Recent Advances in Microbial-Assisted Remediation of Cadmium-Contaminated Soil
Abstract
1. Introduction
2. Review Methodology
3. Sources of Cadmium
4. Cadmium Toxicity and Plants
4.1. Seed Germination and Seedling Growth
4.2. Cadmium-Induced Changes in Growth and Development
4.3. Impact on Amino Acids, Proteins and Organic Osmolytes
4.4. Plant Water Relations
4.5. Impact on Photosynthesis
5. The Role of Microbes in the Bioremediation of Cd-Contaminated Soils
5.1. Remediation of Cd by Bacteria
5.2. Remediation of Cd by Fungi
5.3. Remediation of Cd by Algae
6. Mechanisms Involved in Bioremediation by Microbes
6.1. Direct Mechanisms
6.1.1. Nitrogen Fixation
6.1.2. Phosphate Solubilization
6.1.3. Phytohormone Production
6.1.4. Antagonistic Role of PGPR
6.1.5. Siderophore Secretion
6.1.6. Volatile Organic Compounds
6.2. Indirect Mechanisms
6.2.1. Production of Antibiotics
6.2.2. Production of Exopolysaccharides
6.2.3. Hydrogen Cyanide
6.2.4. Lipo-Chito-Oligosaccharides
7. Factors Affecting PGPR Bioremediation
8. Recent Advancements (Genetic and Metabolic Engineering), (Membrane and Enzyme Technology), (Metagenomics Approaches) and (Nanoparticle Technology)
8.1. Membrane and Enzyme Technology
8.2. Genetic and Metabolic Engineering
8.3. Metagenomics Approaches
8.4. Nanoparticle Technology
9. Challenges and Future Prospects
10. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zulfiqar, U.; Farooq, M.; Hussain, S.; Maqsood, M.; Hussain, M.; Ishfaq, M.; Ahmad, M.; Anjum, M.Z. Lead toxicity in plants: Impacts and remediation. J. Environ. Manag. 2019, 250, 109557. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.; Sharma, A.; Yadav, S.; Jule, L.T.; Krishnaraj, R. Nanomaterials for remediation of environmental pollutants. Bioinorg. Chem. Appl. 2021, 2021, 1764647. [Google Scholar] [CrossRef] [PubMed]
- Fergusson, J.E. (Ed.) The Heavy Elements: Chemistry, Environmental Impact and Health Effects; Pergamon Press: Oxford, UK, 1990. [Google Scholar]
- Haider, F.U.; Wang, X.; Zulfiqar, U.; Farooq, M.; Hussain, S.; Mehmood, T.; Naveed, M.; Li, Y.; Liqun, C.; Saeed, Q.; et al. Biochar application for remediation of organic toxic pollutants in contaminated soils; An update. Ecotoxicol. Environ. Saf. 2022, 248, 114322. [Google Scholar] [CrossRef] [PubMed]
- Zulfiqar, U.; Haider, F.U.; Ahmad, M.; Hussain, D.; Maqsood, M.F.; Ishfaq, M.; Shahzad, B.; Waqas, M.M.; Ali, B.; Tayyab, M.N.; et al. Chromium Toxicity, Speciation, and Remediation Strategies in Soil-Plant Interface: A Critical Review. Front. Plant Sci. 2023, 13, 5468. [Google Scholar] [CrossRef]
- Zulfiqar, U.; Ayub, A.; Hussain, S.; Waraich, E.A.; El-Esawi, M.A.; Ishfaq, M.; Ahmad, M.; Ali, N.; Maqsood, M.F. Cadmium toxicity in plants: Recent progress on morpho-physiological effects and remediation strategies. J. Soil Sci. Plant Nutr. 2021, 22, 212–269. [Google Scholar] [CrossRef]
- Qianqian, M.; Haider, F.U.; Farooq, M.; Adeel, M.; Shakoor, N.; Jun, W.; Jiaying, X.; Wang, X.W.; Panjun, L.; Cai, L. Selenium treated Foliage and biochar treated soil for improved lettuce (Lactuca sativa L.) growth in Cd-polluted soil. J. Cleaner Prod. 2022, 335, 130267. [Google Scholar] [CrossRef]
- Rahimzadeh, M.R.; Rahimzadeh, M.R.; Kazemi, S.; Moghadamnia, A.A. Cadmium toxicity and treatment: An update. Casp. J. Intern. Med. 2017, 8, 135. [Google Scholar]
- Gul, I.; Manzoor, M.; Hashim, N.; Shah, G.M.; Waani, S.P.T.; Shahid, M.; Antoniadis, V.; Rinklebe, J.; Arshad, M. Challenges in microbially and chelate-assisted phytoextraction of cadmium and lead—A review. Environ. Pollut. 2021, 287, 117667. [Google Scholar] [CrossRef]
- Liu, L.; Li, J.W.; Yue, F.X.; Yan, X.W.; Wang, F.Y.; Bloszies, S.; Wang, Y. Effects of arbuscular mycorrhizal inoculation and biochar amendment on maize growth, cadmium uptake and soil cadmium speciation in Cd-contaminated soil. Chemosphere 2018, 194, 495–503. [Google Scholar] [CrossRef]
- Younas, F.; Younas, S.; Bibi, I.; Farooqi, Z.U.R.; Hameed, M.A.; Mohy-Ud-Din, W.; Shehzad, M.T.; Hussain, M.M.; Shakil, Q.; Shahid, M. A critical review on the separation of heavy metal (loid) s from the contaminated water using various agricultural wastes. Int. J. Phytoremed. 2023, 1–20. [Google Scholar] [CrossRef]
- Haider, F.U.; Liqun, C.; Coulter, J.A.; Cheema, S.A.; Wu, J.; Zhang, R.; Wenjun, M.; Farooq, M. Cadmium toxicity in plants: Impacts and remediation strategies. Ecotoxicol. Environ. Saf. 2021, 211, 111887. [Google Scholar] [CrossRef]
- Abbas, T.; Rizwan, M.; Ali, S.; Adrees, M.; Zia-ur-Rehman, M.; Qayyum, M.F.; Ok, Y.S.; Murtaza, G. Effect of Biochar on Alleviation of Cadmium Toxicity in Wheat (Triticum aestivum L.) Grown on Cd-Contaminated Saline Soil. Environ. Sci. Pollut. Res. 2018, 25, 25668–25680. [Google Scholar] [CrossRef]
- Chen, Q.; Lu, X.; Guo, X.; Pan, Y.; Yu, B.; Tang, Z.; Guo, Q. Differential Responses to Cd Stress Induced by Exogenous Application of Cu, Zn or Ca in the Medicinal Plant Catharanthus roseus. Ecotoxicol. Environ. Saf. 2018, 157, 266–275. [Google Scholar] [CrossRef] [PubMed]
- Zulfiqar, U.; Jiang, W.; Xiukang, W.; Hussain, S.; Ahmad, M.; Maqsood, M.F.; Ali, N.; Ishfaq, M.; Kaleem, M.; Haider, F.U.; et al. Cadmium Phytotoxicity, Tolerance, and Advanced Remediation Approaches in Agricultural Soils; A Comprehensive Review. Front. Plant Sci. 2022, 13, 773815. [Google Scholar] [CrossRef] [PubMed]
- Dermont, G.; Bergeron, M.; Mercier, G.; Richer-Laflèche, M. Soil washing for metal removal: A review of physical/chemical technologies and field applications. J. Hazard. Mater. 2008, 152, 1–31. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Liu, C.; Hsu, P.C.; Zhao, J.; Wu, T.; Tang, J.; Liu, K.; Cui, Y. Remediation of heavy metal contaminated soil by asymmetrical alternating current electrochemistry. Nat. Commun. 2019, 10, 2440. [Google Scholar] [CrossRef]
- Kumar, A.; Subrahmanyam, G.; Mondal, R.; Cabral-Pinto, M.M.S.; Shabnam, A.A.; Jigyasu, D.K.; Malyan, S.K.; Kishor-Fagodiya, R.; Khan, S.A.; Kumar, A.; et al. Bio-remediation approaches for alleviation of cadmium contamination in natural resources. Chemosphere 2021, 268, 128855. [Google Scholar] [CrossRef]
- Younas, F.; Mustafa, A.; Farooqi, Z.U.R.; Wang, X.; Younas, S.; Mohy-Ud-Din, W.; Ashir Hameed, M.; Mohsin Abrar, M.; Maitlo, A.A.; Noreen, S. Current and emerging adsorbent technologies for wastewater treatment: Trends, limitations, and environmental implications. Water 2021, 13, 215. [Google Scholar] [CrossRef]
- Ahluwalia, S.S.; Goyal, D. Microbial and plant derived biomass for removal of heavy metals from wastewater. Bioresour. Technol. 2007, 98, 2243–2257. [Google Scholar] [CrossRef]
- Saha, L.; Tiwari, J.; Bauddh, K.; Ma, Y. Recent developments in microbe–plant-based bioremediation for tackling heavy metal-polluted soils. Front. Microbiol. 2021, 12, 731723. [Google Scholar] [CrossRef]
- Ortiz, A.; Sansinenea, E. The role of beneficial microorganisms in soil quality and plant health. Sustainability 2022, 14, 5358. [Google Scholar] [CrossRef]
- Mohy-Ud-Din, W.; Akhtar, M.J.; Bashir, S.; Asghar, H.N.; Nawaz, M.F.; Chen, F. Isolation of Glyphosate-Resistant Bacterial Strains to Improve the Growth of Maize and Degrade Glyphosate under Axenic Condition. Agriculture 2023, 13, 886. [Google Scholar] [CrossRef]
- Kuiper, I.; Bloemberg, G.V.; Lugtenberg, B.J.J. Selection of a plant-bacterium pair as a novel tool for rhizostimulation of polycyclic aromatic hydrocarbon degrading bacteria. Mol. Plant-Microbe Interact. 2001, 14, 1197–1205. [Google Scholar] [CrossRef]
- Radhakrishnan, R.; Hashem, A.; Abd-Allah, E.F. Bacillus: A Biological Tool for Crop Improvement through Bio-Molecular Changes in Adverse Environments. Front. Physiol. 2017, 8, 667. [Google Scholar] [CrossRef]
- Imam, S.S.A.; Rajpoot, I.K.; Gajjar, B.; Sachdeva, A. Comparative Study of Heavy Metal Bioremediation in Soil by Bacillus subtilis and Saccharomyces Cerevisiae. Int. J. Sci. Technol. 2016, 9, 106911. [Google Scholar] [CrossRef]
- Bakiyaraj, R.; Baskaran, L.; Chidambaram, A.L.; Mahakavi, T.; Santhoshkumar, M. Bioremediation of Chromium by Bacillus subtilis and Pseudomonas Aeruginosa. Int. J. Curr. Microbiol. Appl. Sci. 2014, 3, 715–719. [Google Scholar]
- Sabae, S.Z.; Hazaa, M.; Hallim, S.A.; Awny, N.M.; Daboor, S.M. Bioremediation of Zn+2, Cu+2 and Fe+2 Using Bacillus subtilis D215 and Pseudomonas Putida Biovar AD225. Biosci. Res. 2006, 3, 189–204. [Google Scholar]
- Sakthipriya, N.; Doble, M.; Sangwai, J.S. Bioremediation of Coastal and Marine Pollution due to Crude Oil Using a Microorganism Bacillus subtilis. Procedia Eng. 2015, 116, 213–220. [Google Scholar] [CrossRef]
- Leong, Y.K.; Chang, J.S. Bioremediation of Heavy Metals Using Microalgae: Recent Advances and Mechanisms. Bioresour. Technol. 2020, 303, 122886. [Google Scholar] [CrossRef]
- Saeed, M.U.; Hussain, N.; Sumrin, A.; Shahbaz, A.; Noor, S.; Bilal, M.; Aleya, L.; Iqbal, H.M. Microbial Bioremediation Strategies with Wastewater Treatment Potentialities—A Review. Sci. Total Environ. 2022, 818, 151754. [Google Scholar] [CrossRef]
- Cimboláková, I.; Uher, I.; Laktičová, K.V.; Vargová, M.; Kimáková, T.; Papajová, I. Heavy Metals and the Environment. In Environmental Factors Affecting Human Health; IntechOpen: London, UK, 2019; p. 29. [Google Scholar]
- Kubier, A.; Wilkin, R.T.; Pichler, T. Cadmium in Soils and Groundwater: A Review. Appl. Geochem. 2019, 108, 104388. [Google Scholar] [CrossRef] [PubMed]
- Khan, Z.; Elahi, A.; Bukhari, D.A.; Rehman, A. Cadmium Sources, Toxicity, Resistance and Removal by Microorganisms—A Potential Strategy for Cadmium Eradication. J. Saudi Chem. Soc. 2022, 26, 101569. [Google Scholar] [CrossRef]
- Hussain, M.M.; Mohy-Ud-Din, W.; Younas, F.; Niazi, N.K.; Bibi, I.; Yang, X.; Rasheed, F.; Farooqi, Z.U.R. Biochar: A game changer for sustainable agriculture. Sustain. Agric. Tech. Progress. Transit. 2022, 143–157. [Google Scholar]
- Huybrechts, M.; Cuypers, A.; Deckers, J.; Iven, V.; Vandionant, S.; Jozefczak, M.; Hendrix, S. Cadmium and Plant Development: An Agony from Seed to Seed. Int. J. Mol. Sci. 2019, 20, 3971. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; Roychoudhury, A. Gibberellic Acid-Priming Promotes Fluoride Tolerance in a Susceptible Indica Rice Cultivar by Regulating the Antioxidant and Phytohormone Homeostasis. J. Plant Growth Regul. 2020, 39, 1476–1487. [Google Scholar] [CrossRef]
- Reed, R.C.; Bradford, K.J.; Khanday, I. Seed Germination and Vigor: Ensuring Crop Sustainability in a Changing Climate. Heredity 2022, 128, 450–459. [Google Scholar] [CrossRef]
- Shu, K.; Liu, X.-D.; Xie, Q.; He, Z.-H. Two Faces of One Seed: Hormonal Regulation of Dormancy and Germination. Mol. Plant 2016, 9, 34–45. [Google Scholar] [CrossRef]
- Seneviratne, M.; Rajakaruna, N.; Rizwan, M.; Madawala, H.; Ok, Y.S.; Vithanage, M. Heavy Metal-Induced Oxidative Stress on Seed Germination and Seedling Development: A Critical Review. Environ. Geochem. Health 2019, 41, 1813–1831. [Google Scholar] [CrossRef]
- El Rasafi, T.; Oukarroum, A.; Haddioui, A.; Song, H.; Kwon, E.E.; Bolan, N.; Tack, F.M.G.; Sebastian, A.; Prasad, M.; Rinklebe, J. Cadmium Stress in Plants: A Critical Review of the Effects, Mechanisms, and Tolerance Strategies. Crit. Rev. Environ. Sci. Technol. 2022, 52, 675–726. [Google Scholar] [CrossRef]
- Aslam, M.M.; Okal, E.J.; Waseem, M. Cadmium Toxicity Impacts Plant Growth and Plant Remediation Strategies. Plants 2022, 11, 111887. [Google Scholar] [CrossRef]
- Farooqi, Z.U.R.; Khursheed, M.M.; Gulzar, N.; Akram, M.A.; Hussain, M.M.; Qadeer, A.; Mohy-Ud-Din, W.; Younas, S. The Risk of Inorganic Environmental Pollution to Humans. In Nanotechnology for Environmental Pollution Decontamination Tools, Methods, and Approaches for Detection and Remediation; Apple Academic Press: Burlington, ON, Canada, 2022; Volume 39. [Google Scholar]
- Seifikalhor, M.; Hassani, S.B.; Aliniaeifard, S. Seed Priming by Cyanobacteria (Spirulina platensis) and Salep Gum Enhances Tolerance of Maize Plant against Cadmium Toxicity. J. Plant Growth Regul. 2020, 39, 1009–1021. [Google Scholar] [CrossRef]
