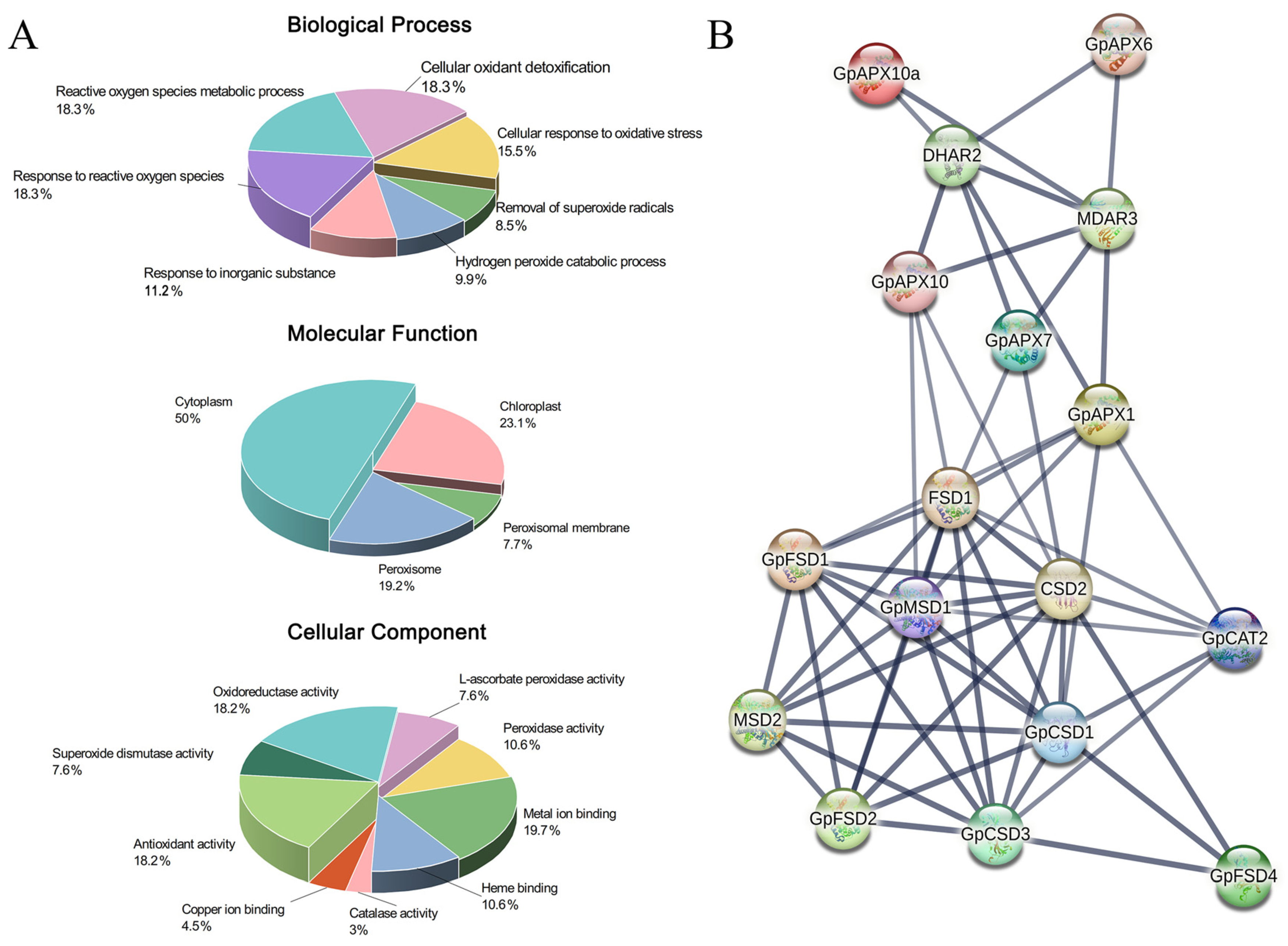
3.3. Gene Function Annotation and Protein Interaction Prediction
3.4. Physical and Chemical Properties and Subcellular Localization of the Proteins
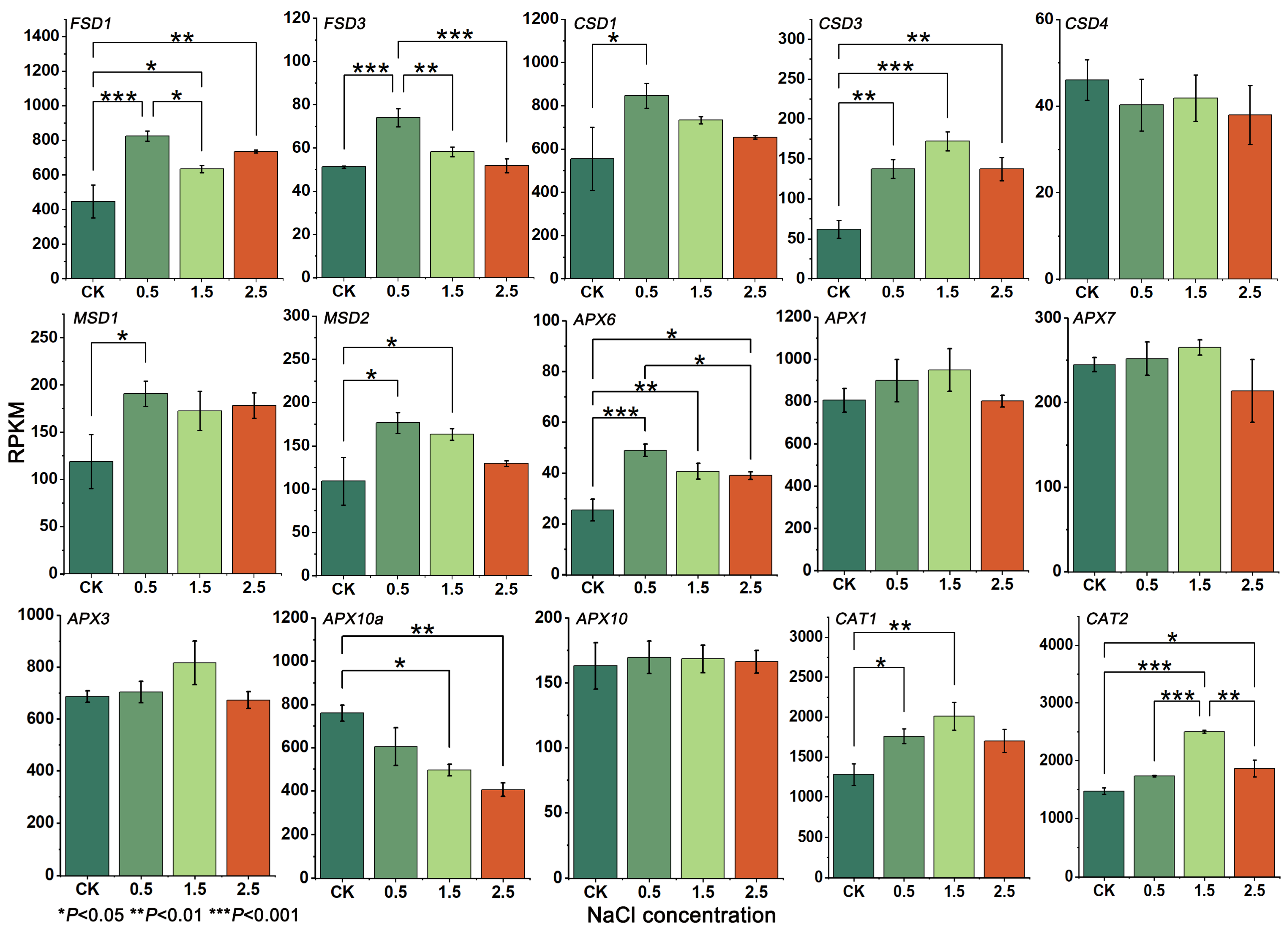
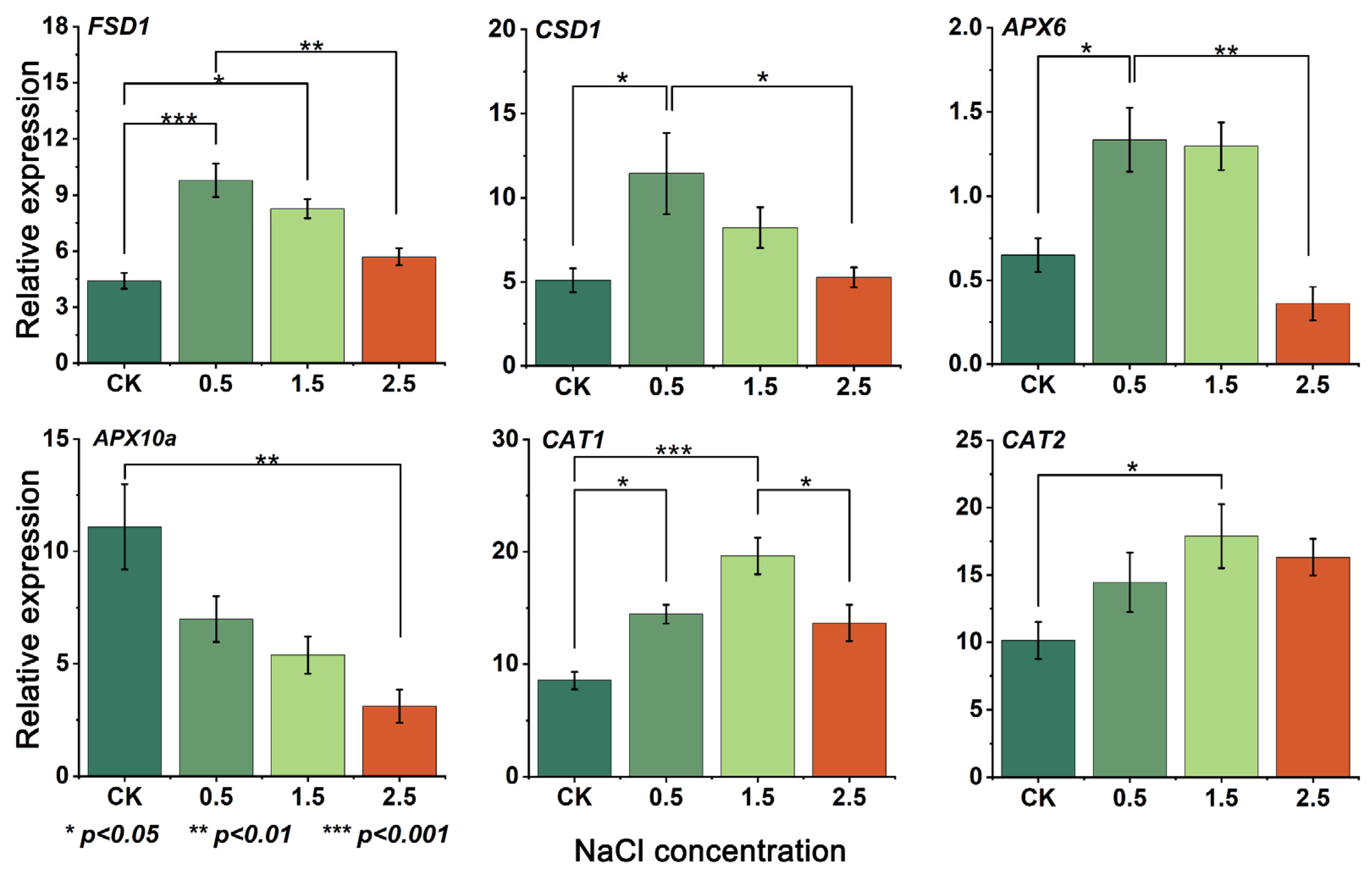
3.5. Gene Expression and RT-qPCR Analysis
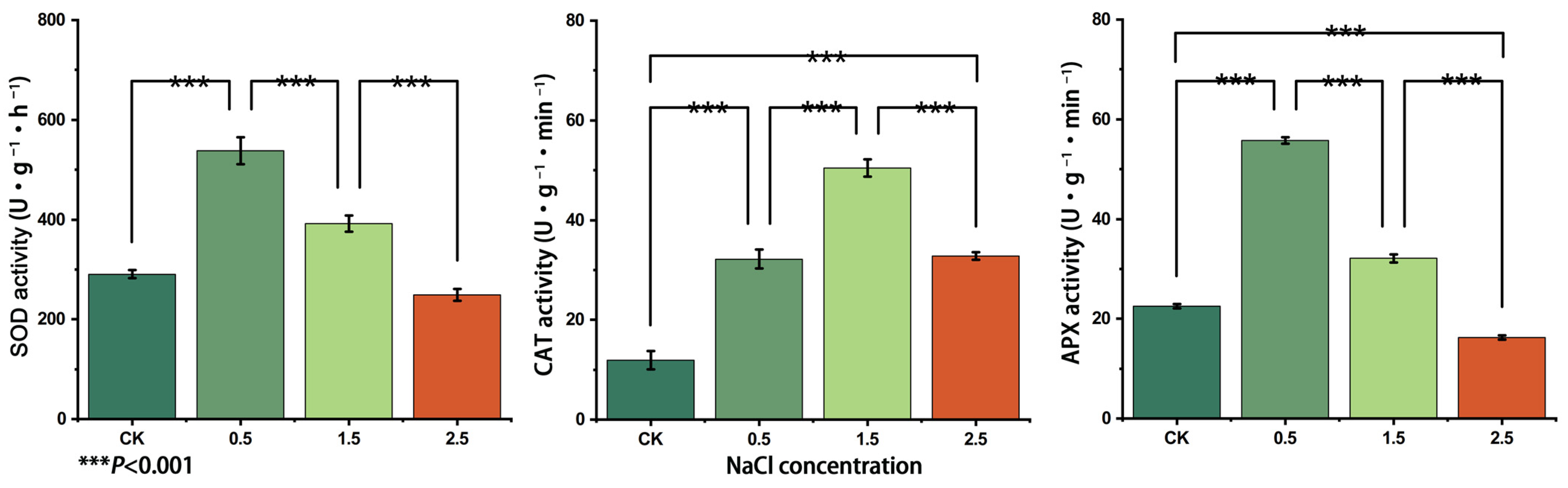
3.6. Changes in SOD, APX and CAT Enzyme Activities
3.7. Molecular Evolutionary Analysis
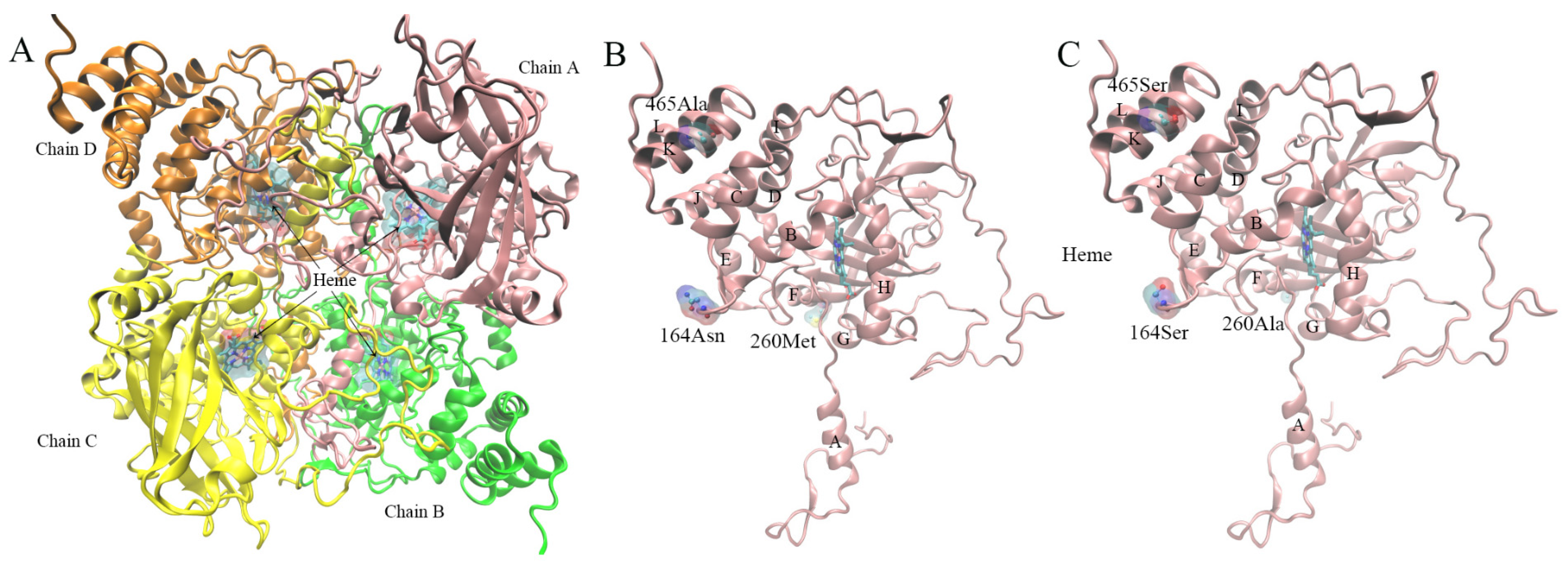
3.8. Protein Model Construction and Quality Evaluation
3.8.1. Comparison of Physical and Chemical Properties of Proteins
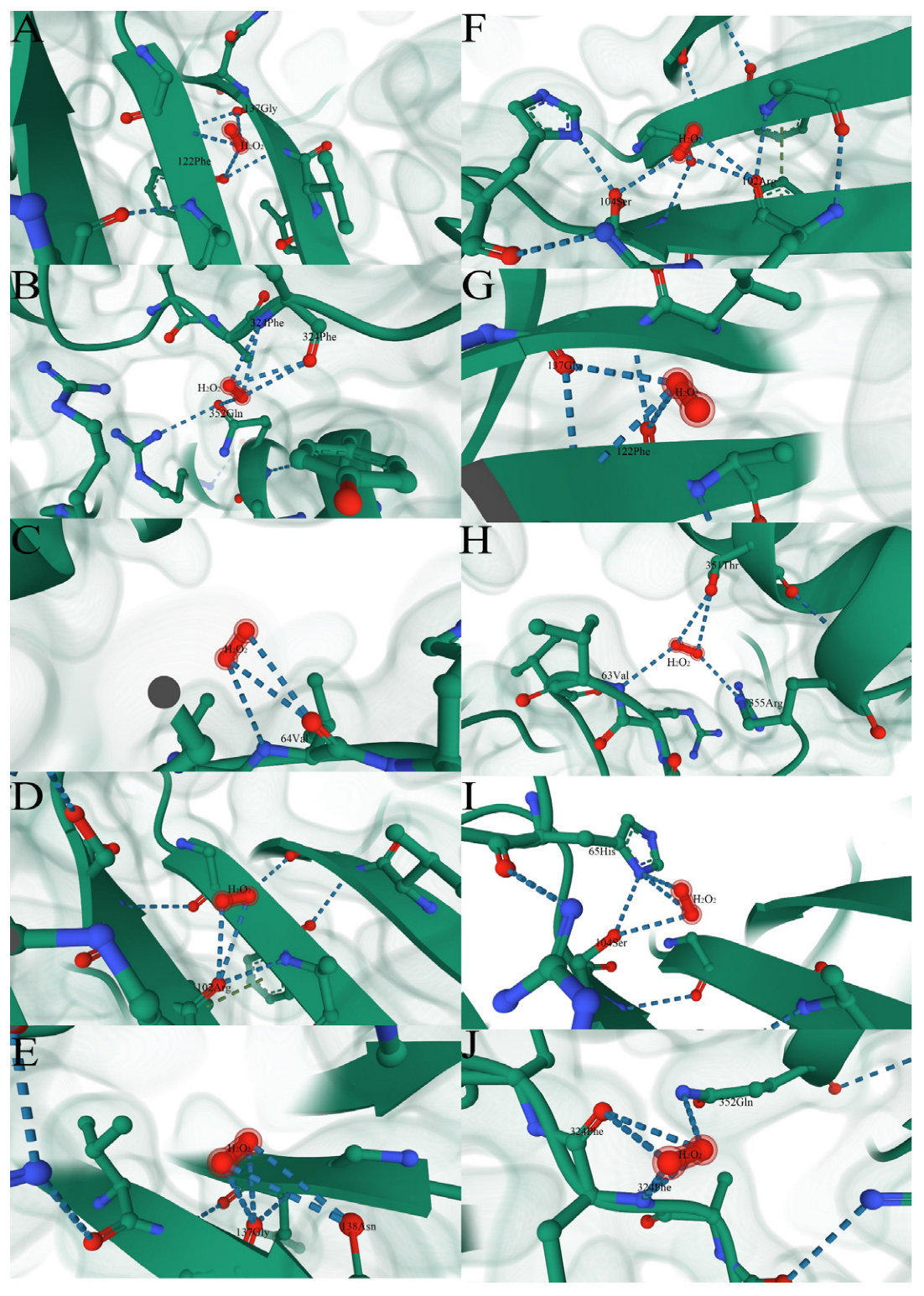
3.8.2. Property Analysis of Active Center
3.8.3. Protein Molecular Docking
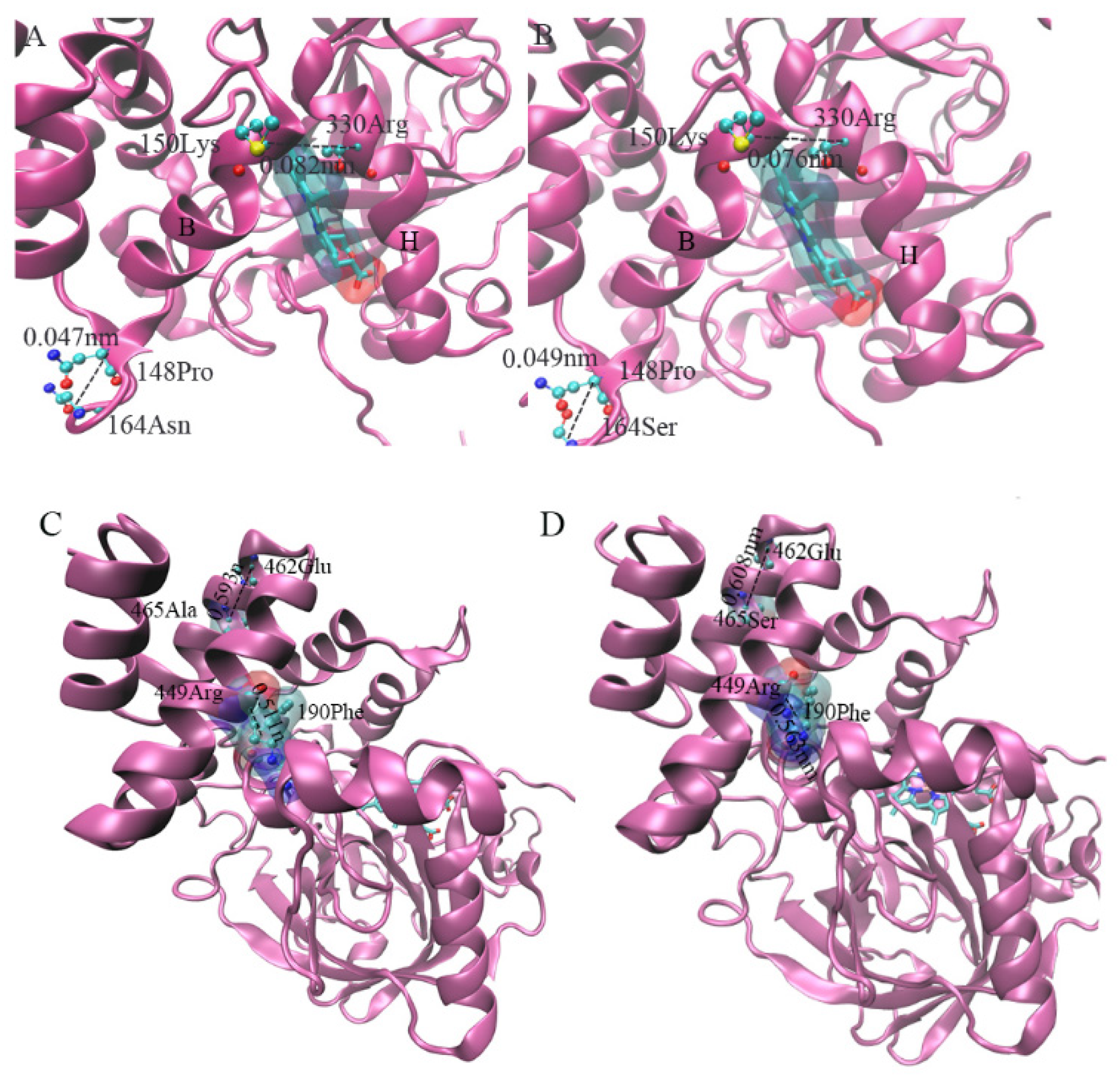
3.8.4. Prediction of Site Mutation and Changes in Protein Conformation
4. Discussion
4.1. Gene identification Results
4.2. Expression Patterns of Antioxidant Enzyme-Related Genes in Leaf Tissues
4.3. Effect of Positive Selection Mutation Site of CAT1 Gene on Protein Structure
4.4. Effect of Positive Selection Mutation Site of CAT1 Gene on Protein Structure
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Hailu, B.; Mehari, H. Impacts of soil salinity/sodicity on soil-water relations and plant growth in dry land areas: A review. J. Nat. Sci. Res. 2021, 12, 1–10. [Google Scholar] [CrossRef]
- Zhang, J. Amelioration and Utilization of Saline–Alkali Land. In Study of Ecological Engineering of Human Settlements; Springer: Singapore, 2020; Volume 3, pp. 83–92. ISBN 978-981-15-1372-5. [Google Scholar] [CrossRef]
- Mahajan, S.; Pandey, G.K.; Tuteja, N. Calcium- and salt-stress signaling in plants: Shedding light on SOS pathway. Arch. Biochem. Biophys. 2008, 471, 146–158. [Google Scholar] [CrossRef]
- Che, Y.; Yao, T.; Wang, H.; Wang, Z.; Zhang, H.; Sun, G.; Zhang, H. Potassium ion regulates hormone, Ca2+ and H2O2 signal transduction and antioxidant activities to improve salt stress resistance in tobacco. Plant Physiol. Biochem. 2022, 186, 40–51. [Google Scholar] [CrossRef]
