Identification and Characterization of Beneficial Soil Microbial Strains for the Formulation of Biofertilizers Based on Native Plant Growth-Promoting Microorganisms Isolated from Northern Mexico
Abstract
:1. Introduction
2. Results
2.1. Isolation of Native Soil Microorganisms and Evaluation of Plant Growth-Promoting Traits
2.2. Preparation of Bio-Formulations and In Vivo Evaluation in a Seedbed Assay
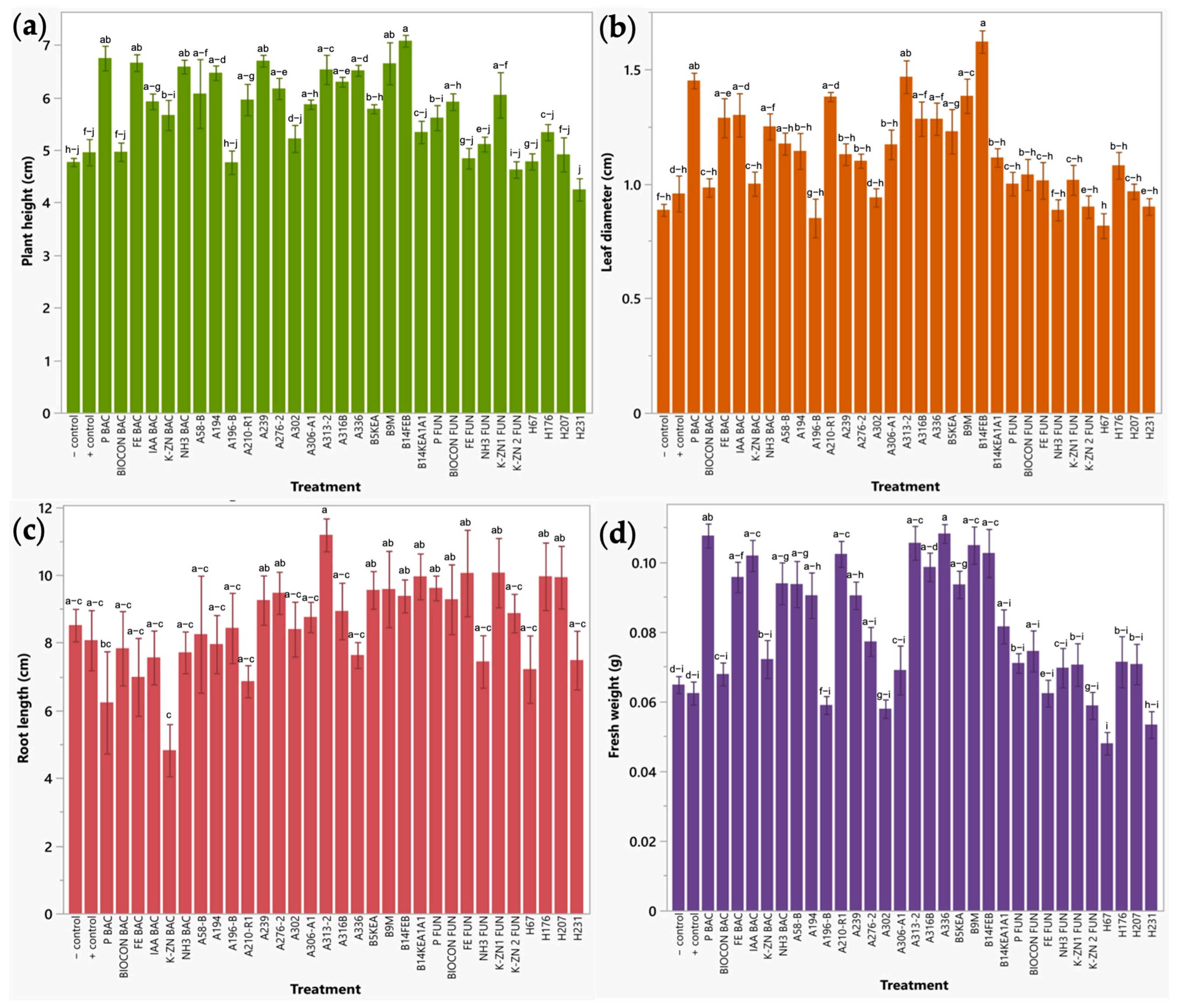
2.2.1. Early Plant Response of Broccoli Sprouts to Bio-Formulations in a Seedbed Assay
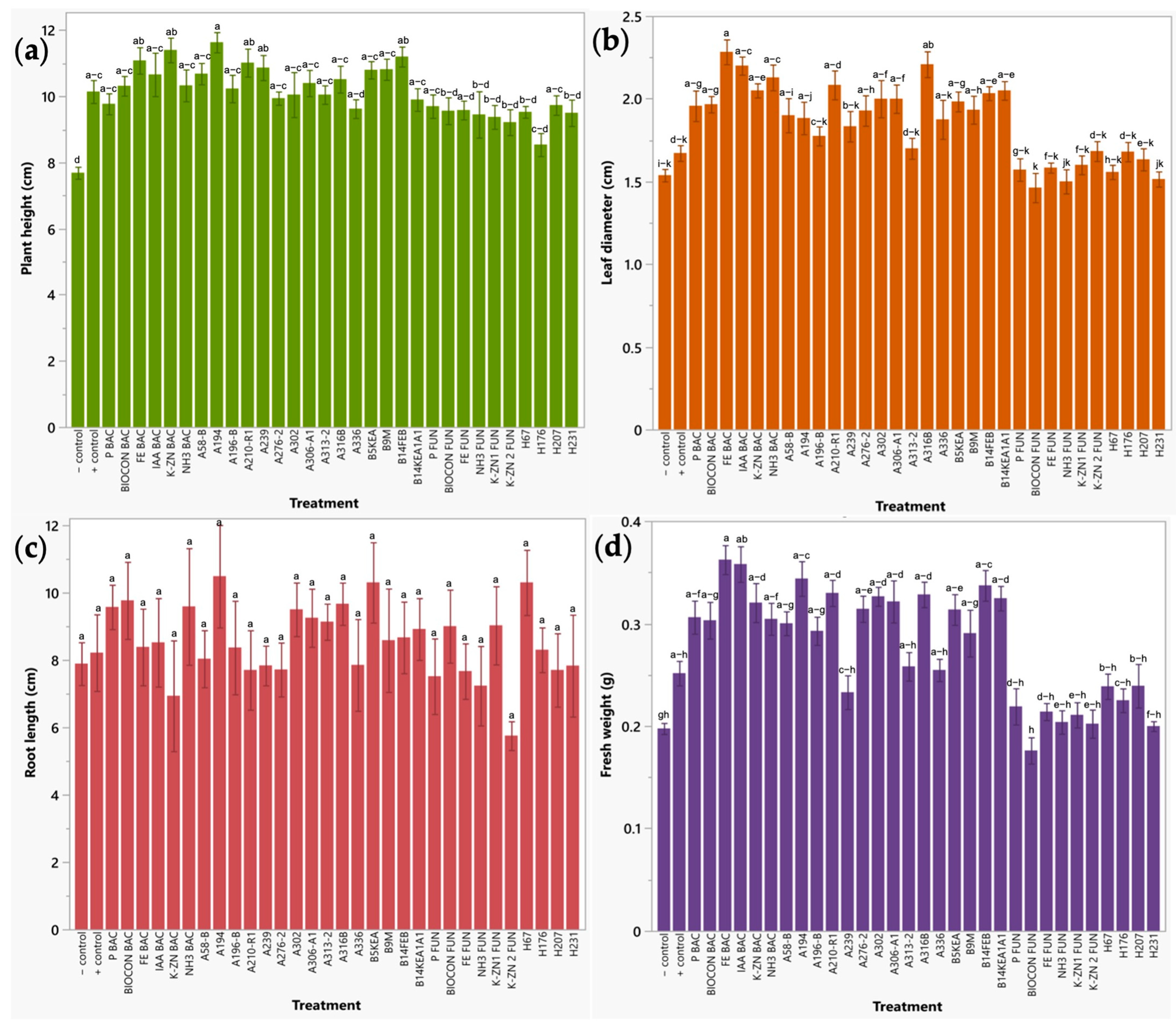
2.2.2. Early Plant Response of Radish Sprouts to Bio-Formulations in a Seedbed Assay
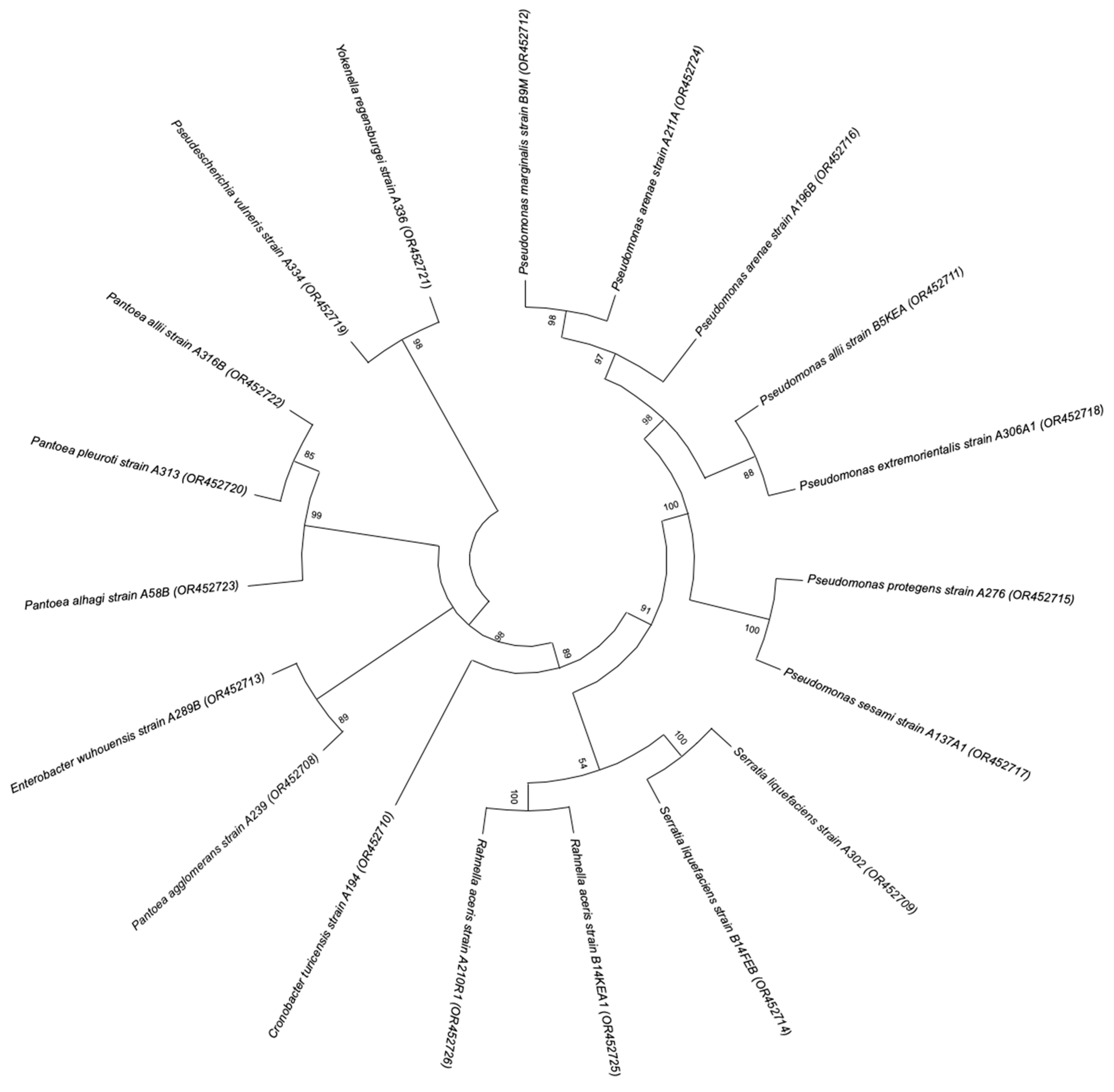
2.3. Identification of Potential PGP Microbial Isolates
3. Discussion
4. Materials and Methods
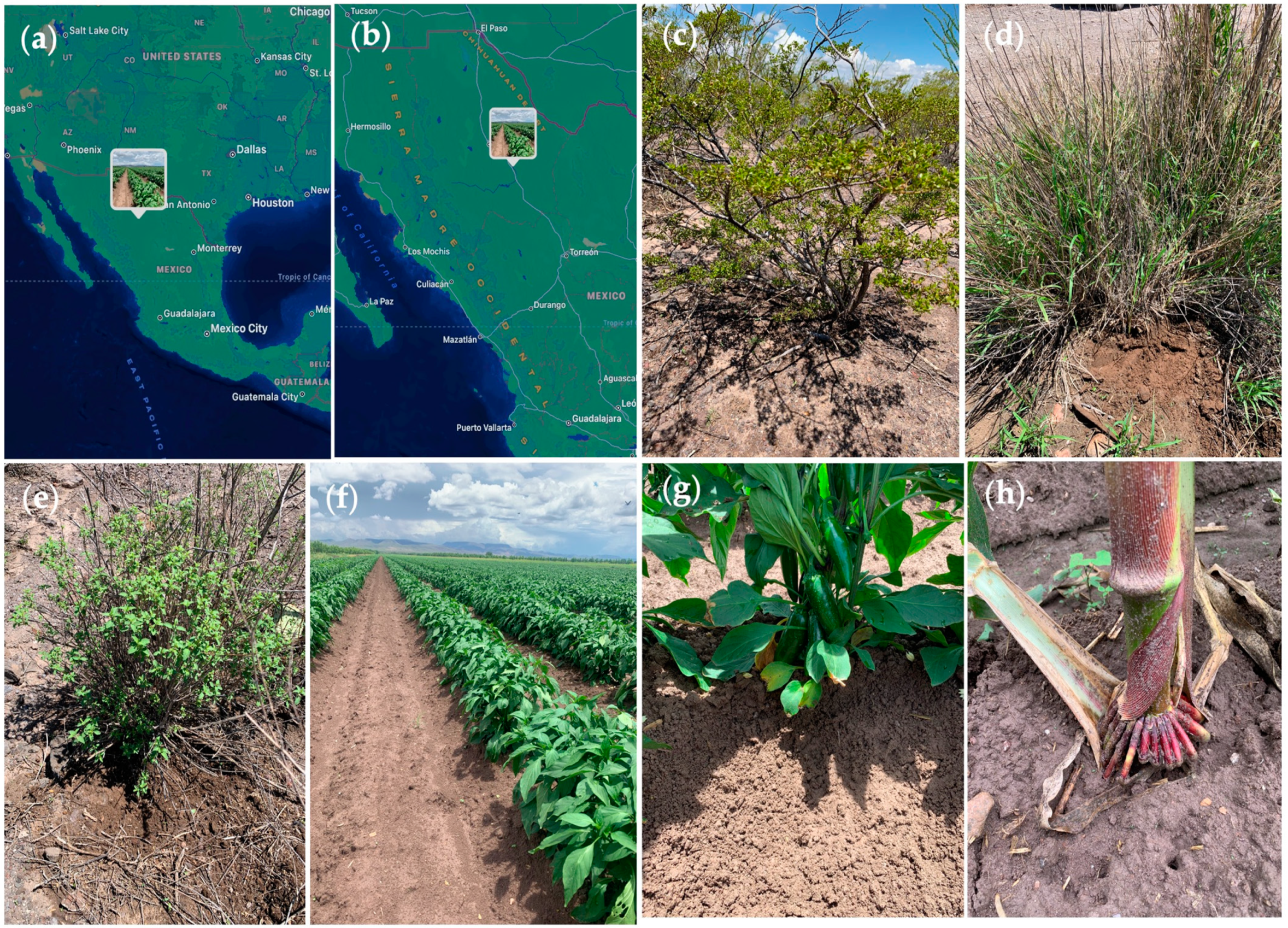
4.1. Sampling
4.2. Isolation of Native Soil Microorganisms
4.3. Evaluation of Plant Growth-Promoting Traits
4.3.1. Potassium, Phosphate, and Zinc Solubilization
4.3.2. Nitrogen Fixation
4.3.3. Ammonia Production
4.3.4. Indole-3-Acetic Acid Secretion
4.3.5. Siderophores Production
4.3.6. Antifungal Activity against Phytopathogenic Fusarium oxysporum
4.4. Preparation of Bacterial and Fungal Bio-Formulations
4.5. In Vivo Evaluation of Bio-Formulations in a Seedbed Assay
4.6. Identification of Potential PGP Microbial Isolates
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Basu, A.; Prasad, P.; Das, S.N.; Kalam, S.; Sayyed, R.Z.; Reddy, M.S.; El Enshasy, H. Plant growth promoting rhizobacteria (PGPR) as green bioinoculants: Recent developments, constraints, and prospects. Sustainability 2021, 13, 1140. [Google Scholar] [CrossRef]
- Mohanty, P.; Singh, P.K.; Chakraborty, D.; Mishra, S.; Pattnaik, R. Insight into the role of PGPR in sustainable agriculture and environment. Front. Sustain. Food Syst. 2021, 5, 667150. [Google Scholar] [CrossRef]
- Hashem, A.; Tabassum, B.; Abd_Allah, E.F. Bacillus subtilis: A plant-growth promoting rhizobacterium that also impacts biotic stress. Saudi J. Biol. Sci. 2019, 26, 1291–1297. [Google Scholar] [CrossRef] [PubMed]
- Kour, D.; Rana, K.L.; Yadav, N.; Yadav, A.N.; Kumar, A.; Meena, V.S.; Singh, B.; Chauhan, V.S.; Dhaliwal, H.S.; Saxena, A.K. Rhizospheric Microbiomes: Biodiversity, Mechanisms of Plant Growth Promotion, and Biotechnological Applications for Sustainable Agriculture. In Plant Growth Promoting Rhizobacteria for Agricultural Sustainability; Kumar, A., Meena, V., Eds.; Springer: Singapore, 2019; pp. 19–65. [Google Scholar] [CrossRef]
- Ramakrishna, W.; Yadav, R.; Li, K. Plant growth promoting bacteria in agriculture: Two sides of a coin. Appl. Soil Ecol. 2019, 138, 10–18. [Google Scholar] [CrossRef]
- Vasseur-Coronado, M.; du Boulois, H.D.; Pertot, I.; Puopolo, G. Selection of plant growth promoting rhizobacteria sharing suitable features to be commercially developed as biostimulant products. Microbiol. Res. 2021, 245, 126672. [Google Scholar] [CrossRef]
- ALKahtani, M.D.; Fouda, A.; Attia, K.A.; Al-Otaibi, F.; Eid, A.M.; Ewais, E.E.D.; Hijri, M.; St-Arnaud, M.; Hassan, S.E.-D.; Khan, N.; et al. Isolation and characterization of plant growth promoting endophytic bacteria from desert plants and their application as bioinoculants for sustainable agriculture. Agronomy 2020, 10, 1325. [Google Scholar] [CrossRef]
- Naziya, B.; Murali, M.; Amruthesh, K.N. Plant growth-promoting fungi (PGPF) instigate plant growth and induce disease resistance in Capsicum annuum L. upon infection with Colletotrichum capsici (Syd.) Butler & Bisby. Biomolecules 2019, 10, 41. [Google Scholar] [CrossRef]
- Guardiola-Márquez, C.E.; Jacobo-Velázquez, D.A. Potential of enhancing anti-obesogenic agriceuticals by applying sustainable fertilizers during plant cultivation. Front. Sustain. Food Syst. 2022, 6, 1034521. [Google Scholar] [CrossRef]