- Sardar, R.; Ahmed, S.; Shah, A.A.; Yasin, N.A. Selenium Nanoparticles Reduced Cadmium Uptake, Regulated Nutritional Homeostasis and Antioxidative System in Coriandrum sativum Grown in Cadmium Toxic Conditions. Chemosphere 2022, 287, 132332. [Google Scholar] [CrossRef] [PubMed]
- Taie, H.A.; Seif El-Yazal, M.A.; Ahmed, S.M.; Rady, M.M. Polyamines Modulate Growth, Antioxidant Activity, and Genomic DNA in Heavy Metal–Stressed Wheat Plant. Environ. Sci. Pollut. Res. 2019, 26, 22338–22350. [Google Scholar] [CrossRef]
- Bailly, C. The Signalling Role of ROS in the Regulation of Seed Germination and Dormancy. Biochem. J. 2019, 476, 3019–3032. [Google Scholar] [CrossRef] [PubMed]
- Larbi, S.; Ben Kaab, S.; Teixeira da Silva, J.; Bettaieb Ben Kaab, L. Cadmium and Copper Stresses Affect Germination and Enzymatic Activities in Chickpea (Cicer arietinum L.). Agrochimica 2020, 2, 191–203. [Google Scholar] [CrossRef]
- Zhu, T.; Li, L.; Duan, Q.; Liu, X.; Chen, M. Progress in Our Understanding of Plant Responses to the Stress of Heavy Metal Cadmium. Plant Signal. Behav. 2021, 16, 1836884. [Google Scholar] [CrossRef]
- Witkowska, D.; Słowik, J.; Chilicka, K. Heavy Metals and Human Health: Possible Exposure Pathways and the Competition for Protein Binding Sites. Molecules 2021, 26, 6060. [Google Scholar] [CrossRef]
- Shanmugaraj, B.M.; Malla, A.; Ramalingam, S. Cadmium Stress and Toxicity in Plants: An Overview. In Cadmium Toxicity and Tolerance in Plants; Elsevier: Amsterdam, The Netherlands, 2019; pp. 1–17. [Google Scholar]
- Kaur, H.; Hussain, S.J. Cadmium: Bioavailability in Soils and Phytotoxicity. In Sustainable Solutions for Elemental Deficiency and Excess in Crop Plants; Springer: Singapore, 2020; pp. 351–391. [Google Scholar]
- Riaz, U.; Aslam, A.; uz Zaman, Q.; Javeid, S.; Gul, R.; Iqbal, S.; Javid, S.; Murtaza, G.; Jamil, M. Cadmium Contamination, Bioavailability, Uptake Mechanism and Remediation Strategies in Soil-Plant-Environment System: A Critical Review. Curr. Anal. Chem. 2021, 17, 49–60. [Google Scholar] [CrossRef]
- Ismael, M.A.; Elyamine, A.M.; Moussa, M.G.; Cai, M.; Zhao, X.; Hu, C. Cadmium in Plants: Uptake, Toxicity, and its Interactions with Selenium Fertilizers. Metallomics 2019, 11, 255–277. [Google Scholar] [CrossRef]
- Wani, K.I.; Zehra, A.; Choudhary, S.; Naeem, M.; Aftab, T. Cadmium, a Nonessential Heavy Metal: Uptake, Translocation, Signaling, Detoxification, and Impact on Amino Acid Metabolism. In Plant Metal and Metalloid Transporters; Springer: Singapore, 2022. [Google Scholar]
- Zhu, G.; Xiao, H.; Guo, Q.; Zhang, Z.; Zhao, J.; Yang, D. Effects of Cadmium Stress on Growth and Amino Acid Metabolism in Two Compositae Plants. Ecotoxicol. Environ. Saf. 2018, 158, 300–308. [Google Scholar] [CrossRef]
- Sakouhi, L.; Kharbech, O.; Massoud, M.B.; Munemasa, S.; Murata, Y.; Chaoui, A. Oxalic Acid Mitigates Cadmium Toxicity in Cicer arietinum L. Germinating Seeds by Maintaining the Cellular Redox Homeostasis. J. Plant Growth Regul. 2022, 41, 697–709. [Google Scholar] [CrossRef]
- Kaleem, M.; Shabir, F.; Hussain, I.; Hameed, M.; Ahmad, M.S.A.; Mehmood, A.; Ashfaq, W.; Riaz, S.; Afzaal, Z.; Maqsood, M.F. Alleviation of Cadmium Toxicity in Zea mays L. through Up-Regulation of Growth, Antioxidant Defense System and Organic Osmolytes under Calcium Supplementation. PLoS ONE 2022, 17, e0269162. [Google Scholar] [CrossRef] [PubMed]
- Alzahrani, Y.; Rady, M.M. Compared to Antioxidants and Polyamines, the Role of Maize Grain-Derived Organic Biostimulants in Improving Cadmium Tolerance in Wheat Plants. Ecotoxicol. Environ. Saf. 2019, 182, 109378. [Google Scholar] [CrossRef]
- Kudoyarova, G.R.; Dodd, I.C.; Veselov, D.S.; Rothwell, S.A.; Veselov, Y. Common and Specific Responses to Availability of Mineral Nutrients and Water. J. Exp. Bot. 2015, 66, 2133–2144. [Google Scholar] [CrossRef]
- Kul, R.; Ekinci, M.; Turan, M.; Ors, S.; Yildirim, E. How Abiotic Stress Conditions Affects Plant Roots. In Plant Roots; IntechOpen: London, UK, 2020; pp. 6–10. [Google Scholar]
- Ghori, N.-H.; Ghori, T.; Hayat, M.; Imadi, S.; Gul, A.; Altay, V.; Ozturk, M. Heavy Metal Stress and Responses in Plants. Int. J. Environ. Sci. Technol. 2019, 16, 1807–1828. [Google Scholar] [CrossRef]
- Zhao, H.; Guan, J.; Liang, Q.; Zhang, X.; Hu, H.; Zhang, J. Effects of Cadmium Stress on Growth and Physiological Characteristics of Sassafras Seedlings. Sci. Rep. 2021, 11, 9913. [Google Scholar] [CrossRef]
- Carvalho, M.E.; Castro, P.R.; Kozak, M.; Azevedo, R.A. The Sweet Side of Misbalanced Nutrients in Cadmium–Stressed Plants. Ann. Appl. Biol. 2020, 176, 275–284. [Google Scholar] [CrossRef]
- Guo, J.; Chen, T.; Zheng, G.; Yang, J.; Qian, T.; Liu, X.; Meng, X.; Li, Y. Cadmium Accumulation Responses in Hylotelephium spectabile: The Role of Photosynthetic Characteristics under Different Nitrogen, Moisture, and Light Conditions. Chemosphere 2023, 319, 138019. [Google Scholar] [CrossRef]
- Zhu, Z.; Huang, Y.; Wu, X.; Liu, Z.; Zou, J.; Chen, Y.; Su, N.; Cui, J. Increased Antioxidative Capacity and Decreased Cadmium Uptake Contribute to Hemin-Induced Alleviation of Cadmium Toxicity in Chinese Cabbage Seedlings. Ecotoxicol. Environ. Saf. 2019, 177, 47–57. [Google Scholar] [CrossRef]
- Giannakoula, A.; Therios, I.; Chatzissavvidis, C. Effect of Lead and Copper on Photosynthetic Apparatus in Citrus (Citrus aurantium L.) Plants. The Role of Antioxidants in Oxidative Damage as a Response to Heavy Metal Stress. Plants 2021, 10, 155. [Google Scholar] [CrossRef]
- Moustakas, M.; Moustaka, J.; Sperdouli, I. Hormesis in Photosystem II: A Mechanistic Understanding. Curr. Opin. Toxicol. 2022, 29, 57–64. [Google Scholar] [CrossRef]
- Singha, K.T.; Sebastian, A.; Prasad, M.N.V. Iron Plaque Formation in the Roots of Pistia stratiotes L.: Importance in Phytoremediation of Cadmium. Int. J. Phytoremediat. 2019, 21, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Kumar, V.; Shahzad, B.; Ramakrishnan, M.; Singh Sidhu, G.P.; Bali, A.S.; Handa, N.; Kapoor, D.; Yadav, P.; Khanna, K. Photosynthetic Response of Plants under Different Abiotic Stresses: A Review. J. Plant Growth Regul. 2020, 39, 509–531. [Google Scholar] [CrossRef]
- Rathi, S.; Mittal, N.; Kumar, D. Photosynthetic Response of Plants Against Heavy Metals. In Heavy Metals in Plants: Physiological to Molecular Approach; CRC Press: Boca Raton, FL, USA, 2022; pp. 1–23. [Google Scholar]
- Mir, A.R.; Pichtel, J.; Hayat, S. Copper: Uptake, Toxicity and Tolerance in Plants and Management of Cu-Contaminated Soil. Biometals 2021, 34, 737–759. [Google Scholar] [CrossRef] [PubMed]
- Jaouani, K.; Karmous, I.; Ostrowski, M.; El Ferjani, E.; Jakubowska, A.; Chaoui, A. Cadmium Effects on Embryo Growth of Pea Seeds during Germination: Investigation of the Mechanisms of Interference of the Heavy Metal with Protein Mobilization-Related Factors. J. Plant Physiol. 2018, 226, 64–76. [Google Scholar] [CrossRef]
- Fattahi, B.; Arzani, K.; Souri, M.K.; Barzegar, M. Effects of Cadmium and Lead on Seed Germination, Morphological Traits, and Essential Oil Composition of Sweet Basil (Ocimum basilicum L.). Ind. Crops Prod. 2019, 138, 111584. [Google Scholar] [CrossRef]
- Shah, A.A.; Khan, W.U.; Yasin, N.A.; Akram, W.; Ahmad, A.; Abbas, M.; Ali, A.; Safdar, M.N. Butanolide alleviated cadmium stress by improving plant growth, photosynthetic parameters and antioxidant defense system of Brassica oleracea. Chemosphere 2020, 261, 127728. [Google Scholar] [CrossRef]
- Chen, L.; Long, C.; Wang, D.; Yang, J. Phytoremediation of cadmium (Cd) and uranium (U) contaminated soils by Brassica juncea L. enhanced with exogenous application of plant growth regulators. Chemosphere 2020, 242, 125112. [Google Scholar] [CrossRef]
- Rizwan, M.; Ali, S.; ur Rehman, M.Z.; Adrees, M.; Arshad, M.; Qayyum, M.F.; Ali, L.; Hussain, A.; Chatha, S.A.S.; Imran, M. Alleviation of cadmium accumulation in maize (Zea mays L.) by foliar spray of zinc oxide nanoparticles and biochar to contaminated soil. Environ. Pollut. 2019, 248, 358–367. [Google Scholar] [CrossRef]
- Ullah, S.; Khan, J.; Hayat, K.; Abdelfattah Elateeq, A.; Salam, U.; Yu, B.; Ma, Y.; Wang, H.; Tang, Z.-H. Comparative study of growth, cadmium accumulation and tolerance of three chickpea (Cicer arietinum L.) cultivars. Plants 2020, 9, 310. [Google Scholar] [CrossRef]
- Tang, L.; Hamid, Y.; Liu, D.; Shohag, M.J.I.; Zehra, A.; He, Z.; Feng, Y.; Yang, X. Foliar application of zinc and selenium alleviates cadmium and lead toxicity of water spinach—Bioavailability/cytotoxicity study with human cell lines. Environ. Int. 2020, 145, 106122. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.S.; Abdelrahman, M.; Tran, C.D.; Nguyen, K.H.; Chu, H.D.; Watanabe, Y.; Fujita, M.; Tran, L.-S.P. Modulation of osmoprotection and antioxidant defense by exogenously applied acetate enhances cadmium stress tolerance in lentil seedlings. Environ. Pollut. 2022, 308, 119687. [Google Scholar] [CrossRef] [PubMed]
- Gutsch, A.; Keunen, E.; Guerriero, G.; Renaut, J.; Cuypers, A.; Hausman, J.-F.; Sergeant, K. Long-term cadmium exposure influences the abundance of proteins that impact the cell wall structure in Medicago sativa stems. Plant Biol. 2018, 20, 1023–1035. [Google Scholar] [CrossRef] [PubMed]
- Kaur, H.; Garg, N. Interactive effects of zinc-arbuscular mycorrhizal (AM) fungi on cadmium uptake, rubisco, osmolyte synthesis and yield in Cajanus cajan (L.) Millsp. Int. J. Sustain. Agric. Res. 2021, 8, 17–42. [Google Scholar] [CrossRef]
- Ahanger, M.A.; Aziz, U.; Alsahli, A.A.; Alyemeni, M.N.; Ahmad, P. Combined kinetin and spermidine treatments ameliorate growth and photosynthetic inhibition in Vigna angularis by up-regulating antioxidant and nitrogen metabolism under cadmium stress. Biomolecules 2020, 10, 147. [Google Scholar] [CrossRef]
- Hasan, N.; Choudhary, S.; Laskar, R.A.; Naaz, N.; Sharma, N. Comparative study of cadmium nitrate and lead nitrate [Cd(NO3)2 and Pb(NO3)2] stress in cyto-physiological parameters of Capsicum annuum L. Horticul. Environ. Biotechnol. 2022, 63, 627–641. [Google Scholar] [CrossRef]
- Zaid, A.; Mohammad, F. Methyl jasmonate and nitrogen interact to alleviate cadmium stress in Mentha arvensis by regulating physio-biochemical damages and ROS detoxification. J. Plant Growth Regul. 2018, 37, 1331–1348. [Google Scholar] [CrossRef]
- Alvarez, A.; Saez, J.M.; Costa, J.S.; Colin, V.L.; Fuentes, M.S.; Cuozzo, S.A.; Benimeli, C.S.; Polti, M.A.; Amoroso, M.J. Actinobacteria: Current research and perspectives for bioremediation of pesticides and heavy metals. Chemosphere 2017, 166, 41–62. [Google Scholar] [CrossRef]
- Mustafa, A.; Zulfiqar, U.; Mumtaz, M.Z.; Radziemska, M.; Haider, F.U.; Holatko, J.; Hammershmiedt, T.; Naveed, M.; Ali, H.; Kintl, A.; et al. Nickel (Ni) phytotoxicity and detoxification mechanisms: A review. Chemosphere 2023, 328, 138574. [Google Scholar] [CrossRef]
- Dixit, R.; Malaviya, D.; Pandiyan, K.; Singh, U.B.; Sahu, A.; Shukla, R.; Singh, B.P.; Rai, J.P.; Sharma, P.K.; Lade, H.; et al. Bioremediation of heavy metals from soil and aquatic environment: An overview of principles and criteria of fundamental processes. Sustainability 2015, 7, 2189–2212. [Google Scholar] [CrossRef]
- Ahemad, M. Remediation of metalliferous soils through the heavy metal resistant plant growth promoting bacteria: Paradigms and prospects. Arab. J. Chem. 2019, 12, 1365–1377. [Google Scholar] [CrossRef]
- Shabayev, V.P.; Bocharnikova, E.A.; Ostroumov, V.E. Remediation of cadmium-polluted soil using plant growth-promoting rhizobacteria and natural zeolite. Euras. Soil Sci. 2020, 53, 809–819. [Google Scholar] [CrossRef]
- He, X.; Xu, M.; Wei, Q.; Tang, M.; Guan, L.; Lou, L.; Xia, Y. Promotion of Growth and Phytoextraction of Cadmium and Lead in Solanum nigrum L. Mediated by Plant-Growth-Promoting Rhizobacteria. Ecotoxicol. Environ. Saf. 2020, 205, 111333. [Google Scholar] [CrossRef] [PubMed]