- Chen, L.; Meng, Y.; Bai, Y.; Yu, H.; Qian, Y.; Zhang, D.; Zhou, Y. Starch and sucrose metabolism and plant hormone signaling pathways play crucial roles in aquilegia salt stress adaption. Int. J. Mol. Sci. 2023, 24, 3948. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Fu, C.; Li, G.; Khan, M.N.; Wu, H. Ros homeostasis and plant salt tolerance: Plant nanobiotechnology updates. Sustainability 2021, 13, 3552. [Google Scholar] [CrossRef]
- Liu, C.; Mao, B.; Yuan, D.; Chu, C.; Duan, M. Salt tolerance in rice: Physiological responses and molecular mechanisms. Crop. J. 2022, 10, 13–25. [Google Scholar] [CrossRef]
- Rajput, V.D.; Harish; Singh, R.K.; Verma, K.K.; Sharma, L.; Quiroz-Figueroa, F.R.; Meena, M.; Gour, V.S.; Minkina, T.; Sushkova, S.; et al. Recent developments in enzymatic antioxidant defence mechanism in plants with special reference to abiotic stress. Biology 2021, 10, 267. [Google Scholar] [CrossRef]
- Dumanović, J.; Nepovimova, E.; Natić, M.; Kuča, K.; Jaćević, V. The significance of reactive oxygen species and antioxidant defense system in plants: A concise overview. Front. Plant Sci. 2021, 11, 552969. [Google Scholar] [CrossRef]
- Wang, W.; Xia, M.X.; Chen, J.; Deng, F.N.; Shen, F.F. Gene expression characteristics and regulation mechanisms of superoxide dismutase and its physiological roles in plants under stress. Biochemistry 2016, 81, 465–480. [Google Scholar] [CrossRef]
- Nyuk, L.M.; Wan, A.C.L.; Nisrin, A.K.; Mohamad, M.; Zaidah, R.; Aziz, A.; Su, D.L.; Mohd, R.I. Susceptibility and tolerance of rice crop to salt threat: Physiological and metabolic inspections. PLoS ONE 2018, 13, e0192732. [Google Scholar] [CrossRef]
- Chew, O.; Whelan, J.; Millar, A.H. Molecular definition of the ascorbate-glutathione cycle in Arabidopsis mitochondria reveals dual targeting of antioxidant defenses in plants. J. Biol. Chem. 2003, 278, 46869–46877. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, F.K.; Menezes-benavente, L.; Margis, R.; Margis-Pinheir, M. Analysis of the molecular evolutionary history of the ascorbate peroxidase gene family: Inferences from the rice genome. J. Mol. Evol. 2004, 59, 761–770. [Google Scholar] [CrossRef] [PubMed]
