Camellia oleifera CoSWEET10 Is Crucial for Seed Development and Drought Resistance by Mediating Sugar Transport in Transgenic Arabidopsis
Abstract
1. Introduction
2. Results
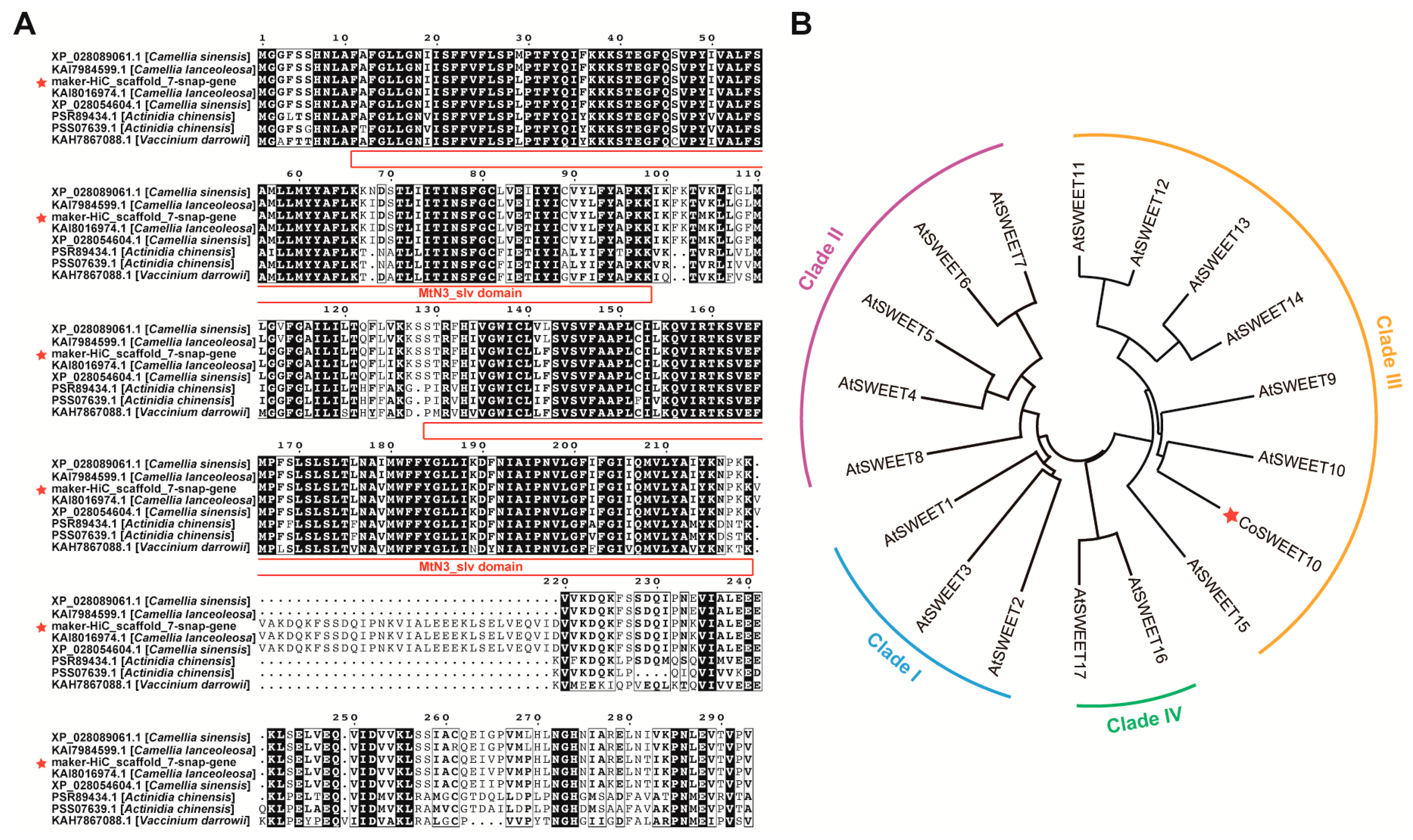
2.1. Identification and Phylogenetic Analysis of CoSWEET10
2.2. Expression Pattern and Localization of CoSWEET10
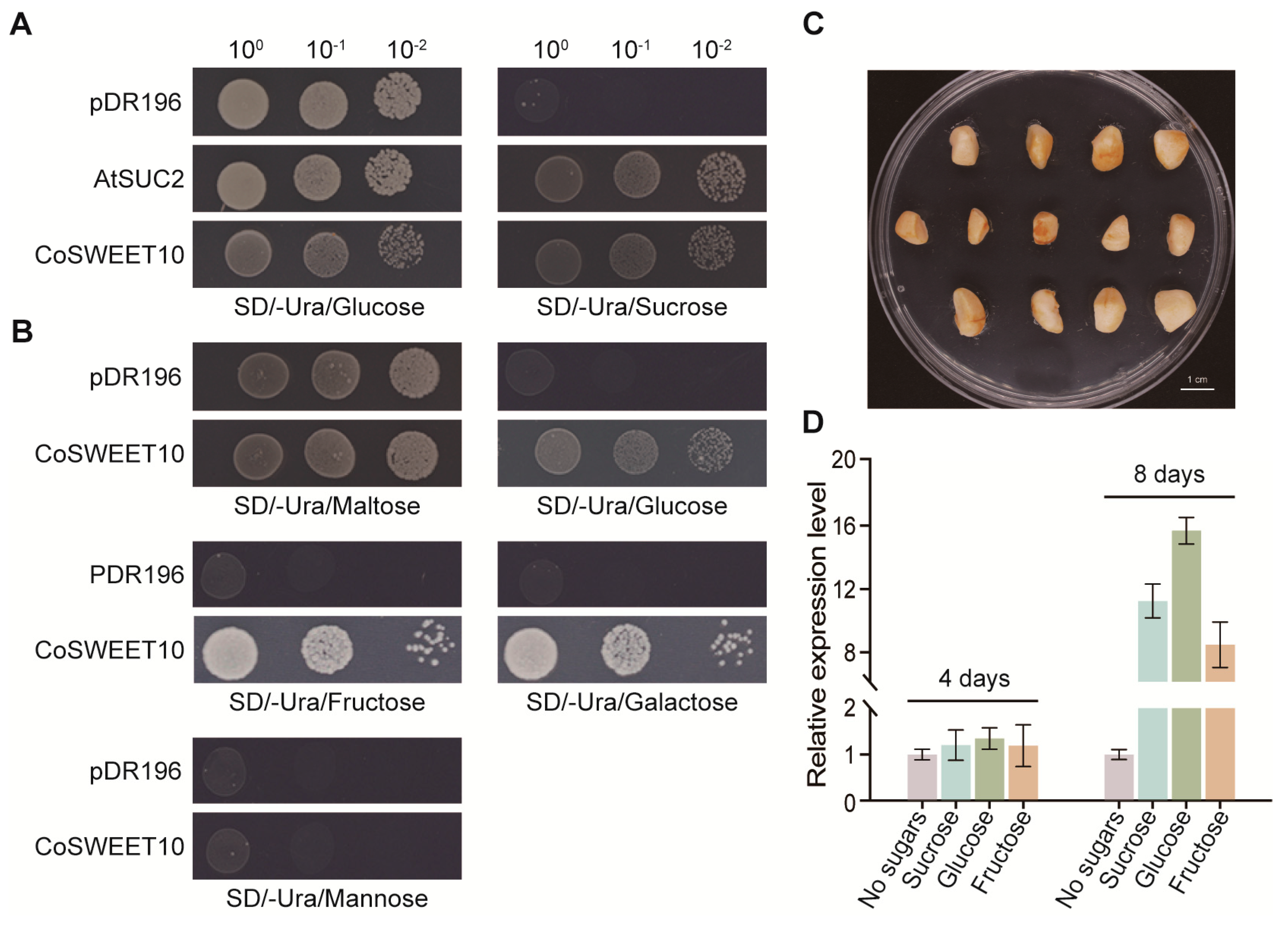
2.3. CoSWEET10 Transport Substrate Specificity in Yeast Cells and Sugar-Induced Analysis in C. oleifera
2.4. Identification of Potential Self-Interaction of CoSWEET10
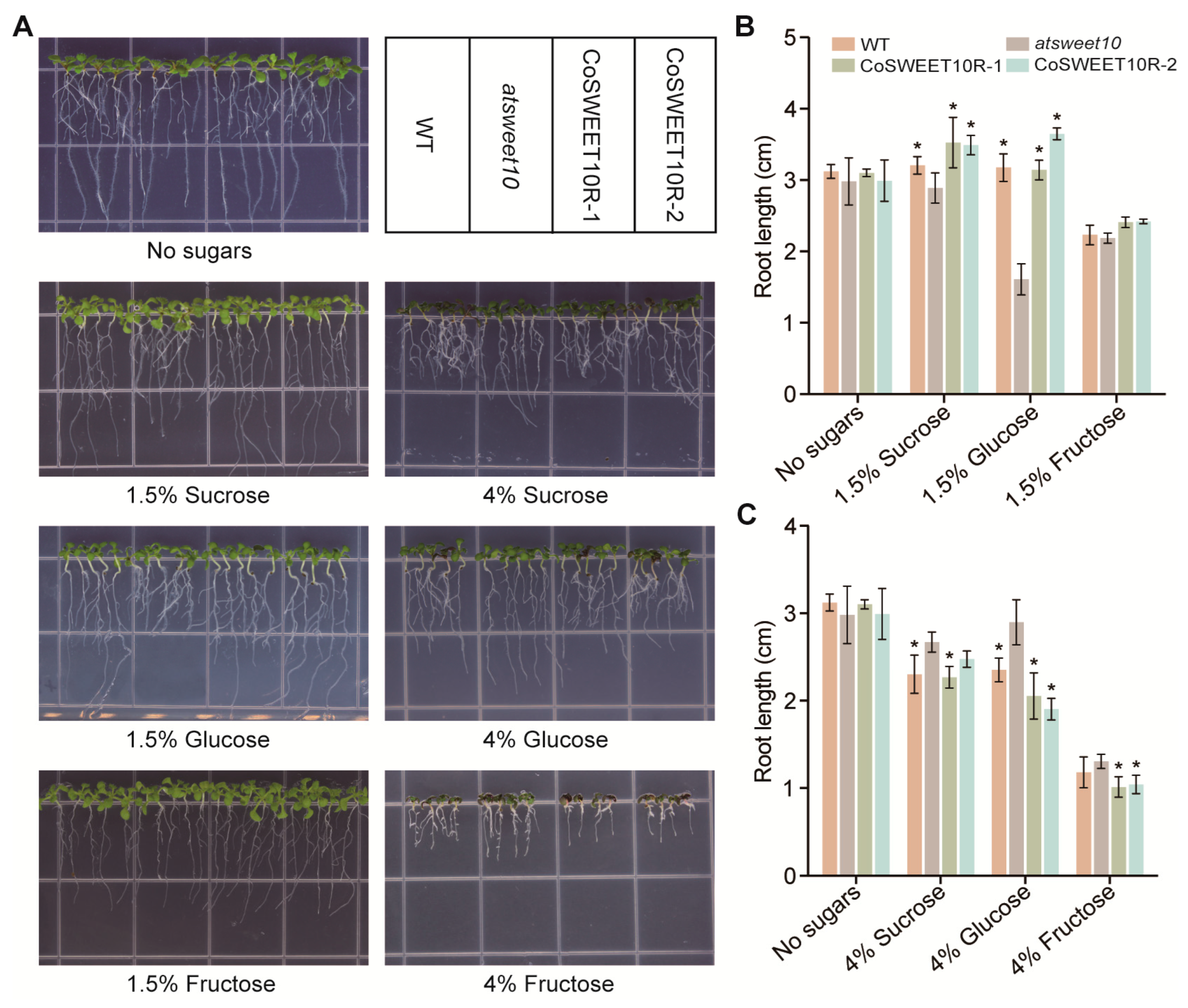
2.5. CoSWEET10 Functions as Sucrose and Hexose Transporter in Arabidopsis
2.6. CoSWEET10 Is Essential for Seed Development in Transgenic Arabidopsis
2.7. CoSWEET10 Enhances Drought Tolerance in Transgenic Arabidopsis
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Protein Sequence and Phylogenetic Analysis
4.3. RNA Extraction and qRT-PCR (Quantitative Real-Time PCR) Experiments
4.4. Subcellular Localization of CoSWEET10
4.5. Bimolecular Fluorescence Complementation Assays
4.6. Complementation Assays for CoSWEET10 in Yeast Cells
4.7. C. oleifera Seed In Vitro Culture on Medium Containing Different Sugars
4.8. Paraffin Embedding and Sectioning
4.9. Sugar Quantification of Arabidopsis Siliques
4.10. Oil Content Measurement of Arabidopsis Seeds
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zhuang, R. Camellia oleifera in China; China Forestry Press: Beijing, China, 2008; p. 366. (In Chinese) [Google Scholar]
- Yang, C.Y.; Liu, X.M.; Chen, Z.Y.; Lin, Y.S.; Wang, S.Y. Comparison of oil content and fatty acid profile of ten new Camellia oleifera cultivars. J. Lipids 2016, 2016, 3982486. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.P.; Shih, P.H.; Hsu, C.L.; Yen, G.C. Hepatoprotection of tea seed oil (Camellia oleifera Abel.) against CCl4-induced oxidative damage in rats. Food Chem. Toxicol. 2007, 45, 888–895. [Google Scholar] [CrossRef]