- Mitra, S.; Purkait, T.; Pramanik, K.; Maiti, T.K.; Dey, R.S. Three-Dimensional Graphene for Electrochemical Detection of Cadmium in Klebsiella michiganensis to Study the Influence of Cadmium Uptake in Rice Plant. Mater. Sci. Eng. C 2019, 103, 109802. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yan, Z.; Gu, D.; Li, D.; Tao, Y.; Zhang, D.; Ao, Y. Characterization of Cadmium-Resistant Rhizobacteria and Their Promotion Effects on Brassica napus Growth and Cadmium Uptake. J. Basic Microbiol. 2019, 59, 579–590. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yu, X.; Cui, Y.; Tu, W.; Shen, T.; Yan, M.; Ma, M. The Potential of Cadmium Ion-Immobilized Rhizobium pusense KG2 to Prevent Soybean Root from Absorbing Cadmium in Cadmium-Contaminated Soil. J. Appl. Microbiol. 2019, 126, 919–930. [Google Scholar] [CrossRef]
- Yankey, R.; Karanja, J.K.; Okal, E.J.; Omoor, I.N.A.; Lin, H.; Bodjremou, D.M.; Lin, Z.X. A Consortium of Plant Growth-Promoting Rhizobacteria Strains Synergistically Assists Jujuncao (Pennisetum giganteum) to Remediate Cadmium Contaminated Soils. Appl. Ecol. Environ. Res. 2021, 19, 2425–2442. [Google Scholar] [CrossRef]
- Wu, B.; He, T.; Wang, Z.; Qiao, S.; Wang, Y.; Xu, F.; Xu, H. Insight into the Mechanisms of Plant Growth Promoting Strain SNB6 on Enhancing the Phytoextraction in Cadmium Contaminated Soil. J. Hazard. Mater. 2020, 385, 121587. [Google Scholar] [CrossRef]
- Abdelkrim, S.; Jebara, S.H.; Saadani, O.; Abid, G.; Taamalli, W.; Zemni, H.; Jebara, M. In Situ Effects of Lathyrus sativus-PGPR to Remediate and Restore Quality and Fertility of Pb and Cd Polluted Soils. Ecotoxicol. Environ. Saf. 2020, 192, 110260. [Google Scholar] [CrossRef]
- Li, Y.; Zeng, J.; Wang, S.; Lin, Q.; Ruan, D.; Chi, H.; Qiu, R.; Yang, Y. Effects of Cadmium-Resistant Plant Growth-Promoting Rhizobacteria and Funneliformis mosseae on the Cadmium Tolerance of Tomato (Lycopersicon esculentum L.). Int. J. Phytoremed. 2020, 22, 451–458. [Google Scholar] [CrossRef]
- Kotoky, R.; Nath, S.; Kumar Maheshwari, D.; Pandey, P. Cadmium Resistant Plant Growth Promoting Rhizobacteria Serratia marcescens S2I7 Associated with the Growth Promotion of Rice Plant. Environ. Sustain. 2019, 2, 135–144. [Google Scholar] [CrossRef]
- Liaquat, F.; Munis, M.F.H.; Arif, S.; Haroon, U.; Shengquan, C.; Qunlu, L. Cd-Tolerant SY-2 Strain of Stenotrophomonas maltophilia: A Potential PGPR, Isolated from the Nanjing Mining Area in China. 3 Biotech 2020, 10, 519. [Google Scholar] [CrossRef] [PubMed]
- Belimov, A.A.; Shaposhnikov, A.I.; Azarova, T.S.; Makarova, N.M.; Safronova, V.I.; Litvinskiy, V.A.; Nosikov, V.N.; Zavalin, A.A.; Tikhonovich, I.A. Microbial Consortium of PGPR, Rhizobia and Arbuscular Mycorrhizal Fungus Makes Pea Mutant SGECdt Comparable with Indian Mustard in Cadmium Tolerance and Accumulation. Plants 2020, 9, 975. [Google Scholar] [CrossRef] [PubMed]
- You, M.; Wang, L.; Zhou, G.; Wang, Y.; Wang, K.; Zou, R.; Cao, W.; Fan, H. Effects of Microbial Agents on Cadmium Uptake in Solanum nigrum L. and Rhizosphere Microbial Communities in Cadmium-Contaminated Soil. Front. Microbiol. 2022, 13, 1106254. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.; Wang, W.; Zheng, X.; Chen, X.; Fu, W.; Wang, G.; Ji, J.; Jin, C.; Guan, C. Improvement of the Cd and Zn Phytoremediation Efficiency of Rice (Oryza sativa) through the Inoculation of a Metal-Resistant PGPR Strain. Chemosphere 2022, 302, 134900. [Google Scholar] [CrossRef] [PubMed]
- Sumranwanich, T.; Leartsiwawinyu, W.; Meeinkuirt, W.; Chayapan, P. Application of Plant Growth-Promoting Rhizobacteria (PGPR) Associated with Energy Plant, Pennisetum purpurenum, in Cadmium and Zinc Contaminated Soil. Res. Sq. 2022. [Google Scholar] [CrossRef]
- Xu, Z.; Wang, D.; Tang, W.; Wang, L.; Li, Q.; Lu, Z.; Guo, S. Phytoremediation of Cadmium-Polluted Soil Assisted by D-gluconate-Enhanced Enterobacter cloacae Colonization in the Solanum nigrum L. Rhizosphere. Sci. Total Environ. 2020, 732, 139265. [Google Scholar] [CrossRef]
- Ali, Q.; Ayaz, M.; Yu, C.; Wang, Y.; Gu, Q.; Wu, H.; Gao, X. Cadmium Tolerant Microbial Strains Possess Different Mechanisms for Cadmium Biosorption and Immobilization in Rice Seedlings. Chemosphere 2022, 303, 135206. [Google Scholar] [CrossRef]
- Khanna, K.; Jamwal, V.L.; Sharma, A.; Gandhi, S.G.; Ohri, P.; Bhardwaj, R.; Ahmad, P. Supplementation with Plant Growth Promoting Rhizobacteria (PGPR) Alleviates Cadmium Toxicity in Solanum lycopersicum by Modulating the Expression of Secondary Metabolites. Chemosphere 2019, 230, 628–639. [Google Scholar] [CrossRef]
- Ghosh, A.; Pramanik, K.; Bhattacharya, S.; Mondal, S.; Ghosh, S.K.; Maiti, T.K. A Potent Cadmium Bioaccumulating Enterobacter cloacae Strain Displays Phytobeneficial Property in Cd-Exposed Rice Seedlings. Curr. Res. Microb. Sci. 2022, 3, 100101. [Google Scholar] [CrossRef]
- Patel, M.; Patel, K.; Al-Keridis, L.A.; Alshammari, N.; Badraoui, R.; Elasbali, A.M.; Adnan, M. Cadmium-Tolerant Plant Growth-Promoting Bacteria Curtobacterium oceanosedimentum Improves Growth Attributes and Strengthens Antioxidant System in Chili (Capsicum frutescens). Sustainability 2022, 14, 4335. [Google Scholar] [CrossRef]
- Cheng, X.; Cao, X.; Tan, C.; Liu, L.; Bai, J.; Liang, Y.; Cai, R. Effects of Four Endophytic Bacteria on Cadmium Speciation and Remediation Efficiency of Sedum plumbizincicola in Farmland Soil. Environ. Sci. Pollut. Res. 2022, 29, 89557–89569. [Google Scholar] [CrossRef] [PubMed]
- Chellaiah, E.R. Cadmium (Heavy Metals) Bioremediation by Pseudomonas aeruginosa: A Mini review. Appl. Water Sci. 2018, 8, 154. [Google Scholar] [CrossRef]
- Smieljan, A.; Wilkinson, K.J.; Rossier, C. Cd Bioaccumulation by a Freshwater Bacterium, Rhodospirillum rubrum. Environ. Sci. Technol. 2003, 37, 701–706. [Google Scholar] [CrossRef] [PubMed]
- Arivalagan, P.; Singaraj, D.; Haridass, V.; Kaliannan, T. Removal of Cadmium from Aqueous Solution by Batch Studies Using Bacillus cereus. Ecol. Eng. 2014, 71, 728–735. [Google Scholar] [CrossRef]
- Bagot, D.; Lebeau, T.; Jezequel, K. Microorganisms for Remediation of Cadmium-Contaminated Soils. Environ. Chem. Lett. 2006, 4, 207–211. [Google Scholar] [CrossRef]
- Ahmad, I.; Akhtar, M.J.; Zahir, Z.A.; Naveed, M.; Mitter, B.; Sessitsch, A. Cadmium-Tolerant Bacteria Induce Metal Stress Tolerance in Cereals. Environ. Sci. Pollut. Res. 2014, 21, 11054–11065. [Google Scholar] [CrossRef]
- Brunetti, G.; Farrag, K.; Soler-Rovira, P.; Ferrara, M.; Nigro, F.; Senesi, N. The Effect of Compost and Bacillus licheniformis on the Phytoextraction of Cr, Cu, Pb and Zn by Three Brassicaceae Species from Contaminated Soils in the Apulia Region, Southern Italy. Geoderma 2012, 170, 322–330. [Google Scholar] [CrossRef]
- Awan, S.A.; Ilyas, N.; Khan, I.; Raza, M.A.; Rehman, A.U.; Rizwan, M.; Rastogi, A.; Tariq, R.; Brestic, M. Bacillus siamensis Reduces Cadmium Accumulation and Improves Growth and Antioxidant Defense System in Two Wheat (Triticum aestivum L.) Varieties. Plants 2020, 9, 878. [Google Scholar] [CrossRef]
- Belimov, A.A.; Hontzeas, N.; Safronova, V.I.; Demchinskaya, S.V.; Piluzza, G.; Bullitta, S.; Glick, B.R. Cadmium-Tolerant Plant Growth-Promoting Bacteria Associated with the Roots of Indian Mustard (Brassica juncea L. Czern.). Soil Biol. Biochem. 2005, 37, 241–250. [Google Scholar] [CrossRef]
- Dell’Amico, E.; Cavalca, L.; Andreoni, V. Analysis of Rhizobacterial Communities in Perennial Graminaceae from Polluted Water Meadow Soil, and Screening of Metal-Resistant, Potentially Plant Growth-Promoting Bacteria. FEMS Microbiol. Ecol. 2005, 52, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Ganesan, V. Rhizoremediation of Cadmium Soil Using a Cadmium-Resistant Plant Growth-Promoting Rhizopseudomonad. Curr. Microbiol. 2008, 56, 403–407. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S.; Mukherjee, S.K. Pseudomonas aeruginosa KUCd1, a Possible Candidate for Cadmium Bioremediation. Braz. J. Microbiol. 2009, 40, 655–662. [Google Scholar] [CrossRef]
- Dary, M.; Chamber-Pérez, M.A.; Palomares, A.J.; Pajuelo, E. In Situ Phytostabilisation of Heavy Metal Polluted Soils Using Lupinus luteus Inoculated with Metal Resistant Plant-Growth Promoting Rhizobacteria. J. Hazard. Mater. 2010, 177, 323–330. [Google Scholar] [CrossRef]
- Jeong, S.; Moon, H.S.; Nam, K.; Kim, J.Y.; Kim, T.S. Application of Phosphate-Solubilizing Bacteria for Enhancing Bioavailability and Phytoextraction of Cadmium (Cd) from Polluted Soil. Chemosphere 2012, 88, 204–210. [Google Scholar] [CrossRef] [PubMed]
- Ahemad, M.; Kibret, M. Mechanisms and Applications of Plant Growth Promoting Rhizobacteria: Current Perspective. J. King Saud Univ. Sci. 2014, 26, 1–20. [Google Scholar] [CrossRef]
- Antoniadis, V.; Levizou, E.; Shaheen, S.M.; Ok, Y.S.; Sebastian, A.; Baum, C.; Prasad, M.N.V.; Wenzel, W.W.; Rinklebe, J. Trace Elements in the Soil-Plant Interface: Phytoavailability, Translocation, and Phytoremediation—A Review. Earth-Science Rev. 2017, 171, 621–645. [Google Scholar] [CrossRef]
- Satyanarayana, T.; Deshmukh, S.K.; Deshpande, M.V. Advancing Frontiers in Mycology & Mycotechnology: Basic and Applied Aspects of Fungi, 1st ed.; Springer: Singapore, 2019; pp. 1–675. [Google Scholar]
- Ali, A.; Guo, D.; Mahar, A.; Wang, P.; Shen, F.; Li, R.; Zhang, Z. Mycoremediation of Potentially Toxic Trace Elements—A Biological Tool for Soil Cleanup: A Review. Pedosphere 2017, 27, 205–222. [Google Scholar] [CrossRef]
- Harms, H.; Schlosser, D.; Wick, L.Y. Untapped Potential: Exploiting Fungi in Bioremediation of Hazardous Chemicals. Nat. Rev. Microbiol. 2011, 9, 177–192. [Google Scholar] [CrossRef]
- Jacob, J.M.; Karthik, C.; Saratale, R.G.; Kumar, S.S.; Prabakar, D.; Kadirvelu, K.; Pugazhendhi, A. Biological Approaches to Tackle Heavy Metal Pollution: A Survey of Literature. J. Environ. Manag. 2018, 217, 56–70. [Google Scholar] [CrossRef]
- Igiri, B.E.; Okoduwa, S.I.; Idoko, G.O.; Akabuogu, E.P.; Adeyi, A.O.; Ejiogu, I.K. Toxicity and bioremediation of heavy metals contaminated ecosystem from tannery wastewater: A review. J. Toxicol. 2018, 2018, 2568038. [Google Scholar] [CrossRef] [PubMed]
- Boswell, G.P.; Jacobs, H.; Ritz, K.; Gadd, G.M.; Davidson, F.A. The development of fungal networks in complex environments. Bull. Math. Biol. 2007, 69, 605–634. [Google Scholar] [CrossRef] [PubMed]
- Mani, D.; Kumar, C. Biotechnological advances in bioremediation of heavy metals contaminated ecosystems: An overview with special reference to phytoremediation. Int. J. Environ. Sci. Technol. 2014, 11, 843–872. [Google Scholar] [CrossRef]
- Kumar, S.; Prasad, S.; Yadav, K.K.; Shrivastava, M.; Gupta, N.; Nagar, S.; Bach, Q.V.; Kamyab, H.; Khan, S.A.; Yadav, S.; et al. Hazardous heavy metals contamination of vegetables and food chain: Role of sustainable remediation approaches—A review. Environ. Res. 2019, 179, 108792. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Liang, Y.; Han, X.; Chiu, T.Y.; Ghosh, A.; Chen, H.; Tang, M. The roles of arbuscular mycorrhizal fungi (AMF) in phytoremediation and tree-herb interactions in Pb contaminated soil. Sci. Rep. 2016, 6, 20469. [Google Scholar] [CrossRef]
- Begum, N.; Qin, C.; Ahanger, M.A.; Raza, S.; Khan, M.I.; Ashraf, M.; Ahmed, N.; Zhang, L. Role of Arbuscular Mycorrhizal Fungi in Plant Growth Regulation: Implications in Abiotic Stress Tolerance. Front. Plant Sci. 2019, 10, 1068. [Google Scholar] [CrossRef]
- Khan, A.G. Producing Mycorrhizal Inoculum for Pytoremedation. In Phytoremediation. Methods in Biotechnology; Humana Press: Totowa, NJ, USA, 2005; pp. 89–97. [Google Scholar]
- Xiao, X.; Luo, S.; Zeng, G.; Wei, W.; Wan, Y.; Chen, L.; Guo, H.; Cao, Z.; Yang, L.; Chen, J.; et al. Biosorption of cadmium by endophytic fungus (EF) Microsphaeropsis sp. LSE10 isolated from cadmium hyperaccumulator Solanum nigrum L. Bioresour. Technol. 2010, 101, 1668–1674. [Google Scholar] [CrossRef]
- Cabral, L.; Soares, C.R.; Giachini, A.J.; Siqueira, J.O. Arbuscular mycorrhizal fungi in phytoremediation of contaminated areas by trace elements: Mechanisms and major benefits of their applications. World J. Microbiol. Biotechnol. 2015, 31, 1655–1664. [Google Scholar] [CrossRef]
- Zhang, F.; Liu, M.; Li, Y.; Che, Y.; Xiao, Y. Effects of arbuscular mycorrhizal fungi, biochar and cadmium on the yield and element uptake of Medicago sativa. Sci. Total Environ. 2019, 655, 1150–1158. [Google Scholar] [CrossRef]