- Saxena, S.C.; Salvi, P.; Kamble, N.U.; Majee, M.; Arora, S. Ectopic overexpression of cytosolic Ascorbate peroxidase gene (APX1) improves salinity stress tolerance in Brassica juncea by strengthening antioxidative defense mechanism. Acta Physiol. Plant. 2020, 42, 45. [Google Scholar] [CrossRef]
- Shalata, A.; Mittova, V.; Volokita, M.; Guy, M.; Tal, M. Response of the cultivated tomato and its wild salt-tolerant relative Lycopersicon pennellii to salt-dependent oxidative stress: The root antioxidative system. Plant Physiol. 2001, 112, 487–494. [Google Scholar] [CrossRef]
- Guan, Q.; Takano, T.; Liu, S. Genetic transformation and analysis of rice Osapx2 gene in Medicago sativa. PLoS ONE 2012, 7, e41233. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Yang, Y.; Jiang, L.; Liu, S. The catalase gene family in cucumber: Genome-wide identification and organization. Genet. Mol. Biol. 2016, 39, 408–415. [Google Scholar] [CrossRef]
- Gondim, F.A.; Gomes-Filho, E.; Costa, J.H.; Alencar, N.L.M.; Prisco, J.T. Catalase plays a key role in salt stress acclimation induced by hydrogen peroxide pretreatment in maize. Plant Physiol. Biochem. 2012, 56, 62–71. [Google Scholar] [CrossRef]
- Ma, S.M.; Zhang, M.L.; Sanderson, S.C. Phylogeography of the rare Gymnocarpos przewalskii (Caryophyllaceae): Indications of multiple glacial refugia in north-western China. Aust. J. Bot. 2012, 60, 20–31. [Google Scholar] [CrossRef]
- Wang, L.; Huang, H.X.; Shan, L.S.; Gao, T.P.; Zhang, X.H. Population structure and spatial distribution pattern of Gymnocarpos przewalskii in Anxi extra-arid desert national nature reserve. J. Lanzhou Univ. (Nat. Sci.) 2015, 51, 690–698, (In Chinese with English Summary). [Google Scholar] [CrossRef]
- Huang, H.X.; Yang, Q.Q.; Cui, P.; Lu, G.; Han, G.J. Changes in morphological and physiological characteristics of Gymnocarpos przewalskii roots in response to water stress. Acta Prataculturae Sin. 2021, 30, 197–207, (In Chinese with English Summary). [Google Scholar] [CrossRef]