- Mahdi, I.; Fahsi, N.; Hafidi, M.; Allaoui, A.; Biskri, L. Plant growth enhancement using rhizospheric halotolerant phosphate solubilizing bacterium Bacillus licheniformis QA1 and Enterobacter asburiae QF11 isolated from Chenopodium quinoa willd. Microorganisms 2020, 8, 948. [Google Scholar] [CrossRef]
- Guardiola-Márquez, C.E.; Santos-Ramírez, M.T.; Segura-Jiménez, M.E.; Figueroa-Montes, M.L.; Jacobo-Velázquez, D.A. Fighting Obesity-Related Micronutrient Deficiencies through Biofortification of Agri-Food Crops with Sustainable Fertilization Practices. Plants 2022, 11, 3477. [Google Scholar] [CrossRef]
- Guardiola-Marquez, C.E.; Pacheco, A.; Mora-Godinez, S.; Schüßler, A.; Gradilla-Hernández, M.S.; Senés-Guerrero, C. Septoglomus species dominate the arbuscular mycorrhiza of five crop plants in an arid region of northern Mexico. Symbiosis 2022, 87, 93–106. [Google Scholar] [CrossRef]
- Guardiola-Márquez, C.E.; Figueroa-Montes, M.L.; Pacheco Moscoa, A.; Senés-Guerrero, C. Native microbial consortia improve maize shoot and root systems at early developmental stages in a seedbed assay. Sci. Fungorum 2021, 51, e1329. [Google Scholar] [CrossRef]
- Miljaković, D.; Marinković, J.; Balešević-Tubić, S. The significance of Bacillus spp. in disease suppression and growth promotion of field and vegetable crops. Microorganisms 2020, 8, 1037. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.J.; Lilly, M.D.; Fox, R.I. The effect of agitation on the morphology and penicillin production of Penicillium chrysogenum. Biotechnol. Bioeng. 1990, 35, 1011–1023. [Google Scholar] [CrossRef]
- Diba, K.; Kordbacheh, P.; Mirhendi, S.H.; Rezaie, S.; Mahmoudi, M. Identification of Aspergillus species using morphological characteristics. Pak. J. Med. Sci. 2007, 23, 867. [Google Scholar]
- Chaudhary, D.K.; Khulan, A.; Kim, J. Development of a novel cultivation technique for uncultured soil bacteria. Sci. Rep. 2019, 9, 6666. [Google Scholar] [CrossRef]
- Lobo, C.B.; Tomás, M.S.J.; Viruel, E.; Ferrero, M.A.; Lucca, M.E. Development of low-cost formulations of plant growth-promoting bacteria to be used as inoculants in beneficial agricultural technologies. Microbiol. Res. 2019, 219, 12–25. [Google Scholar] [CrossRef]
- Adomako, M.O.; Roiloa, S.; Yu, F.H. Potential Roles of Soil Microorganisms in Regulating the Effect of Soil Nutrient Heterogeneity on Plant Performance. Microorganisms 2022, 10, 2399. [Google Scholar] [CrossRef]
- Santoyo, G.; Urtis-Flores, C.A.; Loeza-Lara, P.D.; Orozco-Mosqueda, M.D.C.; Glick, B.R. Rhizosphere colonization determinants by plant growth-promoting rhizobacteria (PGPR). Biology 2021, 10, 475. [Google Scholar] [CrossRef]
- Pascale, A.; Proietti, S.; Pantelides, I.S.; Stringlis, I.A. Modulation of the root microbiome by plant molecules: The basis for targeted disease suppression and plant growth promotion. Front. Plant Sci. 2020, 10, 1741. [Google Scholar] [CrossRef]
- Widawati, S. Isolation of Indole Acetic Acid (IAA) producing Bacillus siamensis from peat and optimization of the culture conditions for maximum IAA production. In Proceedings of the IOP Conference Series: Earth and Environmental Science, Bogor, Indonesia, 28–29 October 2019; IOP Publishing: Bristol, UK, 2020; Volume 572, p. 012025. [Google Scholar] [CrossRef]
- Marwanto, B.; Bustaman, H.; Handajaningsih, M.; Murcitro, B.G. Qualitative In Vitro Evaluation of Plant Growth Promoting Activity of Selected Microbial Isolates Used for Biofertilizer Application. In International Seminar on Promoting Local Resources for Sustainable Agriculture and Development (ISPLRSAD 2020); Advances in Biological Sciences Research; Atlantis Press: Amsterdam, The Netherlands, 2021; pp. 299–309. [Google Scholar] [CrossRef]
- Abedinzadeh, M.; Etesami, H.; Alikhani, H. Characterization of rhizosphere and endophytic bacteria from roots of maize (Zea mays L.) plant irrigated with wastewater with biotechnological potential in agriculture. Biotechnol. Rep. J. 2019, 21, e00305. [Google Scholar] [CrossRef] [PubMed]