- Lee, C.P.; Yen, G.C. Antioxidant activity and bloactive compounds of tea seed (Camellia oleifera Abel.) oil. J. Agric. Food Chem. 2006, 54, 779–784. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.G.; Long, S.P.; Ort, D.R. Improving photosynthetic efficiency for greater yield. Annu. Rev. Plant Biol. 2010, 61, 235–261. [Google Scholar] [CrossRef]
- Paterson, A.H.; Li, Z.K. Paleo-green revolution for rice. Proc. Natl. Acad. Sci. USA 2011, 108, 10931–10932. [Google Scholar] [CrossRef] [PubMed]
- Lalonde, S.; Wipf, D.; Frommer, W.B. Transport mechanisms for organic forms of carbon and nitrogen between source and sink. Annu. Rev. Plant Biol. 2004, 55, 341–372. [Google Scholar] [CrossRef]
- Ayre, B.G. Membrane-transport systems for sucrose in relation to whole-plant carbon partitioning. Mol. Plant 2011, 4, 377–394. [Google Scholar] [CrossRef]
- Chen, L.Q.; Qu, X.Q.; Hou, B.H.; Sosso, D.; Osorio, S.; Fernie, A.R.; Frommer, W.B. Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science 2012, 335, 207–211. [Google Scholar] [CrossRef]
- Julius, B.T.; Leach, K.A.; Tran, T.M.; Mertz, R.A.; Braun, D.M. Sugar transporters in plants: New insights and discoveries. Plant Cell Physiol. 2017, 58, 1442–1460. [Google Scholar] [CrossRef]
- Chen, L.Q.; Hou, B.H.; Lalonde, S.; Takanaga, H.; Hartung, M.L.; Qu, X.Q.; Guo, W.J.; Kim, J.G.; Underwood, W.; Chaudhuri, B.; et al. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 2010, 468, 527–532. [Google Scholar] [CrossRef]
- Jia, B.; Zhu, X.F.; Pu, Z.J.; Duan, Y.X.; Hao, L.J.; Zhang, J.; Chen, L.Q.; Jeon, C.O.; Xuan, Y.H. Integrative view of the diversity and evolution of SWEET and semiSWEET sugar transporters. Front. Plant Sci. 2017, 8, 2178. [Google Scholar] [CrossRef]