- Wang, F.; Liu, X.; Shi, Z.; Tong, R.; Adams, C.A.; Shi, X. Arbuscular mycorrhizae alleviate negative effects of zinc oxide nanoparticle and zinc accumulation in maize plants—A soil microcosm experiment. Chemosphere 2016, 147, 88–97. [Google Scholar] [CrossRef]
- Garg, N.; Bhandari, P. Cadmium toxicity in crop plants and its alleviation by arbuscular mycorrhizal (AM) fungi: An overview. Plant Biosyst. 2014, 148, 609–621. [Google Scholar] [CrossRef]
- Varma, A.; Sherameti, I.; Tripathi, S.; Prasad, R.; Das, A.; Sharma, M.; Bakshi, M.; Johnson, J.M.; Bhardwaj, S.; Arora, M.; et al. The symbiotic fungus Piriformosporaindica: Review. In Fungal Associations; Springer: Berlin/Heidelberg, Germany, 2012; pp. 231–254. [Google Scholar]
- Sahu, A.; Mandal, A.; Thakur, J.; Manna, M.; Rao, A.S. Exploring bioaccumulation efficacy of Trichoderma viride: An alternative bioremediation of cadmium and lead. Natl. Acad. Sci. Lett. 2012, 35, 299–302. [Google Scholar] [CrossRef]
- Yaghoubian, Y.; Siadat, S.A.; Telavat, M.R.M.; Pirdashti, H.; Yaghoubia, I. Bioremoval of cadmium from aqueous solutions by filamentous fungi: Trichoderma spp. and Piriformospora indica. Environ. Sci. Pollut. Res. 2019, 26, 7863–7872. [Google Scholar] [CrossRef]
- Barros, L.; Macedo, G.; Duarte, M.; Silva, E.; Lobato, A.K.C.L. Biosorption of cadmium using the fungus Aspergillus niger. Braz. J. Chem. Eng. 2003, 20, 229–239. [Google Scholar] [CrossRef]
- Tripathi, P.; Singh, P.C.; Mishra, A.; Chauhan, P.S.; Dwivedi, S.; Bais, R.T.; Tripathi, R.D. Trichoderma: A Potential Bioremediator for Environmental Clean up. Clean Technol. Environ. Policy 2013, 15, 541–550. [Google Scholar] [CrossRef]
- Wang, B.; Liu, L.; Gao, Y.; Chen, J. Improved phytoremediation of oilseed rape (Brassica napus) by Trichoderma mutant constructed by restriction enzyme-mediated integration (REMI) in cadmium polluted soil. Chemosphere 2009, 74, 1400–1403. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zhou, Q. Single and Joint Toxicity of Chlorimuron-Ethyl, Cadmium, and Copper Acting on Wheat Triticum aestivum. Ecotoxicol. Environ. Saf. 2005, 60, 169–175. [Google Scholar] [CrossRef]
- Aksu, Z. Equilibrium and Kinetic Modeling of Cadmium (II) Biosorption by C. vulgaris in a Batch System. Purify Technol. 2002, 21, 285–294. [Google Scholar] [CrossRef]
- Mohsenzadeh, F.; Shahrokhi, F. Biological Removing of Cadmium from Contaminated Media by Fungal Biomass of Trichoderma Species. J. Environ. Health Sci. Eng. 2014, 12, 102. [Google Scholar] [CrossRef]
- Olguín, E.J.; Sánchez-Galván, G. Heavy Metal Removal in Phytofiltration and Phycoremediation: The Need to Differentiate between Bioadsorption and Bioaccumulation. New Biotechnol. 2012, 30, 3–8. [Google Scholar] [CrossRef]
- Sattayawat, P.; Yunus, I.S.; Noirungsee, N.; Mukjang, N.; Pathom-Aree, W.; Pekkoh, J.; Pumas, C. Synthetic Biology-Based Approaches for Microalgal Bio-removal of Heavy Metals from Wastewater Effluents. Front. Environ. Sci. 2021, 9, 562. [Google Scholar] [CrossRef]
- Kumar, S.S.; Kadier, A.; Malyan, S.K.; Ahmad, A.; Bishnoi, N.R. Phytoremediation and Rhizoremediation: Uptake, Mobilization and Sequestration of Heavy Metals by Plants, Plant-Microbe Interactions in Agro-Ecological Perspectives; Springer: Singapore, 2017; pp. 309–332. [Google Scholar]
- Chen, H.; Arocena, J.M.; Li, J.; Thring, R.W.; Zhou, J. Assessments of Chromium (and Other Metals) in Vegetables and Potential Bio-accumulations in Humans Living in Areas Affected by Tannery Wastes. Chemosphere 2014, 112, 412–419. [Google Scholar] [CrossRef] [PubMed]
- Davis, T.A.; Volesky, B.; Mucci, A. A Review of the Biochemistry of Heavy Metal Biosorption by Brown Algae. Water Res. 2003, 37, 4311–4330. [Google Scholar] [CrossRef] [PubMed]
- Rybak, A.; Messyasz, B.; Łeska, B. Freshwater Ulva (Chlorophyta) as a Bioaccumulator of Selected Heavy Metals (Cd, Ni and Pb) and Alkaline Earth Metals (Ca and Mg). Chemosphere 2012, 89, 1066–1076. [Google Scholar] [CrossRef]
- Soeprobowati, T.R.; Hariyati, R. Bioaccumulation of Pb, Cd, Cu, and Cr by Porphyridium cruentum (S.F. Gray) Nägeli. Int. J. Mar. Sci. 2013, 3, 212–218. [Google Scholar] [CrossRef]
- Saunders, R.J.; Paul, N.A.; Hu, Y.; de Nys, R. Sustainable sources of biomass for bioremediation of heavy metals in waste water derived from coal-fired power generation. PLoS ONE. 2012, 7, e36470. [Google Scholar] [CrossRef] [PubMed]
- Kumar, J.I.; Oommen, C. Removal of heavy metals by biosorption using freshwater alga Spirogyra hyalina. J. Environ. Biol. 2012, 33, 27–31. [Google Scholar]
- Kotrba, P.; Mackova, M.; Macek, T. Microbial Biosorption of Metals—General Introduction. In Microbial Biosorption of Metals; Springer: Dordrecht, The Netherlands, 2011; pp. 1–329. [Google Scholar]
- Tuzen, M.; Sari, A. Biosorption of Selenium from Aqueous Solution by Green Algae (Cladophora hutchinsiae) Biomass: Equilibrium, Thermodynamic and Kinetic Studies. Chem. Eng. J. 2010, 158, 200–206. [Google Scholar] [CrossRef]
- Sarin, C.; Sarin, S. Removal of Cadmium and Zinc from Soil Using Immobilized Cell of Biosurfactant Producing Bacteria. Environ. Asia 2010, 3, 49–53. [Google Scholar]
- Zhu, Z.; Yang, X.; Wang, K.; Huang, H.; Zhang, X.; Fang, H.; He, Z. Bioremediation of Cd-DDT Co-Contaminated Soil Using the Cd-Hyperaccumulator Sedum alfredii and DDT-Degrading Microbes. J. Hazard. Mater. 2012, 235–236, 144–151. [Google Scholar] [CrossRef]
- Haq, F.; Butt, M.; Ali, H.; Chaudhary, H.J. Biosorption of Cadmium and Chromium from Water by Endophytic Kocuria rhizophila: Equilibrium and Kinetic Studies. Desalination Water Treat. 2015, 57, 19946–19958. [Google Scholar] [CrossRef]
- Mitra, S.; Pramanik, K.; Ghosh, P.K.; Soren, T.; Sarkar, A.; Dey, R.S.; Pandey, S.; Maiti, T.K. Characterization of Cd-Resistant Klebsiella michiganensis MCC3089 and Its Potential for Rice Seedling Growth Promotion under Cd Stress. Microbiol. Res. 2018, 210, 12–25. [Google Scholar] [CrossRef] [PubMed]
- Mitra, S.; Pramanik, K.; Sarkar, A.; Ghosh, P.K.; Soren, T.; Maiti, T.K. Bioaccumulation of Cadmium by Enterobacter sp. and Enhancement of Rice Seedling Growth under Cadmium Stress. Ecotoxicol. Environ. Saf. 2018, 156, 183–196. [Google Scholar] [CrossRef] [PubMed]
- Peng, W.; Li, X.; Song, J.; Jiang, W.; Liu, Y.; Fan, W. Bioremediation of cadmium-and zinc-contaminated soil using Rhodobacter sphaeroides. Chemosphere 2018, 197, 33–41. [Google Scholar] [CrossRef]
- Khadim, H.J.; Ammar, S.H.; Ebrahim, S.E. Biomineralization based remediation of cadmium and nickel contaminated wastewater by ureolytic bacteria isolated from barn horses soil. Environ. Technol. Innov. 2019, 14, 100315. [Google Scholar] [CrossRef]
- Zhang, C.; Tao, Y.; Li, S.; Ke, T.; Wang, P.; Wei, S.; Chen, L. Bioremediation of cadmium-trichlorfon co-contaminated soil by Indian mustard (Brassica juncea) associated with the trichlorfon-degrading microbe Aspergillus sydowii: Related physiological responses and soil enzyme activities. Ecotoxicol. Environ. Saf. 2019, 179, 109756. [Google Scholar] [CrossRef]
- Zeng, X.; Xu, H.; Lu, J.; Chen, Q.; Li, W.; Wu, L.; Tang, J.; Ma, L. The Immobilization of Soil Cadmium by the Combined Amendment of Bacteria and Hydroxyapatite. Sci. Rep. 2020, 10, 20473. [Google Scholar] [CrossRef]
- Wang, Y.; Luo, Y.; Zeng, G.; Wu, X.; Wu, B.; Li, X.; Xu, H. Characteristics and in situ remediation effects of heavy metal immobilizing bacteria on cadmium and nickel co-contaminated soil. Ecotoxicol. Environ. Saf. 2020, 192, 110294. [Google Scholar] [CrossRef]
- Ma, H.; Wei, M.; Wang, Z.; Hou, S.; Li, X.; Heng, X. Bioremediation of cadmium polluted soil using a novel cadmium immobilizing plant growth promotion strain Bacillus sp. TZ5 loaded on biochar. J. Hazard. Mater. 2020, 388, 124065. [Google Scholar] [CrossRef]
- Xu, M.; Liu, Y.; Deng, Y.; Zhang, S.; Hao, X.; Zhu, P.; Zhou, J.; Yin, H.; Liang, Y.; Jiang, H.; et al. Bioremediation of cadmium-contaminated paddy soil using an autotrophic and heterotrophic mixture. RSC Adv. 2020, 10, 26090–26101. [Google Scholar] [CrossRef]
- Minari, G.D.; Saran, L.M.; Lima Constancio, M.T.; Correia da Silva, R.; Rosalen, D.L.; José de Melo, W.; Carareto Alves, L.M. Bioremediation potential of new cadmium, chromium, and nickel-resistant bacteria isolated from tropical agricultural soil. Ecotoxicol. Environ. Saf. 2020, 204, 111038. [Google Scholar] [CrossRef] [PubMed]
- Oziegbe, O.; Oluduro, A.O.; Oziegbe, E.J.; Ahuekwe, E.F.; Olorunsola, S.J. Assessment of heavy metal bioremediation potential of bacterial isolates from landfill soils. Saudi J. Biol. Sci. 2021, 28, 3948–3956. [Google Scholar] [CrossRef] [PubMed]
- Haider, F.U.; Coulter, J.A.; Cheema, S.A.; Farooq, M.; Wu, J.; Zhang, R.; Shuaijie, G.; Liqun, C. Co-application of Biochar and Microorganisms Improves Soybean Performance and Remediate Cadmium-Contaminated Soil. Ecotoxicol. Environ. Saf. 2021, 214, 112112. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Zhao, J.; Liu, X.; Sun, L.; Tian, J.; Wu, N. Cadmium Pollution Impact on the Bacterial Community Structure of Arable Soil and the Isolation of the Cadmium Resistant Bacteria. Front. Microbiol. 2021, 12, 698834. [Google Scholar] [CrossRef] [PubMed]
- Mo, T.; Jiang, D.; Shi, D.; Xu, S.; Huang, X.; Huang, Z. Remediation mechanism of “double-resistant” bacteria—Sedum alfredii Hance on Pb- and Cd-contaminated soil. Ecol. Process 2022, 11, 20. [Google Scholar] [CrossRef]
- Haider, F.U.; Farooq, M.; Naveed, M.; Cheema, S.A.; ul Ain, N.; Salim, M.A.; Liqun, C.; Mustafa, A. Influence of Biochar and Microorganism Co-Application on Stabilization of Cadmium (Cd) and Improved Maize Growth in Cd-Contaminated Soil. Front. Plant Sci. 2022, 13, 983830. [Google Scholar] [CrossRef]
- Yuan, B.; Huang, L.; Liu, X.; Bai, L.; Liu, H.; Jiang, H.; Zhu, P.; Xiao, Y.; Geng, J.; Liu, Q.; et al. Application of Mixotrophic Acidophiles for the Bioremediation of Cadmium-Contaminated Soils Elevates Cadmium Removal, Soil Nutrient Availability, and Rice Growth. Ecotoxicol. Environ. Saf. 2022, 236, 113499. [Google Scholar] [CrossRef]
- Lu, H.; Xia, C.; Chinnathambi, A.; Nasif, O.; Narayanan, M.; Shanmugam, S.; Chi, N.T.L.; Pugazhendhi, A.; On-Uma, R.; Jutamas, K.; et al. Evaluation of Cadmium Tolerance and Remediated Efficacy of Wild and Mutated Enterobacter Species Isolated from Potassium Nitrate (KNO₃) Processing Unit Contaminated Soil. Chemosphere 2023, 311, 136899. [Google Scholar] [CrossRef]
- Xu, T.; Xi, J.; Ke, J.; Wang, Y.; Chen, X.; Zhang, Z.; Lin, Y. Deciphering soil amendments and actinomycetes for remediation of cadmium (Cd) contaminated farmland. Ecotoxicol. Environ. Saf. 2023, 249, 114388. [Google Scholar] [CrossRef]
- Peng, C.; Zhao, X.; Ji, X.; Wu, J.; Liang, W.; Song, H.; Zhang, W.; Wang, X. Mixed Bacteria Passivation for the Remediation of Arsenic, Lead, and Cadmium: Medium Optimization and Mechanisms. Process Saf. Environ. Prot. 2023, 170, 720–727. [Google Scholar] [CrossRef]
- Khan, I.; Aftab, M.; Shakir, S.U.; Ali, M.; Qayyum, S.; Rehman, M.U.; Haleem, K.S.; Touseef, I. Mycoremediation of heavy metal (Cd and Cr)–polluted soil through indigenous metallotolerant fungal isolates. Environ. Monit. Assess. 2019, 191, 585. [Google Scholar] [CrossRef] [PubMed]
- Jin, Z.; Deng, S.; Wen, Y.; Jin, Y.; Pan, L.; Zhang, Y.; Black, T.; Jones, K.C.; Zhang, H.; Zhang, D. Application of Simplicillium chinense for Cd and Pb biosorption and enhancing heavy metal phytoremediation of soils. Sci. Total Environ. 2019, 697, 134148. [Google Scholar] [CrossRef] [PubMed]
- Bano, A.; Hussain, J.; Akbar, A.; Mehmood, K.; Anwar, M.; Hasni, M.S.; Ullah, S.; Sajid, S.; Ali, I. Biosorption of heavy metals by obligate halophilic fungi. Chemosphere 2018, 199, 218–222. [Google Scholar] [CrossRef] [PubMed]
- Noormohamadi, H.R.; Fat’hi, M.R.; Ghaedi, M.; Ghezelbash, G.R. Potentiality of white-rot fungi in biosorption of nickel and cadmium: Modeling optimization and kinetics study. Chemosphere 2019, 216, 124–130. [Google Scholar] [CrossRef]