- Wang, Z.B.; Gao, Q.X.; Sun, J.Z.; Ma, Q.L. Study on biological characteristics of rare endangered plant Gymnocarpos przewalskii. Resour. Dev. Mark. 2009, 25, 481–509, (In Chinese with English Summary). [Google Scholar]
- Lian, Z.H.; Yang, H.L.; Wu, F.M.; Huang, H.X. Effect of NaCl or PEG stress on seed germination of Gymnocarpos przewalskii. Sci. Soil Water Conserv. 2017, 15, 89–96, (In Chinese with English Summary). [Google Scholar] [CrossRef]
- Zhou, X.J.; Huang, H.X.; Zhang, J.X.; Ma, B.D.; Lu, G.; Qi, J.W.; Zhang, T.; Zhu, Z. Effects of salt stress on photosynthetic characteristics of Gymnocarpos przewalskii seedlings. Acta Prataculturae Sin. 2023, 32, 75–83, (In Chinese with English Summary). [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.D.; et al. Trinity: Reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 2013, 29, 644. [Google Scholar] [CrossRef]
- Subin, M.P.; Francis, S. Phytotoxic effects of cadmium on seed germination, early seedling growth and antioxidant enzyme activities in Cucurbita maxima duchesne. Int. Res. J. Biol. Sci. 2013, 2, 40–47. [Google Scholar]
- Sarwar, M.; Anjum, S.; Alam, M.W.; Ali, Q.; Ayyub, C.M.; Haider, M.S.; Ashraf, M.I.; Mahboob, W. Triacontanol regulates morphological traits and enzymatic activities of salinity affected hot pepper plants. Sci. Rep. 2022, 12, 3736. [Google Scholar] [CrossRef]
- Pereira, G.J.G.; Molina, S.M.G.; Lea, P.J.; Azevedo, R.A. Activity of antioxidant enzymes in response to cadmium in Crotalaria juncea. Plant Soil 2022, 239, 123–132. [Google Scholar] [CrossRef]
- Gascuel, O. New algorithms and methods to estimate Maximum-Likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.C.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME suite. Nucleic Acids Res. 2015, 43, 39. [Google Scholar] [CrossRef]
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. eggNOG-mapper v2: Functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Rui, X. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 194–1202. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, 638–646. [Google Scholar] [CrossRef]