- Anjali, A.; Fatima, U.; Manu, M.S.; Ramasamy, S.; Senthil-Kumar, M. Structure and regulation of SWEET transporters in plants: An update. Plant Physiol. Biochem. 2020, 156, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.; Wang, S. Rice MtN3/Saliva/SWEET family genes and their homologs in cellular organisms. Mol. Plant 2013, 6, 665–674. [Google Scholar] [CrossRef] [PubMed]
- Sosso, D.; Luo, D.; Li, Q.B.; Sasse, J.; Yang, J.; Gendrot, G.; Suzuki, M.; Koch, K.E.; McCarty, D.R.; Chourey, P.S.; et al. Seed filling in domesticated maize and rice depends on SWEET-mediated hexose transport. Nat. Genet. 2015, 47, 1489–1493. [Google Scholar] [CrossRef]
- Ma, L.; Zhang, D.; Miao, Q.; Yang, J.; Xuan, Y.; Hu, Y. Essential role of sugar transporter OsSWEET11 during the early stage of rice grain filling. Plant Cell Physiol. 2017, 58, 863–873. [Google Scholar] [CrossRef]
- Yang, J.; Luo, D.; Yang, B.; Frommer, W.B.; Eom, J.S. SWEET11 and 15 as key players in seed filling in rice. New Phytol. 2018, 218, 604–615. [Google Scholar] [CrossRef]
- Wang, S.; Liu, S.; Wang, J.; Yokosho, K.; Zhou, B.; Yu, Y.C.; Liu, Z.; Frommer, W.B.; Ma, J.F.; Chen, L.Q.; et al. Simultaneous changes in seed size, oil content and protein content driven by selection of SWEET homologues during soybean domestication. Natl. Sci. Rev. 2020, 7, 1776–1786. [Google Scholar] [CrossRef]
- Zhang, Z.; Zou, L.; Ren, C.; Ren, F.; Wang, Y.; Fan, P.; Li, S.; Liang, Z. VvSWEET10 mediates sugar accumulation in grapes. Genes 2019, 10, 255. [Google Scholar] [CrossRef] [PubMed]
- Ren, R.; Yue, X.; Li, J.; Xie, S.; Guo, S.; Zhang, Z. Coexpression of sucrose synthase and the SWEET transporter, which are associated with sugar hydrolysis and transport, respectively, increases the hexose content in Vitis vinifera L. grape berries. Front. Plant Sci. 2020, 11, 321. [Google Scholar] [CrossRef]
- Ko, H.Y.; Ho, L.H.; Neuhaus, H.E.; Guo, W.J. Transporter SISWEET15 unloads sucrose from phloem and seed coat for fruit and seed development in tomato. Plant Physiol. 2021, 187, 2230–2245. [Google Scholar] [CrossRef]