- Mpanga, I.K.; Gomez-Genao, N.; Moradtalab, N.; Wanke, D.; Chrobaczek, V.; Ahmed, A.; Windisch, S.; Geistlinger, J.; Hafiz, F.B.; Walker, F.; et al. The role of N form supply for PGPM-host plant interactions in maize. J. Plant Nutr. Soil Sci. 2019, 182, 908–920. [Google Scholar] [CrossRef]
- Bhattacharyya, C.; Banerjee, S.; Acharya, U.; Mitra, A.; Mallick, I.; Haldar, A.; Haldar, S.; Ghosh, A.; Ghosh, A. Evaluation of plant growth promotion properties and induction of antioxidative defense mechanism by tea rhizobacteria of Darjeeling, India. Sci. Rep. 2020, 10, 15536. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Cruz, R.; Tapia Vázquez, I.; Batista-García, R.; Méndez-Santiago, E.; Sánchez-Carbente, M.; Leija, A.; Lira-Ruan, V.; Hernández, G.; Wong-Villarreal, A.; Folch-Mallol, J. Isolation and characterization of endophytes from nodules of Mimosa pudica with biotechnological potential. Microbiol. Res. 2019, 218, 76–86. [Google Scholar] [CrossRef] [PubMed]
- Hassan, M.K.; McInroy, J.A.; Kloepper, J.W. The interactions of rhizodeposits with plant growth-promoting rhizobacteria in the rhizosphere: A review. Agriculture 2019, 9, 142. [Google Scholar] [CrossRef]
- Azuddin, N.F.; Mohamad Noor Azmy, M.S.; Zakaria, L. Molecular identification of endophytic fungi in lawn grass (Axonopus compressus) and their pathogenic ability. Sci. Rep. 2023, 13, 4239. [Google Scholar] [CrossRef]
- Burks, C.; Darby, A.; Gómez Londoño, L.; Momany, M.; Brewer, M.T. Azole-resistant Aspergillus fumigatus in the environment: Identifying key reservoirs and hotspots of antifungal resistance. PLoS Pathog. 2021, 17, e1009711. [Google Scholar] [CrossRef]
- Abdel-Motaal, F.; Kamel, N.; El-Zayat, S.; Abou-Ellail, M. Early blight suppression and plant growth promotion potential of the endophyte Aspergillus flavus in tomato plant. Ann. Agric. Sci. 2020, 65, 117–123. [Google Scholar] [CrossRef]
- Tannous, J.; Barda, O.; Luciano-Rosario, D.; Prusky, D.B.; Sionov, E.; Keller, N.P. New insight into pathogenicity and secondary metabolism of the plant pathogen Penicillium expansum through deletion of the epigenetic reader SntB. Front. Microbiol. 2020, 11, 610. [Google Scholar] [CrossRef]
- Konappa, N.; Krishnamurthy, S.; Arakere, U.C.; Chowdappa, S.; Ramachandrappa, N.S. Efficacy of indigenous plant growth-promoting rhizobacteria and Trichoderma strains in eliciting resistance against bacterial wilt in a tomato. Egypt. J. Biol. Pest. Control 2020, 30, 106. [Google Scholar] [CrossRef]
- Agustiyani, D.; Purwaningsih, S.; Dewi, T.K.; Nditasari, A.; Nugroho, A.A.; Sutisna, E.; Mulyani, N.; Antonius, S. Characterization of PGPR isolated from rhizospheric soils of various plant and its effect on growth of radish (Raphanus sativus L.). In Proceedings of the IOP Conference Series: Earth and Environmental Science, Banjarbaru City, Indonesia, 23–24 October 2021; IOP Publishing: Bristol, UK, 2022; Volume 976, p. 012037. [Google Scholar] [CrossRef]
- Mohamed, M.H.; Ali, M.; Eid, R.S.; El-Desouky, H.S.; Petropoulos, S.A.; Sami, R.; Al-Mushhin, A.A.M.; Ismail, K.A.; Zewail, R.M.Y. Phosphorus and biofertilizer application effects on growth parameters, yield and chemical constituents of broccoli. Agronomy 2021, 11, 2210. [Google Scholar] [CrossRef]
- Teja, G.S.; Nayak, M.H.; Prasanth, P.; Mamatha, A.; Praneeth, S. Influence of inorganic nutrients and biofertilizers on growth and yield of tropical radish (Raphanus sativus L.) Cv. Pusa Chetki. Pharma Innov. J. 2021, 10, 1810–1812. [Google Scholar]
- Abro, M.A.; Sun, X.; Li, X.; Jatoi, G.H.; Guo, L.D. Biocontrol potential of fungal endophytes against Fusarium oxysporum f. sp. cucumerinum causing wilt in cucumber. Plant Pathol. J. 2019, 35, 598. [Google Scholar] [CrossRef] [PubMed]
- Kabir, M.H.; Unban, K.; Kodchasee, P.; Govindarajan, R.K.; Lumyong, S.; Suwannarach, N.; Wongputtisin, P.; Shetty, K.; Khanongnuch, C. Endophytic Bacteria Isolated from Tea Leaves (Camellia sinensis var. assamica) Enhanced Plant-Growth-Promoting Activity. Agriculture 2023, 13, 533. [Google Scholar] [CrossRef]