- Lamrood, P.Y.; Ralegankar, S.D. Biosorption of Cu, Zn, Fe, Cd, Pb and Ni by non-treated biomass of some edible mushrooms. Asian J. Exp. Biol. Sci. 2013, 4, 190195. [Google Scholar]
- Nagy, B.; Maicaneanu, A.; Indolean, C.; Manzatu, C.; Silaghi-Dumitrescu, L.; Majdik, C. Comparative study of Cd(II) biosorption on cultivated Agaricus bisporus and wild Lactarius piperatus based biocomposites. Linear and nonlinear equilibrium modelling and kinetics. J. Taiwan Inst. Chem. Eng. 2014, 45, 921–929. [Google Scholar] [CrossRef]
- Hussain, B.; Ashraf, M.N.; ur Rahman, S.; Abbas, A.; Li, J.; Farooq, M. Cadmium stress in paddy fields: Effects of soil conditions and remediation strategies. Sci. Total Environ. 2021, 754, 142188. [Google Scholar] [CrossRef]
- Romera, E.; González, F.; Ballester, A.; Blázquez, M.L.; Munoz, J.A. Comparative study of biosorption of heavy metals using different types of algae. Bioresour. Technol. 2007, 98, 3344–3353. [Google Scholar] [CrossRef]
- Sjahrul, M.; Arifin, D. Phytoremediation of Cd2+ by marine phytoplanktons, Tetracelmis chuii and Chaetoceros calcitrans. Int. J. Chem. 2012, 4, 69–74. [Google Scholar] [CrossRef]
- Ibrahim, W.M.; Hassan, A.F.; Azab, Y.A. Biosorption of toxic heavy metals from aqueous solution by Ulva lactuca activated carbon. Egypt. J. Basic Appl. Sci. 2016, 3, 241–249. [Google Scholar] [CrossRef]
- Sooksawat, N.; Meetam, M.; Kruatrachue, M.; Pokethitiyook, P.; Inthorn, D. Equilibrium and kinetic studies on biosorption potential of charophyte biomass to remove heavy metals from synthetic metal solution and municipal wastewater. Bioremed. J. 2016, 20, 240–251. [Google Scholar] [CrossRef]
- Chandrashekharaiah, P.; Debanjan, S.; Santanu, D.; Avishek, B. Cadmium biosorption and biomass production by two freshwater microalgae Scenedesmus acutus and Chlorella pyrenoidosa: An integrated approach. Chemosphere 2021, 269, 128755. [Google Scholar]
- Ahanger, M.A.; Qin, C.; Begum, N.; Maodong, Q.; Dong, X.X.; El-Esawi, M.; El-Sheikh, M.-A.; Alatar, A.A.; Zhang, L. Nitrogen availability prevents oxidative effects of salinity on wheat growth and photosynthesis by up-regulating the antioxidants and osmolytes metabolism, and secondary metabolite accumulation. BMC Plant Biol. 2019, 19, 479. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-González, C.; Rodríguez-Pie, L.; Maister, O.; Rodellas, V.; Alorda-Keinglass, A.; Diego-Feliu, M.; Folch, A.; Garcia-Orellana, J.; Gasol, J.M. High spatial heterogeneity and low connectivity of bacterial communities along a Mediterranean subterranean estuary. Mol. Ecol. 2022, 31, 5745–5764. [Google Scholar] [CrossRef] [PubMed]
- Plett, D.C.; Ranathunge, K.; Melino, V.J.; Kuya, N.; Uga, Y.; Kronzucker, H.J. The intersection of nitrogen nutrition and water use in plants: New paths toward improved crop productivity. J. Exp. Bot. 2020, 71, 4452–4468. [Google Scholar] [CrossRef] [PubMed]
- Raza, A.; Zahra, N.; Hafeez, M.B.; Ahmad, M.; Iqbal, S.; Shaukat, K.; Ahmad, G. Nitrogen fixation of legumes: Biology and Physiology. In The Plant Family Fabaceae: Biology and Physiological Responses to Environmental Stresses; Springer: Singapore, 2020; pp. 43–74. [Google Scholar]
- Dutta, S.; Bhattacharjya, D.; Sinha, S.; Mandal, A.K. Salt-tolerant and plant growth-promoting Rhizobacteria: A new-fangled approach for improving crop yield. In Harsh Environment and Plant Resilience: Molecular and Functional Aspects; Springer: Singapore, 2021; pp. 367–385. [Google Scholar]
- Hussain, M.M.; Farooqi, Z.U.R.; Rasheed, F.; Din, W.M.U. Role of microorganisms as climate engineers: Mitigation and adaptations to climate change. In Climate Change and Microbes: Impacts and Vulnerability; Springer: Singapore, 2021; pp. 1–18. [Google Scholar]
- Singh, R.K.; Singh, P.; Li, H.-B.; Song, Q.-Q.; Guo, D.-J.; Solanki, M.K.; Verma, K.K.; Malviya, M.K.; Song, X.-P.; Lakshmanan, P.; et al. Diversity of nitrogen-fixing rhizobacteria associated with sugarcane: A comprehensive study of plant-microbe interactions for growth enhancement in Saccharum spp. BMC Plant Biol. 2020, 20, 220. [Google Scholar] [CrossRef]
- Leakey, R.R. The Role of Trees in Agroecology. In Routledge Handbook of Agricultural Biodiversity; Routledge: London, UK, 2017; pp. 238–252. [Google Scholar]
- Gómez-Sagasti, M.T.; Marino, D. PGPRs and Nitrogen-Fixing Legumes: A Perfect Team for Efficient Cd Phytoremediation? Front. Plant Sci. 2015, 6, 81. [Google Scholar]
- Cao, Z.; Kühn, P.; He, J.-S.; Bauhus, J.; Guan, Z.-H.; Scholten, T. Calibration of Near-Infrared Spectra for Phosphorus Fractions in Grassland Soils on the Tibetan Plateau. Agronomy 2022, 12, 783. [Google Scholar] [CrossRef]
- Li, Y.; Wei, S.; Chen, X.; Dong, Y.; Zeng, M.; Yan, C.; Hou, L.; Jiao, R. Isolation of cadmium-resistance and siderophore-producing endophytic bacteria and their potential use for soil cadmium remediation. Heliyon 2023, 9, e17661. [Google Scholar] [CrossRef]
- Kalayu, G. Phosphate solubilizing microorganisms: Promising approach as biofertilizers. Int. J. Agron. 2019, 2019, 4917256. [Google Scholar] [CrossRef]
- Gerke, J. Phytate (inositol hexakisphosphate) in soil and phosphate acquisition from inositol phosphates by higher plants. A review. Plants 2015, 4, 253–266. [Google Scholar] [CrossRef] [PubMed]
- Gurdeep, K.; Reddy, M.S. Effects of phosphate-solubilizing bacteria, rock phosphate and chemical fertilizers on maize-wheat cropping cycle and economics. Pedosphere 2015, 25, 428–437. [Google Scholar]
- Yang, P.; Zhou, X.F.; Wang, L.L.; Li, Q.S.; Zhou, T.; Chen, Y.K.; Zhao, Z.Y.; He, B.Y. Effect of phosphate-solubilizing bacteria on the mobility of insoluble cadmium and metabolic analysis. Int. J. Environ. Rese. Public Health 2018, 15, 1330. [Google Scholar] [CrossRef] [PubMed]
- Chibuike, G.U.; Obiora, S.C. Heavy metal polluted soils: Effect on plants and bioremediation methods. Appl. Environ. Soil Sci. 2014, 2014, 752708. [Google Scholar] [CrossRef]
- Ren, Z.; Cheng, R.; Chen, P.; Xue, Y.; Xu, H.; Yin, Y.; Zhang, L. Plant-associated microbe system in treatment of heavy metals–contaminated soil: Mechanisms and applications. Water Air Soil Pollut. 2023, 234, 39. [Google Scholar] [CrossRef]
- Khan, A. Promises and potential of in situ nano-phytoremediation strategy to mycorrhizo-remediate heavy metal contaminated soils using non-food bioenergy crops (Vetiver zizinoides & Cannabis sativa). Int. J. Phytoremed. 2020, 22, 900–915. [Google Scholar]
- Alsamhary, K. Vermi-cyanobacterial remediation of cadmium-contaminated soil with rice husk biochar: An eco-friendly approach. Chemosphere 2023, 311 Pt 1, 136931. [Google Scholar] [CrossRef]
- Fahad, S.; Hussain, S.; Bano, A.; Saud, S.; Hassan, S.; Shan, D.; Wu, C. Potential role of phytohormones and plant growth-promoting rhizobacteria in abiotic stresses: Consequences for changing environment. Environ. Sci. Pollut. Res. 2015, 22, 4907–4921. [Google Scholar] [CrossRef]
- Farooqi, Z.U.R.; Din, W.M.U.; Hussain, M.M. Microbial Responses under Climate Change Scenarios: Adaptation and Mitigations Climate Change and Microbial Diversity; Apple Academic Press: Burlington, ON, Canada, 2023; pp. 1–20. [Google Scholar]
- Pathania, P.; Bhatia, R.; Khatri, M. Cross-competence and affectivity of maize rhizosphere bacteria Bacillus sp. MT7 in tomato rhizosphere. Sci. Horticul. 2020, 272, 109480. [Google Scholar] [CrossRef]
- Pan, W.; Lu, Q.; Xu, Q.R.; Zhang, R.R.; Li, H.Y.; Yang, Y.H.; Liu, H.J.; Du, S.T. Abscisic acid-generating bacteria can reduce Cd concentration in pakchoi grown in Cd-contaminated soil. Ecotoxicol. Environ. Saf. 2019, 177, 100–107. [Google Scholar] [CrossRef]
- Morales-Cedeño, L.R.; del Carmen Orozco-Mosqueda, M.; Loeza-Lara, P.D.; Parra-Cota, F.I.; de Los Santos-Villalobos, S.; Santoyo, G. Plant growth-promoting bacterial endophytes as biocontrol agents of pre-and post-harvest diseases: Fundamentals, methods of application and future perspectives. Microbiol. Res. 2021, 242, 126612. [Google Scholar] [CrossRef] [PubMed]
- Chandra, H.; Kumari, P.; Bisht, R.; Prasad, R.; Yadav, S. Plant growth promoting Pseudomonas aeruginosa from Valeriana wallichii displays antagonistic potential against three phytopathogenic fungi. Mol. Biol. Rep. 2020, 47, 6015–6026. [Google Scholar] [CrossRef] [PubMed]
- Kareem, A.; Farooqi, Z.U.R.; Kalsom, A.; Mohy-Ud-Din, W.; Hussain, M.M.; Raza, M.; Khursheed, M.M. Organic farming for sustainable soil use, management, food production and climate change mitigation. In Sustainable Agriculture: Technical Progressions and Transitions; Springer: Cham, Switzerland, 2022; pp. 39–59. [Google Scholar]
- Sivasakthi, S.; Usharani, G.; Saranraj, P. Biocontrol potentiality of plant growth promoting bacteria (PGPR)-Pseudomonas fluorescens and Bacillus subtilis: A review. Afr. J. Agric. Res. 2014, 9, 1265–1277. [Google Scholar]
- Mahmud, K.; Missaoui, A.; Lee, K.; Ghimire, B.; Presley, H.W.; Makaju, S. Rhizosphere microbiome manipulation for sustainable crop production. Curr. Plant Biol. 2021, 27, 100210. [Google Scholar] [CrossRef]
- Thathana, M.G.; Murage, H.; Abia, A.L.K.; Pillay, M. Morphological Characterization and Determination of Aflatoxin-Production Potentials of Aspergillus flavus Isolated from Maize and Soil in Kenya. Agriculture 2017, 7, 80. [Google Scholar] [CrossRef]
- Vurukonda, S.S.K.P.; Giovanardi, D.; Stefani, E. Plant Growth Promoting and Biocontrol Activity of Streptomyces spp. as Endophytes. Int. J. Mol. Sci. 2018, 19, 952. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Lima, D.; Mtz-Enriquez, A.I.; Carrión, G.; Basurto-Cereceda, S.; Pariona, N. The Bifunctional Role of Copper Nanoparticles in Tomato: Effective Treatment for Fusarium Wilt and Plant Growth Promoter. Sci. Horticul. 2021, 277, 109810. [Google Scholar] [CrossRef]
- Ramadan, E.M.; AbdelHafez, A.A.; Hassan, E.A.; Saber, F.M. Plant growth promoting rhizobacteria and their potential for biocontrol of phytopathogens. Afr. J. Microbiol. Res. 2016, 10, 486–504. [Google Scholar]
- Paul, A.; Bhakta, J.N. Biosorption-driven green technology for the treatment of heavy metal (loids)-contaminated effluents. In Intelligent Environmental Data Monitoring for Pollution Management; Elsevier: Amsterdam, The Netherlands, 2021; pp. 71–91. [Google Scholar]
- Tarfeen, N.; Nisa, K.U.; Hamid, B.; Bashir, Z.; Yatoo, A.M.; Dar, M.A.; Mohiddin, F.A.; Amin, Z.; Ahmad, R.A.A.; Sayyed, R.Z. Microbial remediation: A promising tool for reclamation of contaminated sites with special emphasis on heavy metal and pesticide pollution: A review. Processes 2022, 10, 1358. [Google Scholar] [CrossRef]
- Hou, D.; O’Connor, D.; Igalavithana, A.D.; Alessi, D.S.; Luo, J.; Tsang, D.C.; Cundy, A.B.; Rinklebe, J.; Bolan, N.S.; Ok, Y.S. Metal Contamination and Bioremediation of Agricultural Soils for Food Safety and Sustainability. Nat. Rev. Earth Environ. 2020, 1, 366–381. [Google Scholar] [CrossRef]
- Oliva-Arancibia, B.; Órdenes-Aenishanslins, N.; Bruna, N.; Ibarra, P.S.; Zacconi, F.C.; Pérez-Donoso, J.M.; Poblete-Castro, I. Co-synthesis of Medium-Chain-Length Polyhydroxyalkanoates and CdS Quantum Dots Nanoparticles in Pseudomonas putida KT2440. J. Biotechnol. 2017, 264, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Singh, D. Role of Genetically Modified Microorganisms in Heavy Metal Bioremediation. In Advances in Environmental Biotechnology; Springer: Singapore, 2017; pp. 197–214. [Google Scholar]
- Etesami, H. Bacterial mediated alleviation of heavy metal stress and decreased accumulation of metals in plant tissues: Mechanisms and future prospects. Ecotoxicol. Environ. Saf. 2018, 147, 175–191. [Google Scholar] [CrossRef]
- Gazitúa, M.C.; Morgante, V.; Poupin, M.J.; Ledger, T.; Rodríguez-Valdecantos, G.; Herrera, C.; Jorquera, M.A.; González, B. The Microbial Community from the Early-Plant Colonizer (Baccharis linearis) Is Required for Plant Establishment on Copper Mine Tailings. Sci. Rep. 2021, 11, 10448. [Google Scholar] [CrossRef] [PubMed]
- Kalaivanan, D.; Ganeshamurthy, A.N. Mechanisms of Heavy Metal Toxicity in Plants. In Abiotic Stress Physiology of Horticultural Crops; Springer: Singapore, 2016; pp. 85–102. [Google Scholar]