- Langmead, B.; Wilks, C.; Antonescu, V.; Charles, R. Scaling read aligners to hundreds of threads on general-purpose processors. Bioinformatics 2019, 35, 421–432. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, 296–303. [Google Scholar] [CrossRef] [PubMed]
- Webb, B.; Sali, A. Comparative protein structure modeling using Modeller. Curr. Protoc. Bioinf. 2016, 54, 5.6.1–5.6.37. [Google Scholar] [CrossRef] [PubMed]
- Pontius, J.; Richelle, J.; Wodak, S.J. Deviations from standard atomic volumes as a quality measure for protein crystal structures. J. Mol. Biol. 1996, 264, 121–136. [Google Scholar] [CrossRef]
- Sinha, K.K.; Bilokapic, S.; Du, Y.M.; Malik, D.; Halic, M. Histone modifications regulate pioneer transcription factor cooperativity. Nature 2023, 619, 378–384. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Forli, S.; Huey, R.; Pique, M.E.; Sanner, M.F.; Goodsell, D.S.; Olson, A.J. Computational protein-ligand docking and virtual drug screening with the AutoDock suite. Nat. Protoc. 2016, 11, 905–919. [Google Scholar] [CrossRef]
- Ittisoponpisan, S.; Islam, S.A.; Khanna, T.; Alhuzimi, E.; David, A.; Sternberg, M.J.E. Can predicted protein 3D structures provide reliable insights into whether missense variants are disease associated? J. Mol. Biol. 2019, 431, 2197–2212. [Google Scholar] [CrossRef]
- Chen, Y.; Wu, H.; Hao, Z.; Zhu, L.; Lu, L.; Shi, J.; Chen, J. The Identification and Expression Analysis of the Liriodendron chinense (Hemsl.) Sarg. SOD Gene Family. Forests 2023, 14, 628. [Google Scholar] [CrossRef]
- Hu, X.X.; Hao, C.; Cheng, Z.M.; Zhong, Y. Genome-wide identification, characterization, and expression analysis of the grapevine superoxide dismutase (SOD) family. Int. J. Genom. 2019, 2019, 7350414. [Google Scholar] [CrossRef]
- Guo, D.; Zhang, Y.Y.; Du, M.; Zhou, B.Y.; Gao, Z.H.; Zhao, M.; Yuan, H.F. Bioinformatics analysis of superoxide dismutase gene family in rape. Mol. Plant Breed. 2020, 18, 367–373, (In Chinese with English Summary). [Google Scholar] [CrossRef]