- Chen, L.Q.; Lin, I.W.N.; Qu, X.Q.; Sosso, D.; McFarlane, H.E.; Londono, A.; Samuels, A.L.; Frommer, W.B. A cascade of sequentially expressed sucrose transporters in the seed coat and endosperm provides nutrition for the Arabidopsis embryo. Plant Cell 2015, 27, 607–619. [Google Scholar] [CrossRef]
- Fei, H.; Yang, Z.; Lu, Q.; Wen, X.; Zhang, Y.; Zhang, A.; Lu, C. OsSWEET14 cooperates with OsSWEET11 to contribute to grain filling in rice. Plant Sci. 2021, 306, 110851. [Google Scholar] [CrossRef] [PubMed]
- Eom, J.S.; Luo, D.; Atienza-Grande, G.; Yang, J.; Ji, C.; Van Thi, L.; Huguet-Tapia, J.C.; Char, S.N.; Liu, B.; Nguyen, H.; et al. Diagnostic kit for rice blight resistance. Nat. Biotechnol. 2019, 37, 1372–1379. [Google Scholar] [CrossRef] [PubMed]
- Oliva, R.; Ji, C.; Atienza-Grande, G.; Huguet-Tapia, J.C.; Perez-Quintero, A.; Li, T.; Eom, J.S.; Li, C.; Nguyen, H.; Liu, B.; et al. Broad-spectrum resistance to bacterial blight in rice using genome editing. Nat. Biotechnol. 2019, 37, 1344–1350. [Google Scholar] [CrossRef]
- Xu, Z.; Xu, X.; Gong, Q.; Li, Z.; Li, Y.; Wang, S.; Yang, Y.; Ma, W.; Liu, L.; Zhu, B.; et al. Engineering broad-spectrum bacterial blight resistance by simultaneously disrupting variable TALE-binding elements of multiple susceptibility genes in rice. Mol. Plant 2019, 12, 1434–1446. [Google Scholar] [CrossRef]
- Li, T.; Liu, B.; Spalding, M.H.; Weeks, D.P.; Yang, B. High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat. Biotechnol. 2012, 30, 390–392. [Google Scholar] [CrossRef]
- Ahmad, F.; Singh, A.; Kamal, A. Osmoprotective role of sugar in mitigating abiotic stress in plants. In Protective Chemical Agents in the Amelioration of Plant Abiotic Stress; John Wiley & Sons: Hoboken, NJ, USA, 2020; pp. 53–70. [Google Scholar]