- Kaur, T.; Devi, R.; Kumar, S.; Kour, D.; Yadav, A.N. Plant growth promotion of pearl millet (Pennisetum glaucum L.) by novel bacterial consortium with multifunctional attributes. Biologia 2023, 78, 621–631. [Google Scholar] [CrossRef]
- Patel, J.S.; Selvaraj, V.; More, P.; Bahmani, R.; Borza, T.; Prithiviraj, B. A Plant Biostimulant from Ascophyllum nodosum Potentiates Plant Growth Promotion and Stress Protection Activity of Pseudomonas protegens CHA0. Plants 2023, 12, 1208. [Google Scholar] [CrossRef]
- Aloo, B.N.; Mbega, E.R.; Makumba, B.A.; Hertel, R.; Daniel, R. Molecular identification and in vitro plant growth-promoting activities of culturable Potato (Solanum tuberosum L.) rhizobacteria in Tanzania. Potato Res. 2021, 64, 67–95. [Google Scholar] [CrossRef]
- Andreolli, M.; Zapparoli, G.; Lampis, S.; Santi, C.; Angelini, E.; Bertazzon, N. In vivo endophytic, rhizospheric and epiphytic colonization of Vitis vinifera by the plant-growth promoting and antifungal strain Pseudomonas protegens MP12. Microorganisms 2021, 9, 234. [Google Scholar] [CrossRef]
- Fan, Y.; Niu, X.; Zhang, D.; Lin, Z.; Fu, M.; Zhou, S. Analysis of the characteristics of phosphine production by anaerobic digestion based on microbial community dynamics, metabolic pathways, and isolation of the phosphate-reducing strain. Chemosphere 2021, 262, 128213. [Google Scholar] [CrossRef]
- Seaton, S.; Lemaire, J.; Inderbitzin, P.; Knight-Connoni, V.; White, J.F.; Trujillo, M.E. Pseudomonas arenae sp. nov., Pseudomonas glycinis sp. nov. and Pseudomonas harudinis sp. nov., three novel bacterial species and plant endophytes. bioRxiv 2021. [Google Scholar] [CrossRef]
- Cheng, C.; Nie, Z.W.; He, L.Y.; Sheng, X.F. Rice-derived facultative endophytic Serratia liquefaciens F2 decreases rice grain arsenic accumulation in arsenic-polluted soil. Environ. Pollut. 2020, 259, 113832. [Google Scholar] [CrossRef] [PubMed]
- Luziatelli, F.; Ficca, A.G.; Cardarelli, M.; Melini, F.; Cavalieri, A.; Ruzzi, M. Genome sequencing of Pantoea agglomerans C1 provides insights into molecular and genetic mechanisms of plant growth-promotion and tolerance to heavy metals. Microorganisms 2020, 8, 153. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Lei, P.; Wang, Q.; Ma, J.; Zhan, Y.; Jiang, K.; Xu, Z.; Xu, H. The endophyte Pantoea alhagi NX-11 alleviates salt stress damage to rice seedlings by secreting exopolysaccharides. Front. Microbiol. 2020, 10, 3112. [Google Scholar] [CrossRef]
- Andreolli, M.; Zapparoli, G.; Angelini, E.; Lucchetta, G.; Lampis, S.; Vallini, G. Pseudomonas protegens MP12: A plant growth-promoting endophytic bacterium with broad-spectrum antifungal activity against grapevine phytopathogens. Microbiol. Res. 2019, 219, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Gamez, R.; Cardinale, M.; Montes, M.; Ramirez, S.; Schnell, S.; Rodriguez, F. Screening, plant growth promotion and root colonization pattern of two rhizobacteria (Pseudomonas fluorescens Ps006 and Bacillus amyloliquefaciens Bs006) on banana cv. Williams (Musa acuminata Colla). Microbiol. Res. 2019, 220, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Sawada, H.; Fujikawa, T.; Tsuji, M.; Satou, M. Pseudomonas allii sp. nov., a pathogen causing soft rot of onion in Japan. Int. J. Syst. Evol. Microbiol. 2021, 71, 004582. [Google Scholar] [CrossRef]
- Xu, S.; Zhao, Y.; Peng, Y.; Shi, Y.; Xie, X.; Chai, A.; Li, B.; Li, L. Comparative genomics assisted functional characterization of Rahnella aceris ZF458 as a novel plant growth promoting rhizobacterium. Front. Microbiol. 2022, 13, 850084. [Google Scholar] [CrossRef]
- Gao, X.; Luan, J.; Wang, L.; Li, H.; Wang, Q.; Wang, Z.; Jin, Z.; Yu, F. Effect of the Plant Growth Promoting Rhizobacterium, Cronobacter sp. Y501, for Enhancing Drought Tolerance in Maize (Zea mays L.). J. Soil Sci. Plant Nutr. 2023, 23, 2786–2797. [Google Scholar] [CrossRef]