- Araki, R.; Murata, J.; Murata, Y. A Novel Barley Yellow Stripe 1-Like Transporter (HvYSL2) Localized to the Root Endodermis Transports Metal–Phytosiderophore Complexes. Plant Cell Physiol. 2011, 52, 1931–1940. [Google Scholar] [CrossRef] [PubMed]
- Złoch, M.; Thiem, D.; Gadzała-Kopciuch, R.; Hrynkiewicz, K. Synthesis of Siderophores by Plant-Associated Metallotolerant Bacteria under Exposure to Cd2+. Chemosphere 2016, 156, 312–325. [Google Scholar] [CrossRef]
- Wang, W.; Qiu, Z.; Tan, H.; Cao, L. Siderophore Production by Actinobacteria. Biometals 2014, 27, 623–631. [Google Scholar] [CrossRef]
- Kang, P.; Wu, P.; Jin, Y.; Shi, S.; Gao, D.; Chen, G.; Li, Q. Formation and Emissions of Volatile Organic Compounds from Homo-PP and Co-PP Resins during Manufacturing Process and Accelerated Photoaging Degradation. Molecules 2020, 25, 2761. [Google Scholar] [CrossRef]
- Mhlongo, M.I.; Piater, L.A.; Madala, N.E.; Labuschagne, N.; Dubery, I.A. The Chemistry of Plant–Microbe Interactions in the Rhizosphere and the Potential for Metabolomics to Reveal Signaling Related to Defense Priming and Induced Systemic Resistance. Front. Plant Sci. 2018, 9, 112. [Google Scholar] [CrossRef]
- Montejano-Ramírez, V.; García-Pineda, E.; Valencia-Cantero, E. Bacterial Compound N, N-Dimethylhexadecylamine Modulates Expression of Iron Deficiency and Defense Response Genes in Medicago truncatula Independently of the Jasmonic Acid Pathway. Plants 2020, 9, 624. [Google Scholar] [CrossRef]
- Montejano-Ramírez, V.; Martínez-Cámara, R.; García-Pineda, E.; Valencia-Cantero, E. Rhizobacterium Arthrobacter agilis UMCV2 Increases Organ-Specific Expression of FRO Genes in Conjunction with Genes Associated with the Systemic Resistance Pathways of Medicago truncatula. Acta Physiol. Plant. 2018, 40, 138. [Google Scholar] [CrossRef]
- Kong, H.G.; Shin, T.S.; Kim, T.H.; Ryu, C.M. Stereoisomers of the Bacterial Volatile Compound 2, 3-Butanediol Differently Elicit Systemic Defense Responses of Pepper against Multiple Viruses in the Field. Front. Plant Sci. 2018, 9, 90. [Google Scholar] [CrossRef]
- Rojas-Solis, D.; Vences-Guzmán, M.A.; Sohlenkamp, C.; Santoyo, G. Bacillus toyonensis COPE52 modifies lipid and fatty acid composition, exhibits antifungal activity, and stimulates growth of tomato plants under saline conditions. Curr. Microbiol. 2020, 77, 2735–2744. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P. Role and significance of biofilm-forming microbes in phytoremediation—A review. Environ. Technol. Innov. 2022, 25, 102182. [Google Scholar] [CrossRef]
- Dimkić, I.; Janakiev, T.; Petrović, M.; Degrassi, G.; Fira, D. Plant-associated Bacillus and Pseudomonas antimicrobial activities in plant disease suppression via biological control mechanisms—A review. Physiol. Mol. Plant Pathol. 2022, 117, 101754. [Google Scholar] [CrossRef]
- Imran, M.; Das, K.R.; Naik, M.M. Co-selection of multi-antibiotic resistance in bacterial pathogens in metal and microplastic contaminated environments: An emerging health threat. Chemosphere 2019, 215, 846–857. [Google Scholar] [CrossRef]
- Gelbicova, T.; Florianova, M.; Hluchanova, L.; Kalova, A.; Korena, K.; Strakova, N.; Karpiskova, R. Comparative analysis of genetic determinants encoding cadmium, arsenic, and benzalkonium chloride resistance in Listeria monocytogenes of human, food, and environmental origin. Front. Microbiol. 2021, 11, 599882. [Google Scholar] [CrossRef]
- Kenawy, A.; Dailin, D.J.; Abo-Zaid, G.A.; Malek, R.A.; Ambehabati, K.K.; Zakaria, K.H.N.; El Enshasy, H.A. Biosynthesis of antibiotics by PGPR and their roles in biocontrol of plant diseases. In Plant Growth Promoting Rhizobacteria for Sustainable Stress Management: Volume 2: Rhizobacteria in Biotic Stress Management; Springer: Singapore, 2019; pp. 1–35. [Google Scholar]
- Aydin, M.H. Rhizoctonia Solani and Its Biological Control. Türkiye Tarımsal Araştırmalar Derg. 2022, 9, 118–135. [Google Scholar] [CrossRef]
- Katara, S.; Devki, V.G.; Neelam, D.; Kant, R. Role of bacteria and fungi in antibiotic production. Antibiotics and Antimicrobial Resistance Genes in the Environment. Pharma Innov. J. 2021, 1, 31–42. [Google Scholar]
- Mitra, A.; Chatterjee, S.; Kataki, S.; Rastogi, R.P.; Gupta, D.K. Bacterial tolerance strategies against lead toxicity and their relevance in bioremediation application. Environ. Sci. Pollut. Res. 2021, 28, 14271–14284. [Google Scholar] [CrossRef]
- Mukherjee, P.; Mitra, A.; Roy, M. Halomonas rhizobacteria of Avicennia marina of Indian Sundarbans promote rice growth under saline and heavy metal stresses through exopolysaccharide production. Front. Microbiol. 2019, 10, 1207. [Google Scholar] [CrossRef]
- Pietri, G.P.; Tontini, M.; Brogioni, B.; Oldrini, D.; Robakiewicz, S.; Henriques, P.; Malić, S. Elucidating the structural and minimal protective epitope of the serogroup X meningococcal capsular polysaccharide. Front. Mol. Biosci. 2021, 8, 745360. [Google Scholar] [CrossRef] [PubMed]
- Gupta, P.; Diwan, B. Bacterial exopolysaccharide mediated heavy metal removal: A review on biosynthesis, mechanism and remediation strategies. Biotechnol. Rep. 2017, 13, 58–71. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, S.; Sharma, S. Insight into Exopolysaccharide-Mediated Stress Tolerance in Plants: A Feasible Approach towards the Development of Next-Generation Bioformulations. J. Soil Sci. Plant Nutr. 2023, 23, 22–33. [Google Scholar] [CrossRef]
- Majumdar, A.; Afsal, F.; Pathak, S.; Upadhayay, M.K.; Roychowdhury, T.; Srivastava, S. Molecular Aspects of Arsenic Responsive Microbes in Soil-Plant-Aqueous Triphasic Systems. In Global Arsenic Hazard: Ecotoxicology and Remediation; Springer: Singapore, 2022; pp. 291–312. [Google Scholar]
- Liu, Y.; Ali, A.; Su, J.-F.; Li, K.; Hu, R.-Z.; Wang, Z. Microbial-Induced Calcium Carbonate Precipitation: Influencing Factors, Nucleation Pathways, and Application in Waste Water Remediation. Sci. Total Environ. 2022, 860, 160439. [Google Scholar] [CrossRef] [PubMed]
- Brahim, B.G.; Ouhdouch, Y. Management of Tomato Foot and Root Rot (TFRR) by Biocontrol Agents with Emphasis on Factors Affecting its Effectiveness. In Probiotics and Plant Health; Springer: Singapore, 2017; pp. 1–19. [Google Scholar]
- Asaf, S.; Khan, A.L.; Khan, M.A.; Imran, Q.M.; Yun, B.-W.; Lee, I.-J. Osmoprotective Functions Conferred to Soybean Plants via Inoculation with Sphingomonas sp. LK11 and Exogenous Trehalose. Microbiol. Res. 2017, 205, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Ilangumaran, G.; Smith, D.L. Plant Growth Promoting Rhizobacteria in Amelioration of Salinity Stress: A Systems Biology Perspective. Front. Plant Sci. 2017, 8, 1768. [Google Scholar] [CrossRef] [PubMed]
- John, C.J.; Kumar, S.; Ge, M. Probiotic Prospects of PGPR for Green and Sustainable Agriculture. Arch. Phytopathol. Plant Protec. 2020, 53, 899–914. [Google Scholar] [CrossRef]
- Dakora, F.D.; Matiru, V.N.; Kanu, A.S. Rhizosphere Ecology of Lumichrome and Riboflavin, Two Bacterial Signal Molecules Eliciting Developmental Changes in Plants. Front. Plant Sci. 2015, 6, 700. [Google Scholar] [CrossRef]
- Ayuso-Calles, M.; Flores-Félix, J.D.; Rivas, R. Overview of the Role of Rhizobacteria in Plant Salt Stress Tolerance. Agronomy 2021, 11, 1759. [Google Scholar] [CrossRef]
- Asaf, S.; Jan, R.; Khan, M.A.; Khan, A.L.; Asif, S.; Bilal, S.; Ahmad, W.; Waqas, M.; Kim, K.M.; Ahmed, A.H.; et al. Unraveling the mutualistic interaction between endophytic Curvularia lunata CSL1 and tomato to mitigate cadmium (Cd) toxicity via transcriptomic insights. Sci. Total Environ. 2023, 861, 160542. [Google Scholar] [CrossRef]
- Freitas, E.V.; Nascimento, C.W.; Souza, A.; Silva, F.B. Citric acid-assisted phytoextraction of lead: A field experiment. Chemosphere 2013, 92, 213–217. [Google Scholar] [CrossRef]
- Azubuike, C.C.; Chikere, C.B.; Okpokwasili, G.C. Bioremediation techniques-classification based on site of application: Principles, advantages, limitations and prospects. World J. Microbiol. Biotechnol. 2016, 32, 180. [Google Scholar] [CrossRef] [PubMed]
- Khodaverdiloo, H.; Han, F.X.; Hamzenejad Taghlidabad, R.; Karimi, A.; Moradi, N.; Kazery, J.A. Potentially toxic element contamination of arid and semi-arid soils and its phytoremediation. Arid Land Res. Manag. 2020, 34, 361–391. [Google Scholar] [CrossRef]
- Abatenh, E.; Gizaw, B.; Tsegaye, Z.; Wassie, M. The role of microorganisms in bioremediation—A review. Open J. Environ. Biol. 2017, 2, 38–46. [Google Scholar] [CrossRef]
- Yang, S.Z.; Jin, H.J.; Wei, Z.; He, R.X.; Ji, Y.J.; Lim, X.M.; Shao-Peng, Y.U. Bioremediation of oil spills in cold environments: A review. Pedosphere 2009, 19, 371–381. [Google Scholar] [CrossRef]
- Megharaj, M.; Ramakrishnan, B.; Venkateswarlu, K.; Sethunathan, N.; Naidu, R. Bioremediation Approaches for Organic Pollutants: A Critical Perspective. Environ. Int. 2011, 37, 1362–1375. [Google Scholar] [CrossRef]
- Javanbakht, V.; Alavi, S.A.; Zilouei, H. Mechanisms of Heavy Metal Removal Using Microorganisms as Biosorbent. Water Sci. Technol. 2014, 69, 1775–1787. [Google Scholar] [CrossRef]
- Oka, T.; Sameshima, Y.; Koga, T.; Kim, H.; Goto, M.; Furukawa, K. Protein Omannosyltransferase a of Aspergillus awamori Is Involved in O-Mannosylation of Glucoamylase I. Microbiology 2005, 151, 3657–3667. [Google Scholar] [CrossRef]
- Timková, I.; Sedláková-Kaduková, J.; Pristaš, P. Biosorption and Bioaccumulation Abilities of Actinomycetes/Streptomycetes Isolated from Metal Contaminated Sites. Separations 2018, 5, 54. [Google Scholar] [CrossRef]
- Wang, J.; Li, Q.; Li, M.M.; Chen, T.H.; Zhou, Y.F.; Yue, Z.B. Competitive Adsorption of Heavy Metal by Extracellular Polymeric Substances (EPS) Extracted from Sulfate Reducing Bacteria. Bioresour. Technol. 2014, 163, 374–376. [Google Scholar] [CrossRef]
- Park, J.H.; Chon, H.T. Characterization of Cadmium Biosorption by Exiguobacterium sp. Isolated from Farmland Soil near Cu-Pb-Zn Mine. Environ. Sci. Pollut. Res. 2016, 23, 11814–11822. [Google Scholar] [CrossRef] [PubMed]
- Phulia, V.; Jamwal, A.; Saxena, N.; Chadha, N.K.; Muralidhar, A.P.; Prusty, A.K. Technologies in Aquatic Bioremediation. In Freshwater Ecosystem and Xenobiotics; Discovery Publishing House PVT. Ltd.: New Delhi, India, 2013; pp. 65–91. [Google Scholar]
- Couto, N.; Fritt-Rasmussen, J.; Jensen, P.E.; Højrup, M.; Rodrigo, A.P.; Ribeiro, A.B. Suitability of Oil Bioremediation in an Arctic Soil Using Surplus Heating from an Incineration Facility. Environ. Sci. Pollut. Res. 2014, 21, 6221–6227. [Google Scholar] [CrossRef] [PubMed]
- Varjani, S.J.; Upasani, V.N. A New Look on Factors Affecting Microbial Degradation of Petroleum Hydrocarbon Pollutants. Int. Biodeterior. Biodegrad. 2017, 120, 71–83. [Google Scholar] [CrossRef]
- Sharma, J. Advantages and limitations of in situ methods of bioremediation. Recent Adv. Biol. Med. 2019, 5, 955923. [Google Scholar] [CrossRef]
- Srinath, T.; Verma, T.; Ramteke, P.W.; Garg, S.K. Chromium (VI) biosorption and bioaccumulation by chromate resistant bacteria. Chemosphere 2002, 48, 427–435. [Google Scholar] [CrossRef]
- Vijayaraghavan, K.; Yun, Y.S. Bacterial biosorbents and biosorption. Biotechnol. Adv. 2008, 26, 266–291. [Google Scholar] [CrossRef]
- Issazadeh, K.; Jahanpour, N.; Pourghorbanali, F.; Raeisi, G.; Faekhondeh, J. Heavy Metals Resistance by Bacterial Strains. Ann. Biol. Res. 2013, 4, 60–63. [Google Scholar]
- Babu, S.O.; Hossain, M.B.; Rahman, M.S.; Rahman, M.; Ahmed, A.S.; Hasan, M.M.; Rakib, A.; Emran, T.B.; Xiao, J.; Simal-Gandara, J. Phytoremediation of toxic metals: A sustainable green solution for clean environment. Appl. Sci. 2021, 11, 10348. [Google Scholar] [CrossRef]
- Iram, S.; Shabbir, R.; Zafar, H.; Javaid, M. Biosorption and Bioaccumulation of Copper and Lead by Heavy Metal-Resistant Fungal Isolates. Arab. J. Sci. Eng. 2015, 40, 1867–1873. [Google Scholar] [CrossRef]
- Sarwar, N.; Imran, M.; Shaheen, M.R.; Ishaque, W.; Kamran, M.A.; Matloob, A.; Rehim, A.; Hussain, S. Phytoremediation Strategies for Soils Contaminated with Heavy Metals: Modifications and Future Perspectives. Chemosphere 2017, 171, 710–721. [Google Scholar] [CrossRef]