- Singh, J.; Thakur, J.K. Photosynthesis and Abiotic Stress in Plants. In Biotic and Abiotic Stress Tolerance in Plants; Springer: Cham, Switzerland, 2018; pp. 27–46. [Google Scholar] [CrossRef]
- Breia, R.; Conde, A.; Badim, H.; Fortes, A.M.; Geros, H.; Granell, A. Plant SWEETs: From sugar transport to plant-pathogen interaction and more unexpected physiological roles. Plant Physiol. 2021, 186, 836–852. [Google Scholar] [CrossRef] [PubMed]
- Valifard, M.; Le Hir, R.; Mueller, J.; Scheuring, D.; Neuhaus, H.E.; Pommerrenig, B. Vacuolar fructose transporter SWEET17 is critical for root development and drought tolerance. Plant Physiol. 2021, 187, 2716–2730. [Google Scholar] [CrossRef]
- Mathan, J.; Singh, A.; Ranjan, A. Sucrose transport in response to drought and salt stress involves ABA-mediated induction of OsSWEET13 and OsSWEET15 in rice. Physiol. Plant. 2021, 171, 620–637. [Google Scholar] [CrossRef]
- Durand, M.; Porcheron, B.; Hennion, N.; Maurousset, L.; Lemoine, R.; Pourtau, N. Water deficit enhances C export to the roots in Arabidopsis thaliana plants with contribution of sucrose transporters in both shoot and roots. Plant Physiol. 2016, 170, 1460–1479. [Google Scholar] [CrossRef]
- Wang, X.Y.; You, H.L.; Yuan, Y.H.; Zhang, H.H.; Zhang, L.Y. The cellular pathway and enzymatic activity for phloem-unloading transition in developing Camellia oleifera Abel. Fruit. Acta Physiol. Plant. 2018, 40, 23. [Google Scholar] [CrossRef]
- Zhou, J.; Du, B.S.; Chen, Y.Q.; Cao, Y.B.; Yu, M.X.; Zhang, L.Y. Integrative physiological and transcriptomic analysis reveals the transition mechanism of sugar phloem unloading route in Camellia oleifera Fruit. Int. J. Mol. Sci. 2022, 23, 4590. [Google Scholar] [CrossRef]
- Xuan, Y.H.; Hu, Y.B.; Chen, L.Q.; Sosso, D.; Ducat, D.C.; Hou, B.H.; Frommer, W.B. Functional role of oligomerization for bacterial and plant SWEET sugar transporter family. Proc. Natl. Acad. Sci. USA 2013, 110, E3685–E3694. [Google Scholar] [CrossRef] [PubMed]