- Leis, E.M.; Dziki, S.; Standish, I.; Waller, D.; Richard, J.; Weinzinger, J.; Harris, C.; Knowles, S.; Goldberg, T. A Bacteriological Comparison of the Hemolymph from Healthy and Moribund Unionid Mussel Populations in the Upper Midwestern USA Prompts the Development of Diagnostic Assays to Detect Yokenella regensburgei. Microorganisms 2023, 11, 1068. [Google Scholar] [CrossRef]
- Berthold-Pluta, A.; Stasiak-Różańska, L.; Pluta, A.; Garbowska, M. Antibacterial activities of plant-derived compounds and essential oils against Cronobacter strains. Eur. Food Res. Technol. 2019, 245, 1137–1147. [Google Scholar] [CrossRef]
- Meteored: Clima en Chihuahua. 2021. Available online: https://www.meteored.mx (accessed on 26 March 2023).
- SEMARNAT: Producción Y Rendimiento de Cultivos Orgánicos Por Entidad Federativa. 2020. Available online: http://dgeiawf.semarnat.gob.mx/ (accessed on 26 March 2023).
- National Park Service. Chihuahua Desert Ecoregion. 2018. Available online: https://www.nps.gov/im/chdn/ecoregion.htm (accessed on 26 March 2023).
- Rat, A.; Naranjo, H.D.; Krigas, N.; Grigoriadou, K.; Maloupa, E.; Alonso, A.V.; Schneider, C.; Papageorgiou, V.P.; Assimopoulou, A.N.; Tsafantakis, N.; et al. Endophytic Bacteria From the Roots of the Medicinal Plant Alkanna tinctoria Tausch (Boraginaceae): Exploration of Plant Growth Promoting Properties and Potential Role in the Production of Plant Secondary Metabolites. Front. Microbiol. 2021, 12, 113. [Google Scholar] [CrossRef] [PubMed]
- Kuan, K.B.; Othman, R.; Abdul Rahim, K.; Shamsuddin, Z.H. Plant growth-promoting rhizobacteria inoculation to enhance vegetative growth, nitrogen fixation and nitrogen remobilisation of maize under greenhouse conditions. PLoS ONE 2016, 11, e0152478. [Google Scholar] [CrossRef] [PubMed]
- Bokhari, A.; Essack, M.; Lafi, F.F.; Andres-Barrao, C.; Jalal, R.; Alamoudi, S.; Razali, R.; Alzubaidy, H.; Shah, K.H.; Siddique, S.; et al. Bioprospecting desert plant Bacillus endophytic strains for their potential to enhance plant stress tolerance. Sci. Rep. 2019, 9, 18154. [Google Scholar] [CrossRef] [PubMed]
- Magotra, S.; Bhagat, N.; Ambardar, S.; Ali, T.; Hurek, B.R.; Hurek, T.; Verma, P.K.; Vakhlu, J. Field evaluation of PGP Bacillus sp. strain D5 native to Crocus sativus, in traditional and non traditional areas, and mining of PGP genes from its genome. Sci. Rep. 2021, 11, 5454. [Google Scholar] [CrossRef]
- Rajawat, M.V.S.; Singh, S.; Tyagi, S.P.; Saxena, A.K. A modified plate assay for rapid screening of potassium-solubilizing bacteria. Pedosphere 2016, 26, 768–773. [Google Scholar] [CrossRef]
- Zhang, C.; Kong, F. Isolation and identification of potassium-solubilizing bacteria from tobacco rhizospheric soil and their effect on tobacco plants. Appl. Soil Ecol. 2014, 82, 18–25. [Google Scholar] [CrossRef]
- Jiménez-Gómez, A.; Flores-Félix, J.D.; García-Fraile, P.; Mateos, P.F.; Menéndez, E.; Velázquez, E.; Rivas, R. Probiotic activities of Rhizobium laguerreae on growth and quality of spinach. Sci. Rep. 2018, 8, 295. [Google Scholar] [CrossRef]
- Elias, F.; Woyessa, D.; Muleta, D. Phosphate solubilization potential of rhizosphere fungi isolated from plants in Jimma Zone, Southwest Ethiopia. Int. J. Microbiol. 2016, 2016, 5472601. [Google Scholar] [CrossRef]
- Lima-Rivera, D.L.; Lopez-Lima, D.; Desgarennes, D.; Velazquez-Rodriguez, A.S.; Carrion, G. Phosphate solubilization by fungi with nematicidal potential. J. Soil Sci. Plant Nutr. 2016, 16, 507–524. [Google Scholar] [CrossRef]
- Sharon, J.A.; Hathwaik, L.T.; Glenn, G.M.; Imam, S.H.; Lee, C.C. Isolation of efficient phosphate solubilizing bacteria capable of enhancing tomato plant growth. J. Soil Sci. Plant Nutr. 2016, 16, 525–536. [Google Scholar] [CrossRef]
- Bapiri, A.; Asgharzadeh, A.; Mujallali, H.; Khavazi, K.; Pazira, E. Evaluation of Zinc solubilization potential by different strains of Fluorescent Pseudomonas. J. Appl. Sci. Environ. 2012, 16, 295–298. Available online: https://www.ajol.info/index.php/jasem/article/view/90952 (accessed on 10 January 2022).