- Tyagi, S.; Shumayla; Verma, P.C.; Singh, K.; Upadhyay, S.K. Molecular characterization of ascorbate peroxidase (APX) and APX-related (APX-R) genes in Triticum aestivum L. Genomics 2020, 112, 4208–4223. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.Q.; Li, J.T.; Bing, J.; Zhang, G.F. The role analysis of APX gene family in the growth and developmental processes and in response to abiotic stresses in Arabidopsis thaliana. Hereditas 2019, 41, 534–547. [Google Scholar] [CrossRef]
- Hong, C.Y.; Hsu, Y.T.; Tsai, Y.C.; Kao, C.H. Expression of ascorbate peroxidase 8 in roots of rice (Oryza sativa L.) seedlings in response to NaCl. J. Exp. Bot. 2007, 58, 3273–3283. [Google Scholar] [CrossRef] [PubMed]
- Leng, X.; Wang, H.; Zhang, S.; Qu, C.; Yang, C.; Xu, Z.; Liu, G. Identification and characterization of the APX gene family and its expression pattern under phytohormone treatment and abiotic stress in Populus trichocarpa. Genes 2021, 12, 334. [Google Scholar] [CrossRef]
- Liao, G.L.; Liu, Q.; Li, Y.Q.; Zhong, M.; Huang, C.H.; Jia, D.F.; Xu, X.B. Identification and expression profiling analysis of ascorbate peroxidase gene family in Actinidia chinensis (Hongyang). J. Plant Res. 2020, 133, 715–726. [Google Scholar] [CrossRef]
- Du, Y.Y.; Wang, P.C.; Chen, J.; Song, C.P. Comprehensive functional analysis of the catalase gene family in Arabidopsis thaliana. J. Integr. Plant Biol. 2008, 50, 1318–1326. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.N.; Wei, Y.L.; Zhang, J.J.; Ding, D.; Liu, M.; Shao, W.J.; Li, J.X.; Xu, J.Y. Bioinformatic analysis of catalase gene family of Arabidopsis and Maize. Mol. Plant Breed. 2021, 12, 1–10. [Google Scholar] [CrossRef]
- Jiang, W.X.; Ye, Q.; Wu, Z.; Zhang, Q.Y.; Wang, L.H.; Liu, J.L.; Hu, X.F.; Guo, D.D.; Wang, X.Q.; Zhang, Z.L.; et al. Analysis of CAT Gene Family and Functional Identification of OsCAT3 in Rice. Genes 2023, 14, 138. [Google Scholar] [CrossRef]
- Palatnik, J.F.; Valle, E.M.; Federico, M.L.; Gómez, L.D.; Melchiorre, M.N.; Paleo, A.D.; Carrillo, N.; Acevedo, A. Status of antioxidant metabolites and enzymes in a Catalase-deficient mutant of barley (Hordeum vulgare L.). Plant Sci. 2002, 162, 363–371. [Google Scholar] [CrossRef]