- Singh, J.; Das, S.; Gupta, K.J.; Ranjan, A.; Foyer, C.H.; Thakur, J.K. Physiological implications of SWEETs in plants and their potential applications in improving source-sink relationships for enhanced yield. Plant Biotechnol. J. 2023, 21, 1528–1541. [Google Scholar] [CrossRef]
- Wang, S.; Yokosho, K.; Guo, R.; Whelan, J.; Ruan, Y.L.; Ma, J.F.; Shou, H. The soybean sugar transporter GmSWEET15 mediates sucrose export from endosperm to early embryo. Plant Physiol. 2019, 180, 2133–2141. [Google Scholar] [CrossRef] [PubMed]
- Kanno, Y.; Oikawa, T.; Chiba, Y.; Ishimaru, Y.; Shimizu, T.; Sano, N.; Koshiba, T.; Kamiya, Y.; Ueda, M.; Seo, M. AtSWEET13 and AtSWEET14 regulate gibberellin-mediated physiological processes. Nat. Commun. 2016, 7, 13245. [Google Scholar] [CrossRef] [PubMed]
- Jeena, G.S.; Kumar, S.; Shukla, R.K. Structure, evolution and diverse physiological roles of SWEET sugar transporters in plants. Plant Mol. Biol. 2019, 100, 351–365. [Google Scholar] [CrossRef]
- Zhang, X.S.; Feng, C.Y.; Wang, M.N.; Li, T.L.; Liu, X.; Jiang, J. Plasma membrane-localized SlSWEET7a and SlSWEET14 regulate sugar transport and storage in tomato fruits. Hortic. Res. 2021, 8, 186. [Google Scholar] [CrossRef]
- Wu, L.B.; Eom, J.S.; Isoda, R.; Li, C.; Char, S.N.; Luo, D.; Schepler-Luu, V.; Nakamura, M.; Yang, B.; Frommer, W.B. OsSWEET11b, a potential sixth leaf blight susceptibility gene involved in sugar transport-dependent male fertility. New Phytol. 2022, 234, 975–989. [Google Scholar] [CrossRef]
- Gupta, P.K.; Balyan, H.S.; Gautam, T. SWEET genes and TAL effectors for disease resistance in plants: Present status and future prospects. Mol. Plant Pathol. 2021, 22, 1014–1026. [Google Scholar] [CrossRef]
- Yao, L.; Ding, C.; Hao, X.; Zeng, J.; Yang, Y.; Wang, X.; Wang, L. CsSWEET1a and CsSWEET17 mediate growth and freezing tolerance by promoting sugar transport across the plasma membrane. Plant Cell Physiol. 2020, 61, 1669–1682. [Google Scholar] [CrossRef]