- AlAli, H.A.; Khalifa, A.; Al-Malki, M. Plant growth-promoting rhizobacteria from Ocimum basilicum improve growth of Phaseolus vulgaris and Abelmoschus esculentus. S. Afr. J. Bot. 2021, 139, 200–209. [Google Scholar] [CrossRef]
- Syed-Ab-Rahman, S.F.; Carvalhais, L.C.; Chua, E.; Xiao, Y.; Wass, T.J.; Schenk, P.M. Identification of soil bacterial isolates suppressing different Phytophthora spp. and promoting plant growth. Front. Plant Sci. 2018, 9, 1502. [Google Scholar] [CrossRef] [PubMed]
- Goswami, D.; Parmar, S.; Vaghela, H.; Dhandhukia, P.; Thakker, J.N. Describing Paenibacillus mucilaginosus strain N3 as an efficient plant growth promoting rhizobacteria (PGPR). Cogent Food Agric. 2015, 1, 1000714. [Google Scholar] [CrossRef]
- Gupta, M.; Kiran, S.; Gulati, A.; Singh, B.; Tewari, R. Isolation and identification of phosphate solubilizing bacteria able to enhance the growth and aloin-A biosynthesis of Aloe barbadensis Miller. Microbiol. Res. 2012, 167, 358–363. [Google Scholar] [CrossRef]
- Kumar, P.; Pahal, V.; Gupta, A.; Vadhan, R.; Chandra, H.; Dubey, R.C. Effect of silver nanoparticles and Bacillus cereus LPR2 on the growth of Zea mays. Sci. Rep. 2020, 10, 20409. [Google Scholar] [CrossRef]
- Hartono, H.; Widada, J.; Kabirun, S. 16s rRNA Sequence Analysis and Ammonium Excretion Ability of Nitrogen Fixing Bacteria Isolated from Mineral Acid Soil. Indones. J. Biotechnol. 2009, 14, 1179–1187. [Google Scholar] [CrossRef]
- Hansen, P.A. The detection of ammonia production by bacteria in agar slants. J. Bacteriol. 1930, 9, 223–229. [Google Scholar] [CrossRef]
- Gilbert, S.; Poulev, A.; Chrisler, W.; Acosta, K.; Orr, G.; Lebeis, S.; Lam, E. Auxin-producing bacteria from duckweeds have different colonization patterns and effects on plant morphology. Plants 2022, 11, 721. [Google Scholar] [CrossRef]
- Fahsi, N.; Mahdi, I.; Mesfioui, A.; Biskri, L.; Allaoui, A. Plant Growth-Promoting Rhizobacteria isolated from the Jujube (Ziziphus lotus) plant enhance wheat growth, Zn uptake, and heavy metal tolerance. Agriculture 2021, 11, 316. [Google Scholar] [CrossRef]
- Kamran, S.; Shahid, I.; Baig, D.N.; Rizwan, M.; Malik, K.A.; Mehnaz, S. Contribution of zinc solubilizing bacteria in growth promotion and zinc content of wheat. Front. Microbiol. 2017, 8, 2593. [Google Scholar] [CrossRef] [PubMed]
- Sarwar, M.; Kremer, R.J. Determination of bacterially derived auxins using a microplate method. Lett. Appl. Microbiol. 1995, 20, 282–285. [Google Scholar] [CrossRef]
- Numponsak, T.; Kumla, J.; Suwannarach, N.; Matsui, K.; Lumyong, S. Biosynthetic pathway and optimal conditions for the production of indole-3-acetic acid by an endophytic fungus, Colletotrichum fructicola CMU-A109. PLoS ONE 2018, 13, e0205070. [Google Scholar] [CrossRef] [PubMed]
- Srimathi, K.; Suji, H.A. Siderophores Detection by using Blue Agar CAS Assay Methods. Int. J. Sci. Res. Biol. Sci. 2018, 5, 180–185. [Google Scholar] [CrossRef]
- Rfaki, A.; Zennouhi, O.; Aliyat, F.Z.; Nassiri, L.; Ibijbijen, J. Isolation, Selection and Characterization of Root-Associated Rock Phosphate Solubilizing Bacteria in Moroccan Wheat (Triticum aestivum L.). Geomicrobiol. J. 2019, 37, 230–241. [Google Scholar] [CrossRef]
- Aquino, J.P.A.D.; Macedo, F.B.D.; Antunes, J.E.L.; Figueiredo, M.D.V.B.; Alcântara, F.D.; Araujo, A.S.F.D. Plant growth-promoting endophytic bacteria on maize and sorghum. Pesqui. Agropecu. 2019, 49, e56241. [Google Scholar] [CrossRef]
- Ren, X.M.; Guo, S.J.; Tian, W.; Chen, Y.; Han, H.; Chen, E.; Li, B.-L.; Li, Y.-Y.; Chen, Z.-J. Effects of plant growth-promoting bacteria (PGPB) inoculation on the growth, antioxidant activity, Cu uptake, and bacterial community structure of rape (Brassica napus L.) grown in Cu-contaminated agricultural soil. Front. Microbiol. 2019, 10, 1455. [Google Scholar] [CrossRef]
- Bacilio, M.; Moreno, M.; Lopez-Aguilar, D.R.; Bashan, Y. Scaling from the growth chamber to the greenhouse to the field: Demonstration of diminishing effects of mitigation of salinity in peppers inoculated with plant growth-promoting bacterium and humic acids. Appl. Soil Ecol. 2017, 119, 327–338. [Google Scholar] [CrossRef]
- Alvarez, E.; Sánchez, L.C. Evaluación del crecimiento de cuatro especies del género Bacillus sp., primer paso para entender su efecto biocontrolador sobre Fusarium sp. Nova 2016, 14, 53–62. [Google Scholar] [CrossRef]
- Ochoa-Velasco, C.E.; Valadez-Blanco, R.; Salas-Coronado, R.; Sustaita-Rivera, F.; Hernández-Carlos, B.; García-Ortega, S.; Santos-Sánchez, N.F. Effect of nitrogen fertilization and Bacillus licheniformis biofertilizer addition on the antioxidants compounds and antioxidant activity of greenhouse cultivated tomato fruits (Solanum lycopersicum L. var. Sheva). Sci. Hortic. 2016, 201, 338–345. [Google Scholar] [CrossRef]
- Jaysree, R.C.; Basu, S.; Singh, P.P.; Ghosal, T.; Patra, P.A.; Keerthi, Y.; Rajendran, N. Isolation of biosurfactant producing bacteria from environmental samples. Pharmacologyonline 2011, 63, 1427–1433. [Google Scholar]
- Ji, X.; Lu, G.; Gai, Y.; Zheng, C.; Mu, Z. Biological control against bacterial wilt and colonization of mulberry by an endophytic Bacillus subtilis strain. FEMS Microbiol. Ecol. 2008, 65, 565–573. [Google Scholar] [CrossRef] [PubMed]
- Weibull, C. Characterization of the protoplasmic constituents of Bacillus megaterium. J. Bacteriol. 1953, 66, 696–702. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Yuan, H.; Ji, H.; Liu, H.; Zhang, Y.; Wang, G.; Chen, L.; Guo, Z. Effect of ZnO nanoparticles on the productivity, Zn biofortification, and nutritional quality of rice in a life cycle study. Plant Physiol. Biochem. 2021, 163, 87–94. [Google Scholar] [CrossRef]