- Iwamoto, T.; Nasu, M. Current bioremediation practice and perspective. J. Biosci. Bioeng. 2001, 92, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Favas, P.J.C.; Pratas, J.; Varun, M.; D’Souza, R.; Paul, M.S. Phytoremediation of Soils Contaminated with Metals and Metalloids at Mining Areas: Potential of Native Flora. Environments 2014, 3, 485–516. [Google Scholar]
- Tak, H.I.; Ahmad, F.; Babalola, O.O. Advances in the application of plant growth-promoting rhizobacteria in phytoremediation of heavy metals. In Reviews of Environmental Contamination and Toxicology; Whitacre, D.M., Ed.; Springer: New York, NY, USA, 2013; Volume 223, pp. 33–52. [Google Scholar]
- Nadeem, S.M.; Zahir, Z.A.; Naveed, M.; Nawaz, S. Mitigation of salinity-induced negative impact on the growth and yield of wheat by plant growth-promoting rhizobacteria in naturally saline conditions. Ann. Microbiol. 2013, 63, 225–232. [Google Scholar] [CrossRef]
- Bücker-Neto, L.; Paiva, A.L.S.; Machado, R.D.; Arenhart, R.A.; Margis-Pinheiro, M. Interactions between plant hormones and heavy metals responses. Genet. Mol. Biol. 2017, 40 (Suppl. S1), 373–386. [Google Scholar] [CrossRef] [PubMed]
- Solanki, R.; Dhankhar, R. Biochemical changes and adaptive strategies of plants under heavy metal stress. Biologia 2011, 66, 195–204. [Google Scholar] [CrossRef]
- Jan, S.; Parray, J.A. Approaches to Heavy Metal Tolerance in Plants; Springer: New Delhi, India, 2016. [Google Scholar]
- Emamverdian, A.; Ding, Y.; Mokhberdoran, F.; Xie, Y. Heavy metal stress and some mechanisms of plant defense response. Sci. World J. 2015, 2015, 756120. [Google Scholar] [CrossRef]
- Zhao, Y.; Yao, J.; Yuan, Z.; Wang, T.; Zhang, Y.; Wang, F. Bioremediation of Cd by Strain GZ-22 Isolated from Mine Soil Based on Biosorption and Microbially Induced Carbonate Precipitation. Environ. Sci. Pollut. Res. 2017, 24, 372–380. [Google Scholar] [CrossRef]
- Gong, X. Kinetic and Equilibrium Studies on the Adsorption of Pb (II), Cd (II) and Cu (II) by Rape Straw. Adsorpt. Sci. Technol. 2013, 31, 559–571. [Google Scholar] [CrossRef]
- Wang, X.S. Cd (II) Removal by Marine Arthrobacter Protophormiae Biomass: Mechanism Characterization and Adsorption Performance. Desalin. Water Treat. 2013, 51, 7710–7720. [Google Scholar] [CrossRef]
- Li, L.; Qian, C.; Cheng, L.; Wang, R. A Laboratory Investigation of Microbe-Inducing CdCO3 Precipitate Treatment in Cd2+ Contaminated Soil. J. Soils Sediments 2010, 10, 248–254. [Google Scholar] [CrossRef]
- Burns, J.L.; Ginn, B.R.; Bates, D.J.; Dublin, S.N.; Taylor, J.V.; Apkarian, R.P.; Amaro-Garcia, S.; Neal, A.L.; Dichristina, T.J. Outer Membrane-Associated Serine Protease Involved in Adhesion of Shewanella oneidensis to Fe(III) Oxides. Environ. Sci. Technol. 2010, 44, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Elzinga, E.J.; Brechbühl, Y.; Voegelin, A.; Kretzschmar, R. Impacts of Shewanella Putrefaciens Strain CN-32 Cells and Extracellular Polymeric Substances on the Sorption of as (V) and As (III) on Fe (III)-(Hydr)oxides. Environ. Sci. Technol. 2011, 45, 2804–2810. [Google Scholar] [CrossRef] [PubMed]
- Rascio, N.; Navari-Izzo, F. Heavy Metal Hyperaccumulating Plants: How and Why Do They Do It? And What Makes Them So Interesting? Plants 2011, 180, 169–181. [Google Scholar] [CrossRef] [PubMed]
- Chaperon, S.; Sauvé, S. Toxicity Interactions of Cadmium, Copper, and Lead on Soil Urease and Dehydrogenase Activity in Relation to Chemical Speciation. Ecotoxicol. Environ. Saf. 2008, 70, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Jusselme, M.D.; Miambi, E.; Mora, P.; Diouf, M.; Rouland-Lefèvre, C. Increased Lead Availability and Enzyme Activities in Root-Adhering Soil of Lantana camara during Phytoextraction in the Presence of Earthworms. Sci. Total Environ. 2013, 445, 101–109. [Google Scholar] [CrossRef]
- Ciarkowska, K.; Sołek-Podwika, K.; Wieczorek, J. Enzyme Activity as an Indicator of Soil-Rehabilitation Processes at a Zinc and Lead Ore Mining and Processing Area. J. Environ. Manag. 2014, 132, 250–256. [Google Scholar] [CrossRef]
- Al-Maqdi, K.A.; Elmerhi, N.; Athamneh, K.; Bilal, M.; Alzamly, A.; Ashraf, S.S.; Shah, I. Challenges and Recent Advances in Enzyme-Mediated Wastewater Remediation—A Review. Nanomaterials 2021, 11, 3124. [Google Scholar] [CrossRef]
- Wasilkowski, D.; Śwędzioł, Z.; Mrozik, A. Przydatność Genetycznie Modyfikowanych Mikroorganizmów do Bioremediacji Zanieczyszczonych Środowisk. Chemik 2012, 66, 817–826. (In Polish) [Google Scholar]
- Sharma, B.; Dangi, A.K.; Shukla, P. Contemporary Enzyme Based Technologies for Bioremediation: A Review. J. Environ. Manag. 2018, 210, 10–22. [Google Scholar] [CrossRef]
- Wolejko, E.; Wydro, U.; Loboda, T. The Ways to Increase Efficiency of Soil Bioremediation. Ecol. Chem. Eng. 2016, 23, 155. [Google Scholar] [CrossRef]
- Liang, Y.; Jiao, S.; Wang, M.; Yu, H.; Shen, Z. A CRISPR/Cas9-Based Genome Editing System for Rhodococcus ruber TH. Metab. Eng. 2020, 57, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Azad, M.A.K.; Amin, L.; Sidik, N.M. Genetically engineered organisms for bioremediation of pollutants in contaminated sites. Chin. Sci. Bull. 2014, 59, 703–714. [Google Scholar] [CrossRef]
- Mujawar, S.Y.; Shamim, K.; Vaigankar, D.C.; Dubey, S.K. Arsenite Biotransformation and Bioaccumulation by Klebsiella pneumoniae Strain SSSW7 Possessing Arsenite Oxidase (aioA) Gene. Biometals 2019, 32, 65–76. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Kumar, A.; Sharma, B. Role of Fungal Enzymes for Bioremediation of Hazardous Chemicals. In Recent Advancement in White Biotechnology through Fungi: Volume 3: Perspective for Sustainable Environments; Gupta, V.K., Sharma, G.D., Tuohy, M., Eds.; Springer: Cham, Switzerland, 2019; pp. 237–256. [Google Scholar]
- Ghosal, D.; Ghosh, S.; Dutta, T.K.; Ahn, Y. Current State of Knowledge in Microbial Degradation of Polycyclic Aromatic Hydrocarbons (PAHS): A Review. Front. Microbiol. 2016, 7, 1369. [Google Scholar] [CrossRef]
- Van Nostrand, J.D.; He, Z.; Zhou, J. Use of Functional Gene Arrays for Elucidating In Situ Biodegradation. Front. Microbiol. 2012, 3, 339. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Zheng, Y.; Ding, C.; Ren, X.; Yuan, J.; Sun, F.; Li, Y. Integrated Metagenomics and Molecular Ecological Network Analysis of Bacterial Community Composition during the Phytoremediation of Cadmium-Contaminated Soils by Bioenergy Crops. Ecotoxicol. Environ. Saf. 2017, 145, 111–118. [Google Scholar] [CrossRef]
- Feng, G.; Xie, T.; Wang, X.; Bai, J.; Tang, L.; Zhao, H.; Wei, W.; Wang, M.; Zhao, Y. Metagenomic Analysis of Microbial Community and Function Involved in Cd-Contaminated Soil. BMC Microbiol. 2018, 18, 13. [Google Scholar] [CrossRef]
- Datta, S.; Rajnish, K.N.; Samuel, M.S.; Pugazlendhi, A.; Selvarajan, E. Metagenomic Applications in Microbial Diversity, Bioremediation, Pollution Monitoring, Enzyme, and Drug Discovery: A Review. Environ. Chem. Lett. 2020, 18, 1229–1241. [Google Scholar] [CrossRef]
- Malla, M.A.; Dubey, A.; Yadav, S.; Kumar, A.; Hashem, A.; Abd_Allah, E.F. Understanding and Designing the Strategies for the Microbe-Mediated Remediation of Environmental Contaminants Using Omics Approaches. Front. Microbiol. 2018, 9, 1132. [Google Scholar] [CrossRef]
- Tripathi, M.; Singh, D.N.; Vikram, S.; Singh, V.S.; Kumar, S. Metagenomic approach towards bioprospection of novel biomolecule(s) and environmental bioremediation, Annu. Res. Rev. Biol. 2018, 22, 1–12. [Google Scholar]
- Jackson, S.A.; Borchert, E.; OGara, F.; Dobson, A.D. Metagenomics for the Discovery of Novel Biosurfactants of Environmental Interest from Marine Ecosystems. Curr. Opin. Biotechnol. 2015, 33, 176–182. [Google Scholar] [CrossRef] [PubMed]
- Williams, W.; Trindade, M. Metagenomics for the discovery of novel biosurfactants. In Functional Metagenomics: Tools and Applications; Springer: Cham, Switzerland, 2017; pp. 95–117. [Google Scholar]
- Awasthi, M.K.; Guo, D.; Awasthi, S.K.; Wang, Q.; Chen, H.; Liu, T.; Duan, Y.; Soundari, P.G.; Zhang, Z. Recent Advances in Phytoremediation of Toxic Metals from Contaminated Sites: A Road Map to a Safer Environment. In Bioremediation of Industrial Waste for Environmental Safety; Bharagava, R.N., Saxena, G., Eds.; Springer: Singapore, 2020; pp. 77–112. [Google Scholar]
- Arumugam, M.; Harrington, E.D.; Foerstner, K.U.; Raes, J.; Bork, P. SmashCommunity: A Metagenomic Annotation and Analysis Tool. Bioinformatics 2010, 26, 2977–2978. [Google Scholar] [CrossRef] [PubMed]
- El-Hadidi, M.; Ruscheweyh, H.-J.; Huson, D.H. Improved Metagenome Analysis Using MEGAN5. In Proceedings of the Joint 21st Annual International Conference on Intelligent Systems for Molecular Biology (ISMB) and 12th European Conference on Computational Biology (ECCB), Berlin, Germany, 21–23 July 2013. [Google Scholar]
- Markowitz, V.M.; Ivanova, N.N.; Szeto, E.; Palaniappan, K.; Chu, K.; Dalevi, D.; Chen, I.-M.A.; Grechkin, Y.; Dubchak, I.; Anderson, I.; et al. IMG/M: A Data Management and Analysis System for Metagenomes. Nucleic Acids Res. 2007, 36, D534–D538. [Google Scholar] [CrossRef] [PubMed]
- Rizwan, M.; Singh, M.; Mitra, C.K.; Morve, R.K. Ecofriendly Application of Nanomaterials: Nanobioremediation. J. Nanopart. Res. 2014, 2014, 431787. [Google Scholar] [CrossRef]
- Karimi-Maleh, H.; Karaman, C.; Karaman, O.; Karimi, F.; Vasseghian, Y.; Fu, L.; Mirabi, A. Nanochemistry Approach for the Fabrication of Fe and N Co-Decorated Biomass-Derived Activated Carbon Frameworks: A Promising Oxygen Reduction Reaction Electrocatalyst in Neutral Media. J. Nanostruct. Chem. 2022, 12, 429–439. [Google Scholar] [CrossRef]
- Rafeeq, H.; Hussain, A.; Ambreen, A.; Waqas, M.; Bilal, M.; Iqbal, H. Functionalized nanoparticles and their environmental remediation potential: A review. J. Nanostruct. Chem. 2022, 12, 1007–1031. [Google Scholar] [CrossRef]
- Brunner, T.J.; Wick, P.; Manser, P.; Spohn, P.; Grass, R.N.; Limbach, L.K.; Bruinink, A.; Stark, W.J. In vitro cytotoxicity of oxide nanoparticles: Comparison to asbestos, silica, and the effect of particle solubility. Environ. Sci. Technol. 2006, 40, 4374–4381. [Google Scholar] [CrossRef]
- Kumar, A.; Choudhary, P.; Kumar, A.; Camargo, P.H.; Krishnan, V. Recent advances in plasmonic photocatalysis based on TiO2 and noble metal nanoparticles for energy conversion, environmental remediation, and organic synthesis. Small 2022, 18, 2101638. [Google Scholar] [CrossRef]
- Fernandes, J.P.; Mucha, A.P.; Francisco, T.; Gomes, C.R.; Almeida, C.M.R. Silver nanoparticles uptake by salt marsh plants—Implications for phytoremediation processes and effects in microbial community dynamics. Mar. Pollut. Bull. 2017, 119, 176–183. [Google Scholar] [CrossRef]
- Yadav, K.K.; Singh, J.K.; Gupta, N.; Kumar, V. A review of nanobioremediation technologies for environmental cleanup: A novel biological approach. J. Mater. Environ. Sci. 2017, 8, 740–757. [Google Scholar]
- Ma, X.; Geisler-Lee, J.; Deng, Y.; Kolmakov, A. Interactions between engineered nanoparticles (ENPs) and plants: Phytotoxicity, uptake and accumulation. Sci. Total Environ. 2010, 408, 3053–3061. [Google Scholar] [CrossRef] [PubMed]
- Rossi, L.; Zhang, W.; Schwab, A.P.; Ma, X. Uptake, accumulation, and in planta distribution of coexisting cerium oxide nanoparticles and cadmium in Glycine max (L.) Merr. Environ. Sci. Technol. 2017, 51, 12815–12824. [Google Scholar] [CrossRef] [PubMed]
- López-Luna, J.; Silva-Silva, M.J.; Martinez-Vargas, S.; Mijangos-Ricardez, O.F.; González-Chávez, M.C.; Solís-Domínguez, F.A.; Cuevas-Díaz, M.C. Magnetite nanoparticle (NP) uptake by wheat plants and its effect on cadmium and chromium toxicological behavior. Sci. Total Environ. 2016, 565, 941–950. [Google Scholar] [CrossRef] [PubMed]
- Rossi, L.; Sharifan, H.; Zhang, W.; Schwab, A.P.; Ma, X. Mutual effects and in planta accumulation of co-existing cerium oxide nanoparticles and cadmium in hydroponically grown soybean (Glycine max (L.) Merr.). Environ. Sci. Nano 2018, 5, 150. [Google Scholar] [CrossRef]
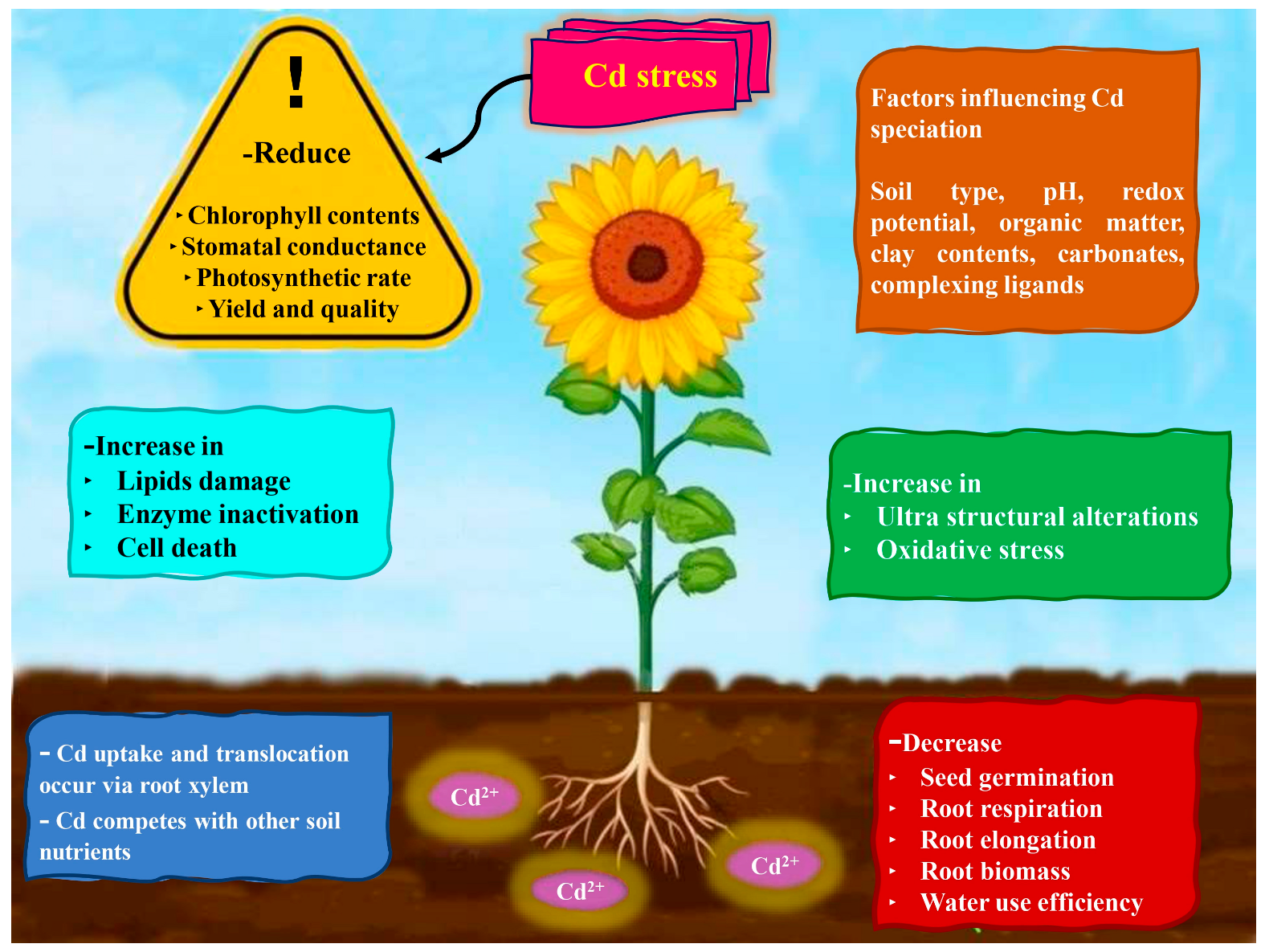