- Phukan, U.J.; Jeena, G.S.; Tripathi, V.; Shukla, R.K. MaRAP2-4, a waterlogging-responsive ERF from Mentha, regulates bidirectional sugar transporter AtSWEET10 to modulate stress response in Arabidopsis. Plant Biotechnol. J. 2018, 16, 221–233. [Google Scholar] [CrossRef]
- Chen, Q.; Hu, T.; Li, X.; Song, C.P.; Zhu, J.K.; Chen, L.; Zhao, Y. Phosphorylation of SWEET sucrose transporters regulates plant root: Shoot ratio under drought. Nat. Plants 2022, 8, 68–77. [Google Scholar] [CrossRef]
- Verslues, P.E.; Agarwal, M.; Katiyar-Agarwal, S.; Zhu, J.H.; Zhu, J.K. Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J. 2006, 45, 523–539. [Google Scholar] [CrossRef]
- Yu, Y.H.; Bi, C.X.; Wang, Q.; Ni, Z.Y. Overexpression of TaSIM provides increased drought stress tolerance in transgenic Arabidopsis. Biochem. Biophys. Res. Commun. 2019, 512, 66–71. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.Y.; Huh, J.H.; Yu, Y.C.; Ho, L.H.; Chen, L.Q.; Tholl, D.; Frommer, W.B.; Guo, W.J. The Arabidopsis vacuolar sugar transporter SWEET2 limits carbon sequestration from roots and restricts Pythium infection. Plant J. 2015, 83, 1046–1058. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.Z.; Beck, M.; Muller, J.; Chen, T.; Wang, X.H.; Wang, F.; Wang, Q.L.; Wang, Y.Q.; Baluska, F.; Logan, D.C.; et al. Actin turnover is required for myosin-dependent mitochondrial movements in Arabidopsis root hairs. PLoS ONE 2009, 4, e5961. [Google Scholar] [CrossRef]
- Li, M.J.; Li, P.M.; Ma, F.W.; Dandekar, A.M.; Cheng, L.L. Sugar metabolism and accumulation in the fruit of transgenic apple trees with decreased sorbitol synthesis. Hortic. Res. 2018, 5, 60. [Google Scholar] [CrossRef]