- Morales, A.; Alvear, M.; Valenzuela, E.; Rubio, R.; Borie, F. Effect of inoculation with Penicillium albidum, a phosphate-solubilizing fungus, on the growth of Trifolium pratense cropped in a volcanic soil. J. Basic Microbiol. 2007, 47, 275–280. [Google Scholar] [CrossRef]
- Saboor, A.; Ali, M.A.; Danish, S.; Ahmed, N.; Fahad, S.; Datta, R.; Ansari, M.J.; Nasif, O.; Rahman, M.H.U.; Glick, B.R. Effect of arbuscular mycorrhizal fungi on the physiological functioning of maize under zinc-deficient soils. Sci. Rep. 2021, 11, 18468. [Google Scholar] [CrossRef]
- Kornarzyński, K.; Sujak, A.; Czernel, G.; Wiącek, D. Effect of Fe3O4 nanoparticles on germination of seeds and concentration of elements in Helianthus annuus L. under constant magnetic field. Sci. Rep. 2020, 10, 8068. [Google Scholar] [CrossRef]
- Afanador-Barajas, L.N.; Navarro-Noya, Y.E.; Luna-Guido, M.L.; Dendooven, L. Impact of a bacterial consortium on the soil bacterial community structure and maize (Zea mays L.) cultivation. Sci. Rep. 2021, 11, 13092. [Google Scholar] [CrossRef]
- Hernández-Montiel, L.G.; Chiquito Contreras, C.J.; Murillo Amador, B.; Vidal Hernández, L.; Quiñones Aguilar, E.E.; Chiquito Contreras, R.G. Efficiency of two inoculation methods of Pseudomonas putida on growth and yield of tomato plants. J. Soil Sci. Plant Nutr. 2017, 17, 1003–1012. [Google Scholar] [CrossRef]
- Naveed, M.; Mitter, B.; Yousaf, S.; Pastar, M.; Afzal, M.; Sessitsch, A. The endophyte Enterobacter sp. FD17: A maize growth enhancer selected based on rigorous testing of plant beneficial traits and colonization characteristics. Biol. Fertil. Soils 2014, 50, 249–262. [Google Scholar] [CrossRef]
- Bononi, L.; Chiaramonte, J.B.; Pansa, C.C.; Moitinho, M.A.; Melo, I.S. Phosphorus-solubilizing Trichoderma spp. from Amazon soils improve soybean plant growth. Sci. Rep. 2020, 10, 2858. [Google Scholar] [CrossRef] [PubMed]
- Acharya, P.; Jayaprakasha, G.K.; Crosby, K.M.; Jifon, J.L.; Patil, B.S. Nanoparticle-mediated seed priming improves germination, growth, yield, and quality of watermelons (Citrullus lanatus) at multi-locations in Texas. Sci. Rep. 2020, 10, 5037. [Google Scholar] [CrossRef] [PubMed]
- James, G. Universal bacterial identification by PCR and DNA sequencing of 16S rRNA gene. In PCR for Clinical Microbiology; Schuller, M., Sloots, T., James, G., Halliday, C., Carter, I., Eds.; Springer: Dordrecht, The Netherlands, 2010; pp. 209–214. [Google Scholar] [CrossRef]

| Selection Level | Number of Isolates | % with Respect to Total Isolates | Criteria |
|---|---|---|---|
| Bacteria | |||
| Total isolates | 798 | 100% | Visual morphological differences. |
| 1st selection | 399 | 50% | P, K, and Zn solubilizing activity, even if it is minimal. |
| 2nd selection | 262 | 33% | Two parameters in level 3 (P, K, Zn) and nitrogen fixation activity. |
| 3rd selection | 137 | 17% | Multi-mechanisms (P, K, and Zn solubilization, N2-fixation, NH3, IAA, and siderophores production). |
| 4th selection | 43 | 5% | High activity levels among different parameters, evaluation of biocontrol activity. |
| 5th selection | 24 | 3% | Best plant growth-promoting traits |
| Fungi | |||
| Total isolates | 209 | 100% | Visual morphological differences. |
| 1st selection | 156 | 75% | P, K, and Zn solubilizing activity, even if it is minimal. |
| 2nd selection | 55 | 26% | P, K, and Zn solubilization, N2-fixation, NH3, IAA, and siderophores production. |
| 3rd selection | 34 | 16% | Second grouping depending on macroscopic morphological differences. |
| 4th selection | 18 | 9% | Best plant growth-promoting traits, including biocontrol activity. |
| PGP Attribute | Level 1 | Level 2 | Level 3 |
|---|---|---|---|
| Phosphate, potassium, and zinc solubilization (clearing halo) | 1 mm | 2–4 mm | ≥5 mm |
| Siderophores production (color change halo) | |||
| Nitrogen fixation (color change halo) | 0–9 mm | 10–14 mm | ≥15 mm |
| Ammonia production (OD425 nm values) | −0.2–1 | 1–1.6 | ≥1.7 |
| Indole-3-acetic acid secretion (hormone concentrations in mg/L) | 0–10 mg/L | 11–25 mg/L | ≥26 mg/L |
| Biocontrol activity (% inhibition in radial growth) | 0–39% | 40–59% | ≥60% |
| Isolate | Phosphate Solubilization (Halo in mm) | Potassium Solubilization (Halo in mm) | Zinc Solubilization (Halo in mm) | Siderophore Production (Halo in mm) | Nitrogen Fixation (Halo in mm) | Ammonia Production (OD425nm Values) | Indole-3-Acetic Acid Production (mg/L) | Biocontrol (Pathogen Growth Inhibition) |
|---|---|---|---|---|---|---|---|---|
| Bacteria | ||||||||
| A302 | 5.0 | 10.0 | 9.0 | 3.0 | 12.0 | 1.0 | 34.4 | 0% |
| A306-A1 | 7.0 | 6.0 | 12.0 | 1.0 | 0.0 | 0.8 | 8.2 | 0% |
| A334-1 | 3.0 | 5.0 | 5.0 | 1.0 | 0.0 | 1.7 | 33.7 | 0% |
| B14FEB | 8.0 | 9.0 | 7.0 | 5.0 | 14.0 | 1.3 | 12.3 | 57% |
| B14KEA1 | 3.0 | 9.0 | 5.0 | 6.0 | 1.0 | 1.4 | 41.2 | 12% |
| B5-KEA | 3.0 | 13.0 | 8.0 | 1.0 | 17.0 | 1.2 | 29.2 | 10% |
| B9M | 7.0 | 6.0 | 8.0 | 2.0 | 13.0 | 1.2 | 40.9 | 0% |
| A276-2 | 5.0 | 7.0 | 10.0 | 1.0 | 7.0 | 1.1 | 5.3 | 0% |
| A137 A1 | 6.0 | 5.0 | 5.0 | 1.0 | 13.0 | 1.2 | 7.4 | 0% |
| A211 A | 2.0 | 4.0 | 5.0 | 3.0 | 14.0 | 1.9 | 24.0 | 15% |
| A239 | 1.0 | 8.0 | 9.0 | 2.0 | 12.0 | 1.9 | 24.1 | 13% |
| A336 | 6.0 | 6.0 | 8.0 | 0.5 | 6.0 | 2.1 | 39.1 | 0% |
| A32-2 | 3.0 | 5.0 | 0.0 | 5.0 | 19.0 | 0.7 | 17.1 | 0% |
| A58 B | 0.0 | 9.0 | 0.0 | 19.0 | 14.0 | 1.1 | 15.9 | 0% |
| A80 | 1.0 | 9.0 | 0.0 | 9.0 | 17.0 | 0.8 | 18.0 | 13% |
| A194 | 0.0 | 11.0 | 0.0 | 7.0 | 5.0 | 2.3 | 10.7 | 18% |
| A210 R1 | 2.0 | 6.0 | 10.0 | 3.0 | 13.0 | 2.0 | 21.1 | 68% |
| A328 B | 2.0 | 7.0 | 0.0 | 2.0 | 11.0 | 1.6 | 12.6 | 50% |
| A316 B | 3.0 | 6.0 | 9.0 | 1.0 | 10.0 | 1.2 | 7.2 | 49% |
| A96 B | 5.0 | 6.0 | 8.0 | 4.0 | 18.0 | 0.9 | 23.1 | 43% |