- Raza, A.; Su, W.; Gao, A.; Mehmood, S.S.; Hussain, M.A.; Nie, W.; Lv, Y.; Zou, X.; Zhang, X. Catalase (CAT) gene family in rapeseed (Brassica napus L.): Genome-wide analysis, identification, and expression pattern in response to multiple hormones and abiotic stress conditions. Int. J. Mol. Sci. 2021, 22, 4281. [Google Scholar] [CrossRef]
- Mansoor, S.; Ali, W.O.; Lone, J.K.; Manhas, S.; Kou, N.; Alam, P.; Ahmad, A.; Ahmad, P. Reactive oxygen species in plants: From source to sink. Antioxidants 2022, 11, 225. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.S.; Webb, R.P.; Holaday, A.S.; Allen, R.D. Overexpression of Superoxide Dismutase Protects Plants from Oxidative Stress (Induction of Ascorbate Peroxidase in Superoxide Dismutase-Overexpressing Plants). Plant Physiol. 1993, 103, 1067–1073. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, L.T.; Chen, Y.Y.; Wang, S.Y.; Fang, Y.X.; Zhang, X.Q.; Wu, Y.H.; Xue, D.W. Genome-wide identification of the SOD gene family and expression analysis under drought and salt stress in barley. Plant Growth Regul. 2021, 94, 49–60. [Google Scholar] [CrossRef]
- Pandey, S.; Fartyal, D.; Agarwal, A.; Shukla, T.; James, D.; Kaul, T.; Negi, Y.K.; Arora, S.; Reddy, M.K. Abiotic stress tolerance in plants: Myriad roles of Ascorbate peroxidase. Plant Sci. 2017, 8, 581. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.J.; Li, G.R.; Huang, F.L.; Li, W.; Cong, A.Q.; Chen, Y.S. Research progress of reactive oxygen species and plant antioxidant system. Anhui Agric. Sci. 2017, 45, 1–3, (In Chinese with English Summary). [Google Scholar] [CrossRef]
- Patrick, F. Darwin’s goldmine is still open: Variation and selection run the world. Front. Cell. Inf. Microbio. 2012, 2, 106. [Google Scholar] [CrossRef][Green Version]
- Alfonso-Prieto, M.; Biarnés, X.; Vidossich, P.; Rovira, C. The Molecular Mechanism of the catalase Reaction. J. Am. Chem. Soc. 2009, 131, 11751–11761. [Google Scholar] [CrossRef]