- Zhang, N.; Shi, J.W.; Zhao, H.Y.; Jiang, J. Activation of small heat shock protein (SlHSP17.7) gene by cell wall invertase inhibitor (SlCIF1) gene involved in sugar metabolism in tomato. Gene 2018, 679, 90–99. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.Q.; Chen, J.; Zheng, H.Y.; Fang, T.; Ogutu, C.; Li, S.H.; Han, Y.P.; Wu, B.H. Comparative assessment of sugar and malic acid composition in cultivated and wild apples. Food Chem. 2015, 172, 86–91. [Google Scholar] [CrossRef]
- Kanai, M.; Hayashi, M.; Kondo, M.; Nishimura, M. The plastidic DEAD-box RNA helicase 22, HS3, is essential for plastid functions both in seed development and in seedling growth. Plant Cell Physiol. 2013, 54, 1431–1440. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.S.; Lv, X.Y.; Xie, G.L.; Zhang, J.; Xu, Y.; Chen, F. Seed-specific overexpression of AtFAX1 increases seed oil content in Arabidopsis. Biochem. Biophys. Res. Commun. 2018, 500, 370–375. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ye, Z.; Du, B.; Zhou, J.; Cao, Y.; Zhang, L. Camellia oleifera CoSWEET10 Is Crucial for Seed Development and Drought Resistance by Mediating Sugar Transport in Transgenic Arabidopsis. Plants 2023, 12, 2818. https://doi.org/10.3390/plants12152818
Ye Z, Du B, Zhou J, Cao Y, Zhang L. Camellia oleifera CoSWEET10 Is Crucial for Seed Development and Drought Resistance by Mediating Sugar Transport in Transgenic Arabidopsis. Plants. 2023; 12(15):2818. https://doi.org/10.3390/plants12152818
Chicago/Turabian StyleYe, Zhihua, Bingshuai Du, Jing Zhou, Yibo Cao, and Lingyun Zhang. 2023. "Camellia oleifera CoSWEET10 Is Crucial for Seed Development and Drought Resistance by Mediating Sugar Transport in Transgenic Arabidopsis" Plants 12, no. 15: 2818. https://doi.org/10.3390/plants12152818
APA StyleYe, Z., Du, B., Zhou, J., Cao, Y., & Zhang, L. (2023). Camellia oleifera CoSWEET10 Is Crucial for Seed Development and Drought Resistance by Mediating Sugar Transport in Transgenic Arabidopsis. Plants, 12(15), 2818. https://doi.org/10.3390/plants12152818