| A313-2 | 3.0 | 5.0 | 0.0 | 2.0 | 7.0 | 1.7 | 55.8 | 21% |
| A289 B | 8.0 | 8.0 | 6.0 | 2.0 | 0.0 | 1.5 | 176.7 | 19% |
| A196 B | 2.0 | 6.0 | 8.0 | 4.0 | 13.0 | 1.3 | 60.6 | 23% |
| A107 A | 6.0 | 8.0 | 0.0 | 1.0 | 11.0 | 1.8 | 64.7 | 0% |
| Fungi | ||||||||
| H97 | 11.0 | 14.0 | 16.0 | 14.0 | 0.0 | 0.9 | 1.2 | 69% |
| H74 | 6.0 | 10.0 | 11.0 | 1.0 | 1.0 | 0.2 | 1.0 | 36% |
| H38 | 5.0 | 13.0 | 11.0 | 0.0 | 0.0 | 0.5 | 1.5 | 64% |
| H132 | 3.0 | 11.0 | 15.0 | 0.0 | 1.0 | 0.2 | 1.0 | 40% |
| H49 | 6.0 | 19.0 | 12.0 | 3.0 | 1.0 | 0.1 | 9.6 | 38% |
| H176 | 15.0 | 15.0 | 14.0 | 23.0 | 0.0 | 0.7 | 5.1 | 51% |
| H133 | 4.0 | 12.0 | 9.0 | 12.0 | 0.0 | 0.3 | 1.4 | 69% |
| H153 | 4.0 | 5.0 | 10.0 | 2.0 | 1.0 | 0.6 | 1.7 | 40% |
| H231 | 6.0 | 20.0 | 15.0 | 0.0 | 1.0 | 0.2 | 1.2 | 33% |
| H176-D | 12.0 | 10.0 | 11.0 | 2.0 | 0.0 | 0.1 | 1.0 | 31% |
| H163 | 4.0 | 9.0 | 14.0 | 0.0 | 1.0 | 0.8 | 2.4 | 47% |
| H207 | 20.0 | 12.0 | 15.0 | 16.0 | 0.0 | 0.9 | 0.5 | 51% |
| H204 | 4.0 | 10.0 | 8.0 | 0.0 | 1.0 | 0.2 | 3.1 | 53% |
| H81 | 4.0 | 4.0 | 12.0 | 1.0 | 1.0 | 0.2 | 1.2 | 36% |
| H202 | 15.0 | 4.0 | 11.0 | 0.0 | 1.0 | 0.6 | 1.5 | 40% |
| H67 | 4.0 | 16.0 | 19.0 | 21.0 | 0.0 | 0.0 | 0.5 | 53% |
| H56 | 5.0 | 5.0 | 10.0 | 0.0 | 1.0 | 0.3 | 2.0 | 38% |
| H56-Zn | 6.0 | 6.0 | 17.0 | 13.0 | 0.0 | 0.7 | 0.9 | 51% |
| Consortia ID | Description | Microbial Consortia |
|---|---|---|
| P BAC | Phosphate-solubilizing bacteria | A302, A306-A1, A334-1, B14FE-B |
| K-ZN BAC | Potassium- and zinc-solubilizing bacteria | B14KEA1-A1, B5KE-A, B9M, A276-2 |
| N-NH3 BAC | Nitrogen(N2)-fixing and ammonia(NH3)-producing bacteria | A137-A1, A211-A, A239, A336 |
| FE BAC | Siderophore-producing bacteria | A32-2, A58-B, A80, A194 |
| BIOCON BAC | Biocontrol bacteria | A210-R, A328-B, A316-B, A96-B |
| IAA BAC | Indole-3-acetic acid-secreting bacteria | A313-2, A289-B, A196-B, A107-A |
| P FUN | Phosphate-solubilizing fungi | H133, H97, H176 |
| K-ZN FUN 1 | Potassium- and zinc-solubilizing fungi 1 | H204, H231, H132, H81, H74, H176-D |
| K-ZN FUN 2 | Potassium- and zinc-solubilizing fungi 2 | H67, H49 |
| N-NH3 FUN | N2-fixing and NH3-producing fungi | H202, H153, H163, H56 |
| FE FUN | Siderophore-producing fungi | H56-Z, H207 |
| BIOCON FUN | Biocontrol fungi | H38 |
| No. | Isolate | Sample Description * | Molecular Identification | Accession Number |
|---|---|---|---|---|
| 1 | A302 | Greasewood, soil, 2 | Serratia liquefaciens strain A302 | OR452709 |
| 2 | A306-A1 | Pepper, root, 3 | Pseudomonas extremorientalis strain A306A1 | OR452718 |
| 3 | A334-1 | Greasewood, soil, 1 | Pseudescherichia vulneris strain A334 | OR452719 |
| 4 | B14FEB | Maize, soil, 2 | Serratia liquefaciens strain B14FEB | OR452714 |
| 5 | B14KEA1 | Pepper, soil, 2 | Rahnella aceris strain B14KEA1 | OR452725 |
| 6 | B5KEA | Pepper, soil, 1 | Pseudomonas allii strain B5KEA | OR452711 |
| 7 | B9M | Maize, root, 3 | Pseudomonas marginalis strain B9M | OR452712 |
| 8 | A276-2 | Pepper, root, 3 | Pseudomonas protegens strain A276 | OR452715 |
| 9 | A137-A1 | Maize, soil, 2 | Pseudomonas sesami strain A137A1 | OR452717 |
| 10 | A211-A | Maize, root, 1 | Pseudomonas arenae strain A211A | OR452724 |
| 11 | A239 | Oregano, soil, 3 | Pantoea agglomerans strain A239 | OR452708 |
| 12 | A336 | Oregano, soil, 2 | Yokenella regensburgei strain A336 | OR452721 |
| 13 | A58B | Greasewood, soil, 1 | Pantoea alhagi strain A58B | OR452723 |
| 14 | A194 | Greasewood, root, 1 | Cronobacter turicensis strain A194 | OR452710 |
| 15 | A210R1 | Maize, root, 1 | Rahnella aceris strain A210R1 | OR452726 |
| 16 | A316B | Pepper, soil, 2 | Pantoea allii strain A316B | OR452722 |
| 17 | A313-2 | Oregano, root, 1 | Pantoea pleuroti strain A313 | OR452720 |
| 18 | A289B | Buffelgrass, soil, 1 | Enterobacter wuhouensis strain A289B | OR452713 |
| 19 | A196-B | Maize root, 3 | Pseudomonas arenae strain A196B | OR452716 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guardiola-Márquez, C.E.; Santos-Ramírez, M.T.; Figueroa-Montes, M.L.; Valencia-de los Cobos, E.O.; Stamatis-Félix, I.J.; Navarro-López, D.E.; Jacobo-Velázquez, D.A. Identification and Characterization of Beneficial Soil Microbial Strains for the Formulation of Biofertilizers Based on Native Plant Growth-Promoting Microorganisms Isolated from Northern Mexico. Plants 2023, 12, 3262. https://doi.org/10.3390/plants12183262
Guardiola-Márquez CE, Santos-Ramírez MT, Figueroa-Montes ML, Valencia-de los Cobos EO, Stamatis-Félix IJ, Navarro-López DE, Jacobo-Velázquez DA. Identification and Characterization of Beneficial Soil Microbial Strains for the Formulation of Biofertilizers Based on Native Plant Growth-Promoting Microorganisms Isolated from Northern Mexico. Plants. 2023; 12(18):3262. https://doi.org/10.3390/plants12183262
Chicago/Turabian StyleGuardiola-Márquez, Carlos Esteban, María Teresa Santos-Ramírez, Melina Lizeth Figueroa-Montes, Eric Oswaldo Valencia-de los Cobos, Iván Jesús Stamatis-Félix, Diego E. Navarro-López, and Daniel A. Jacobo-Velázquez. 2023. "Identification and Characterization of Beneficial Soil Microbial Strains for the Formulation of Biofertilizers Based on Native Plant Growth-Promoting Microorganisms Isolated from Northern Mexico" Plants 12, no. 18: 3262. https://doi.org/10.3390/plants12183262
APA StyleGuardiola-Márquez, C. E., Santos-Ramírez, M. T., Figueroa-Montes, M. L., Valencia-de los Cobos, E. O., Stamatis-Félix, I. J., Navarro-López, D. E., & Jacobo-Velázquez, D. A. (2023). Identification and Characterization of Beneficial Soil Microbial Strains for the Formulation of Biofertilizers Based on Native Plant Growth-Promoting Microorganisms Isolated from Northern Mexico. Plants, 12(18), 3262. https://doi.org/10.3390/plants12183262