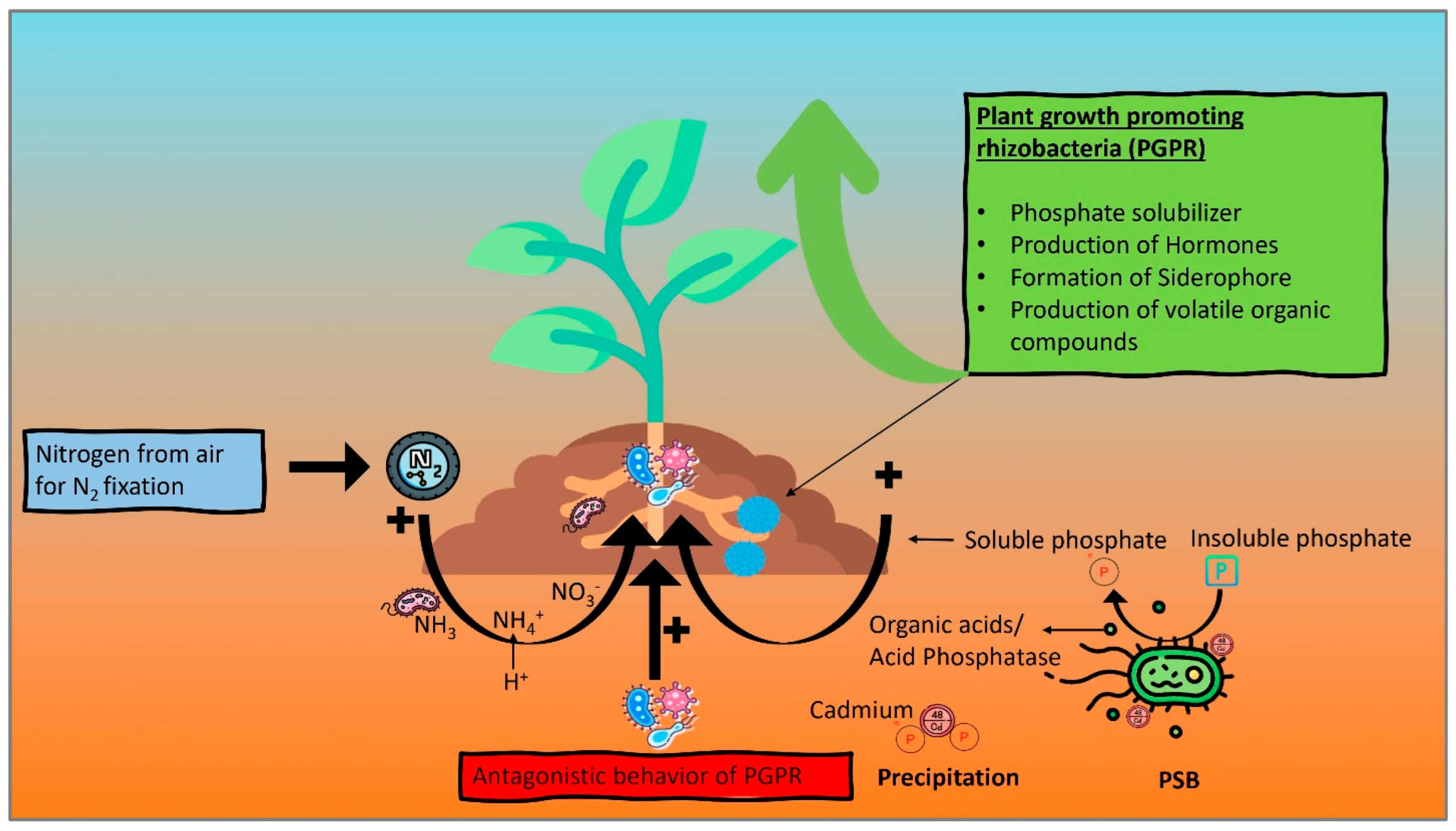
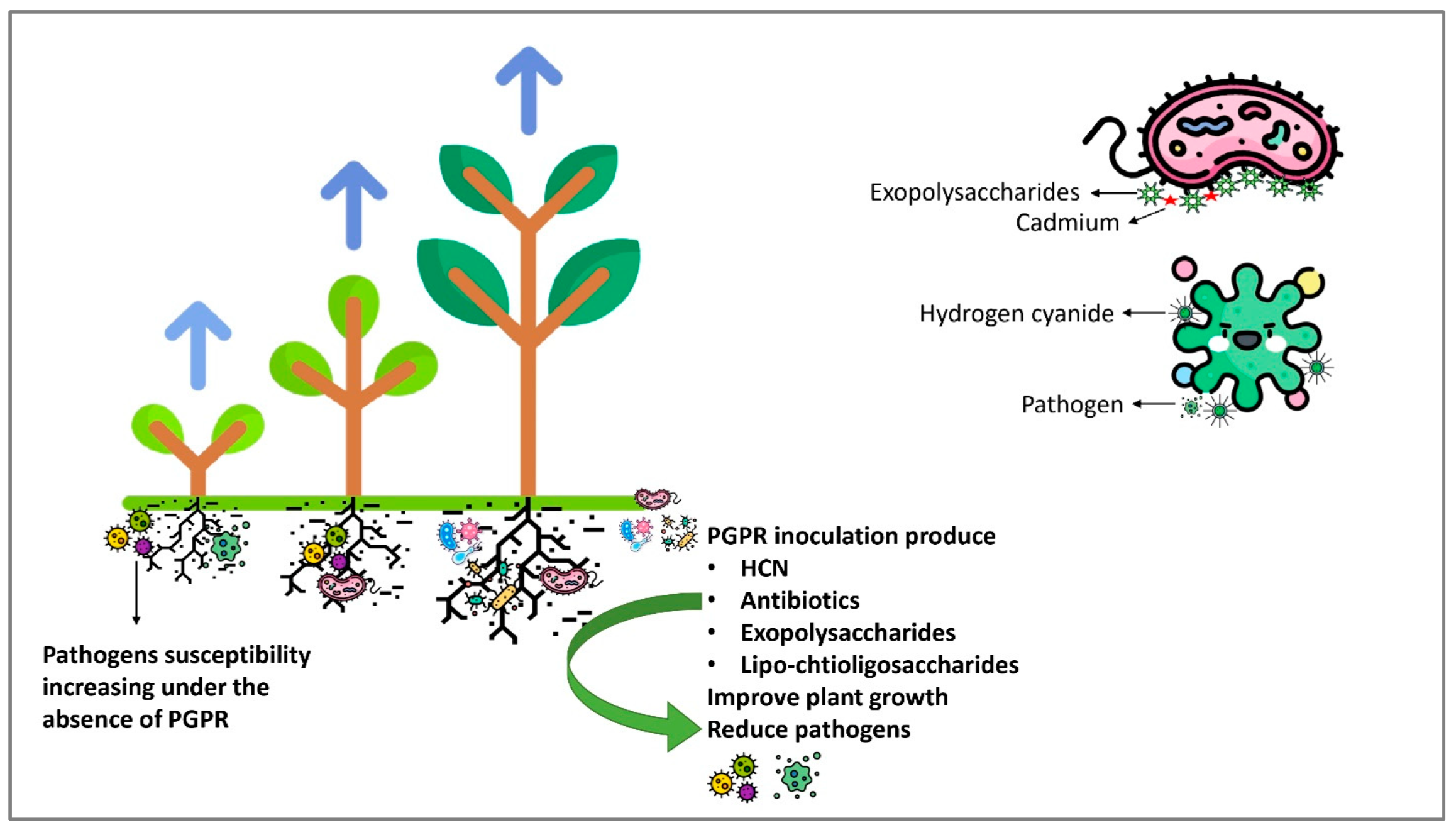
| Plant Species | Level of Cd | Changes/Damages | References |
|---|---|---|---|
| Seed germination and seedling growth | |||
| Pisum sativum L. | 20–500 µM | Inhibition of proteolytic enzymes and restriction of starch metabolism, leading to the failure of protein mobilization in seeds. | [73] |
| Ocimum basilicum L. | 20 mg kg−1 | Alterations in the embryo and reductions in the oil contents of seeds. | [74] |
| Brassica oleracea L. | 5 mg L−1 | Decreased seed germination with an increase in MDA contents, electrolyte leakage and H2O2 contents. | [75] |
| Sassafras tzumu Hemsl. | 100 mg kg−1 | Restricted seedling growth and germination, and impairment of photosynthesis at higher doses. | [63] |
| Brassica juncea L. | 15 mg kg−1 | Disintegration occured in roots and shoots, and levels of ROS increased in plant shoots. | [76] |
| Oryza sativa L. | 50 μM | Lower seed germination rate due to the hyperaccumulation of Cd. | [53] |
| Zea mays L. | 100 mg kg−1 | Reduced seedling growth and activity of cellular antioxidants. | [77] |
| Growth and development | |||
| Cicer arietinum L. | 50 μM | Reduction in growth and appearance of symptoms of necrosis and chlorosis in leaves. | [78] |
| Ipomoea aquatica Forsk | Reduced growth and development of root and shoots. | [79] | |
| Lens culinaris | 50 µg g−1 | Increased electrolyte leakage and ROS production, resulting in lower plant growth. | [80] |
| Medicago sativa L. | 10 mg kg−1 | Higher concentrations damaged proteins, changed cell wall infrastructure and metabolism, and limited growth. | [81] |
| Osmolytes and photosynthesis | |||
| Cajanus cajan L. | 10 mg kg−1 | Lower organic osmolytes ultimately caused a disturbance in osmotic adjustments. | [82] |
| Vigna angularis | 64 mg L−1 | Cellular antioxidants decreased at higher concentrations, resulting in the lower production of low-molecular-weight osmolytes. | [83] |
| Zea mays L. | 150 μM | Reduction in photosynthetic pigments and gas exchange traits. | [58] |
| Coriandrum sativum L. | 20 µM L−1 | Inhibited gas exchange traits and biochemical processes. | [45] |
| Capsicum annuum L. | 500 ppm | Induction of stomatal closure, resulting in decreased photosynthetic pigments, a smaller stomatal size and reduced transpiration. | [84] |
| Mentha arvensis | 150 mg Kg−1 | Reductions in mineral assimilation, photosynthetic attributes and photosynthetic pigments occurred. | [85] |
| Experiment | Contamination Level | Microorganisms | Plant | Results | References |
|---|---|---|---|---|---|
| Pot | 10 mg kg−1 | Pseudomonas fluorescens | Hordeum vulgare | Phyto-stabilization of Cd due to PGPR activity, increased uptake of essential plant nutrients and enhanced plant growth attributes. | [90] |
| Greenhouse pot | 10.7 mg kg−1 Cd | Bacillus spp. | Solanum nigrum | Increased plant growth attributes under Cd stress, enhanced absorption of P and Fe as well as increased Cd contents in aerial plant parts. | [91] |
| Incubation study | 200 μg/mL | Klebsiella michiganensis | Oryza sativa | Cd bioaccumulation by tolerant bacteria with a concurrent decline in its uptake by plants. | [92] |
| Pot | 50 mg kg−1 | Cupriavidus necator, Sphingomonas and Curtobacterium spp. | Brassica napus | Increased plant biomass and growth traits under Cd contamination in inoculated treatments along with enhanced Cd uptake by aerial plant parts. | [93] |
| Pot | 0, 50, and 100 mg L−1 | Rhizobium pusense | Glycine max | Decreased soybean root Cd contents by 45.9 and 35.3%, respectively, at contamination levels of 50 and 100 mg L−1. | [94] |
| Pot | (0, 25, 50, 75, 100, 150 and 200 mg kg−1) | Enterobacter cloacae, Klebsiella pneumonia and Klebsiella spp. | Pennisetum giganteum | Combined application of rhizobacteria increased the bioaccumulation factor of Cd for plants. | [95] |
| Pot | 0, 5, 10, 15 and 20 mg kg−1 | Serratia marcescens | Chrysopogon zizanioides | Increased phytoaccumulation of Cd, soil biological health, as well as antioxidative potential of plants under bacterial inoculation. Maximum Cd phytoextraction in roots (289.47 mg kg−1), leaves (59.38 mg kg−1) and stem (88.33 mg kg−1) with a concomitant increase in plant biomass (9.68–45.99%). | [96] |
| Field | 2.2 mg kg−1 | Rhizobium leguminosarum, Bacillus simplex, Luteibacter sp. + Variovorax sp., Pseudomonas fluorescens | Lathyrus sativus | Increased growth attributes as well as nodule number, and plant nutrient uptake, and phytoaccumulation along with reduced rhizosphere concentration of Cd (61%). | [97] |
| Pot | 50 and 100 mg kg−1 | Fungi “Funneliformis mosseae” and bacteria Enterobacter sp. and Enterobacter ludwigii | Lycopersicon esculentum | Increased dry weights of shoots (119–154%) and roots (91–173%) under combined inoculation. Furthermore, decreased Cd concentrations in shoots as well as translocation factors under inoculated treatments were observed. | [98] |
| Pot | 0, 0.25, 0.5, 0.75 and 1 M CdSO4 | Serratia marcescens | Oryza sativa | Increased Cd removal from soil (66 mg kg−1 after 20 days). | [99] |
| Incubation study | 0, 0.25, and 0.5 mM Cd | Stenotrophomonas maltophilia | Capsicum annuum | Under Cd stress, increased root lengths (1.46 times) in the inoculated treatment compared to the control. | [100] |
| Pot | 15 mg kg−1 | Variovorax paradoxus, Rhizobium leguminosarum and fungus Glomus spp. | Pisum sativum and Brassica juncea | More prominent positive effect of consortium inoculation on Pisum sativum rather than Brassica juncea, in terms of growth, nutrient uptake and increased seed Cd concentration. | [101] |
| Greenhouse pot | 2.12 mg kg−1 | Bacillus megaterium, Glomus mosseae, and Piriformospora indica | Solanum nigrum | Cd accumulation (104%) observed under the combined application of Bacillus megaterium and Glomus mosseae in addition to increased soil biological health under contaminated conditions. | [102] |
| Pot | 100 mg kg−1 | Bacillus sp. | Oryza sativa | Reduced bioavailable Cd concentration (39.3%), increased phytoextraction efficiency of rice for Cd (48.2%) and increased rice growth and yield traits under inoculation compared to the control. | [103] |
| Greenhouse | 0.064 mg L−1 | Klebsiella huaxiensis and Pantoea cypripedii | Pennisetum purpurenum | Enhanced Cd phytoaccumulation in all variants of the tested plant (28.43–38.07 mg kg−1). | [104] |
| Pot | 30 μmol L−1 | Enterobacter cloacae | Solanum nigrum | Increased soil Cd phytoextraction by plants along with increased plant growth under Cd stress. | [105] |
| Pot | 0, 0.25, and 0.50 mg kg−1 | Bacillus spp. | Oryza sativa | Increased Cd immobilization in soil by its surface adsorption concomitant with increased plant growth. | [106] |
| Incubation | 0.4 mM CdCl2 | Pseudomonas aeruginosa and Burkholderia gladioli | Solanum lycopersicum | Alleviation of Cd toxicity in plants was evident by an increase in phenolic compounds, osmolytes and low-molecular-weight organic acids. | [107] |
| Growth room trial | 0–400 μg/mL | Enterobacter cloacae | Oryza sativa | Increased Cd removal efficiency (72.11%) against a contamination level of 400 μg/mL | [108] |
| Pot | 2 g | Curtobacterium oceanosedimentum | Capsicum frutescens | Increased root (58%) and shoot (60%) lengths, enhanced accumulation of Cd in roots compared to shoots under bacterial inoculation. | [109] |
| Pot | 1.68 mg kg−1 | Buttiauxella, Pedobacter, Aeromonas eucrenophila, and Ralstonia pickettii | Sedum plumbizincicola | Inoculation led to reduced reducible and residual Cd and increased Cd availability coefficients by 1.15–6.41 units. Cd contents in shoots (29.63–46.01%) and roots (11.42–84.47%), bioconcentration factor (2.13–2.72) and Cd removal rate (48.25%) compared to the control treatment | [110] |
| Species name | Initial Cd Concentration | Experimental Medium | Cd Remediation Efficiency | References |
|---|---|---|---|---|
| Bacterial species | ||||
| Pseudomonas fluorescens and Bacillus subtilis | 150 mg L−1 | Soil | 16.7 | [162] |
| Pseudomonas sp. DDT-1 | 0.9 mg kg−1 | Soil | 40.3 | [163] |
| Kocuria rhizophila | 150 mg L−1 | Aqueous | 9.07 mg g−1 | [164] |
| Klebsiella michiganensis | 1000 µg ml−1 | Soil | 97% | [165] |
| Enterobacter sp | 3500 µg ml−1 | Soil | 95% | [166] |
| Rhodobacter sphaeroides | 65.33 mg kg−1 | Soil | 30.7 | [167] |
| Bacillus aryabhattai and Bacillus amyloliquefaciens | 250 mg L−1 | Soil | 96% | [168] |
| Aspergillus sydowii | 50 mg kg−1 | Soil | 10.44% | [169] |
| Cupriavidus sp. | 13.82 mg kg−1 | Soil | 58.2% | [170] |
| Paenibacillus sp. and Bacillus sp. | 20 mg L−1 | Soil | 128.50% | [171] |
| Bacillus sp. TZ5 | 150 mg L−1 | Soil | 48.49% | [172] |
| Acidithiobacillus caldus DX, Acidithiobacillus thiooxidans DX, Acidithiobacillus thiooxidans ZJ, Acidithiobacillus thiooxidans AO1, Ferroplasma acidiphilum DX, Acidithiobacillus caldus S1 and Leptospirillum ferriphilum DX | 9.09 mg kg−1 | Soil | 32.09% | [173] |
| Cupriavidus sp. (KU168590), Ensifer sp. (KU168586), Burkholderia sp. (KU168588), and Paenibacillus sp. (KU168587) | 0.21 mg kg−1 | Soil | 33.0% | [174] |
| Enterobacter cloacae, Pseudomonas aeruginosa, and Klebsiella Edwardsii | 50 mg L−1 | Soil | 58.80% | [175] |
| Bacillus subtilis | 147.75 mg kg−1 | Soil | 35.17% | [176] |
| Burkholderia sp. and Bacillus sp. | 5 mM | Soil | 84.17% | [177] |
| Bacillus sp. | 49 mg kg−1 | Soil | 43.53% | [178] |
| Bacillus subtilis | 147.75 mg kg−1 | Soil | 18.56% | [179] |
| Firmicutes sp. and Proteobacteria sp. | 14.9 mg kg−1 | Soil | 40.0 | [180] |
| Enterobacter hormaechei SFC3 | 100 µg ml−1 | Soil | 90.21% | [181] |
| Streptomyces pactum Act12 and Streptomyces Roche D74 | 1.62 mg kg−1 | Soil | 56.39% | [182] |
| Bacillus velezensis | - | Soil | 1.65 μg g−1 | [183] |
| Fungi | ||||
| Aspergillus niger, Aspergillus fumigatus, and Penicillium rubens | 0.6 mg L−1 | Soil | 79% | [184] |
| Simplicillium chinense | 400 mg L−1 | Soil | 88% | [185] |
| Aspergillus flflavus, Aspergillus gracilis, Aspergillus penicillioides, Aspergillus restrictus, and Sterigmatomyces halophilus | 1000 mg L−1 | Soil | 95% | [186] |
| Phanerochaete chrysosporium | 25 mg L−1 | Soil | 96% | [187] |
| Agaricus bisporus, Pleurotus platypus, and Calocybe indica | - | - | 98.97 | [188] |
| Lactarius piperatus and Agaricus bisporus | 265 mg L−1 | Aqueous | 95% | [189] |
| Rhizophagus intraradices | - | Soil | 38% | [190] |
| Trichoderma harzianum | 147.75 mg kg−1 | Soil | 47.69% | [176] |
| Algae/cyanobacteria | ||||
| Asparagopsis armata | 150 mg L−1 | Aqueous | 10.6% | [191] |
| Chaetoceros calcitrans and Tetracelmis chuii | - | Aqueous | - | [192] |
| Ulva lactuca | 80 mg L−1 | Aqueous | 41.0% | [193] |
| Chara aculeolate | - | Aqueous | 23 mg g−1 | [194] |
| Chlorella pyrenoidosa | 1.5 mg L−1 | Aqueous | 45.45 | [195] |
| Scenedesmus acutus | 1.5 mg L−1 | Aqueous | 57.14 | [195] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zulfiqar, U.; Haider, F.U.; Maqsood, M.F.; Mohy-Ud-Din, W.; Shabaan, M.; Ahmad, M.; Kaleem, M.; Ishfaq, M.; Aslam, Z.; Shahzad, B. Recent Advances in Microbial-Assisted Remediation of Cadmium-Contaminated Soil. Plants 2023, 12, 3147. https://doi.org/10.3390/plants12173147
Zulfiqar U, Haider FU, Maqsood MF, Mohy-Ud-Din W, Shabaan M, Ahmad M, Kaleem M, Ishfaq M, Aslam Z, Shahzad B. Recent Advances in Microbial-Assisted Remediation of Cadmium-Contaminated Soil. Plants. 2023; 12(17):3147. https://doi.org/10.3390/plants12173147
Chicago/Turabian StyleZulfiqar, Usman, Fasih Ullah Haider, Muhammad Faisal Maqsood, Waqas Mohy-Ud-Din, Muhammad Shabaan, Muhammad Ahmad, Muhammad Kaleem, Muhammad Ishfaq, Zoya Aslam, and Babar Shahzad. 2023. "Recent Advances in Microbial-Assisted Remediation of Cadmium-Contaminated Soil" Plants 12, no. 17: 3147. https://doi.org/10.3390/plants12173147
APA StyleZulfiqar, U., Haider, F. U., Maqsood, M. F., Mohy-Ud-Din, W., Shabaan, M., Ahmad, M., Kaleem, M., Ishfaq, M., Aslam, Z., & Shahzad, B. (2023). Recent Advances in Microbial-Assisted Remediation of Cadmium-Contaminated Soil. Plants, 12(17), 3147. https://doi.org/10.3390/plants12173147