- Chen, H.L.; Li, M.J.; Liu, C.Q.; Zhang, H.B.; Xian, M.; Liu, H.Z. Enhancement of the catalytic activity of Isopentenyl diphosphate isomerase (IDI) from Saccharomyces cerevisiae through random and site-directed mutagenesis. Microb. Cell Fact. 2018, 17, 65. [Google Scholar] [CrossRef] [PubMed]
- Costa, P.H.A.D.; Neto, A.D.D.A.; Bezerra, M.A.; Prisco, J.T.; Gomes-Filho, E. Antioxidant-enzymatic system of two sorghum genotypes differing in salt tolerance. Braz. J. Plant Physiol. 2005, 17, 353–361. [Google Scholar] [CrossRef]


| Polypeptide | Number of Amino Acids | Molecular Weight | Isoelectric Point | Average Hydrophilicity | Instability Index |
|---|---|---|---|---|---|
| GpCAT1 | 478 | 55,052.26 | 6.75 | −0.522 | 31.32 |
| AsCAT1 | 478 | 55,163.38 | 6.78 | −0.518 | 34.67 |
| Properties | GpCAT1 | AsCAT1 |
|---|---|---|
| Depth | 15.47 Å | 14.4 Å |
| Hydrophobicity | −0.73 | −0.72 |
| Metal | 1 | 1 |
| Surface | 4.18 Å2 | 3.90 Å2 |
| Volume | 227.33 Å3 | 186.88 Å3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qi, J.; Luo, Y.; Huang, H.; Lu, S.; Zhao, F.; Deng, Z.; Qiu, Y. Molecular Mechanism of Response and Adaptation of Antioxidant Enzyme System to Salt Stress in Leaves of Gymnocarpos przewalskii. Plants 2023, 12, 3370. https://doi.org/10.3390/plants12193370
Qi J, Luo Y, Huang H, Lu S, Zhao F, Deng Z, Qiu Y. Molecular Mechanism of Response and Adaptation of Antioxidant Enzyme System to Salt Stress in Leaves of Gymnocarpos przewalskii. Plants. 2023; 12(19):3370. https://doi.org/10.3390/plants12193370
Chicago/Turabian StyleQi, Jianwei, Yongzhong Luo, Haixia Huang, Songsong Lu, Fei Zhao, Zhuo Deng, and Yingde Qiu. 2023. "Molecular Mechanism of Response and Adaptation of Antioxidant Enzyme System to Salt Stress in Leaves of Gymnocarpos przewalskii" Plants 12, no. 19: 3370. https://doi.org/10.3390/plants12193370
APA StyleQi, J., Luo, Y., Huang, H., Lu, S., Zhao, F., Deng, Z., & Qiu, Y. (2023). Molecular Mechanism of Response and Adaptation of Antioxidant Enzyme System to Salt Stress in Leaves of Gymnocarpos przewalskii. Plants, 12(19), 3370. https://doi.org/10.3390/plants12193370





