Single-Bud Expression Analysis of Bud Dormancy Factors in Peach
Abstract
1. Introduction
2. Results
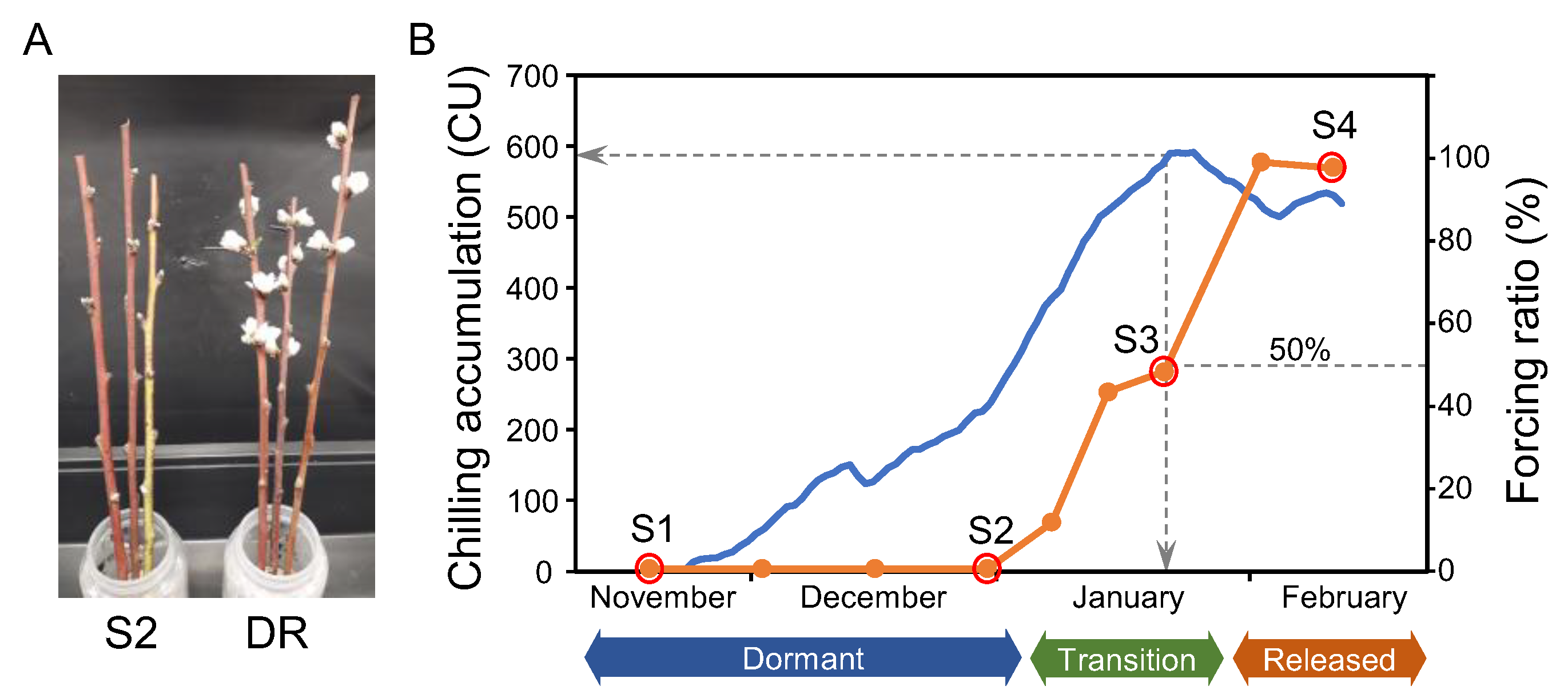
2.1. Assessment of Bud Dormancy Stages by Budbreak Forcing
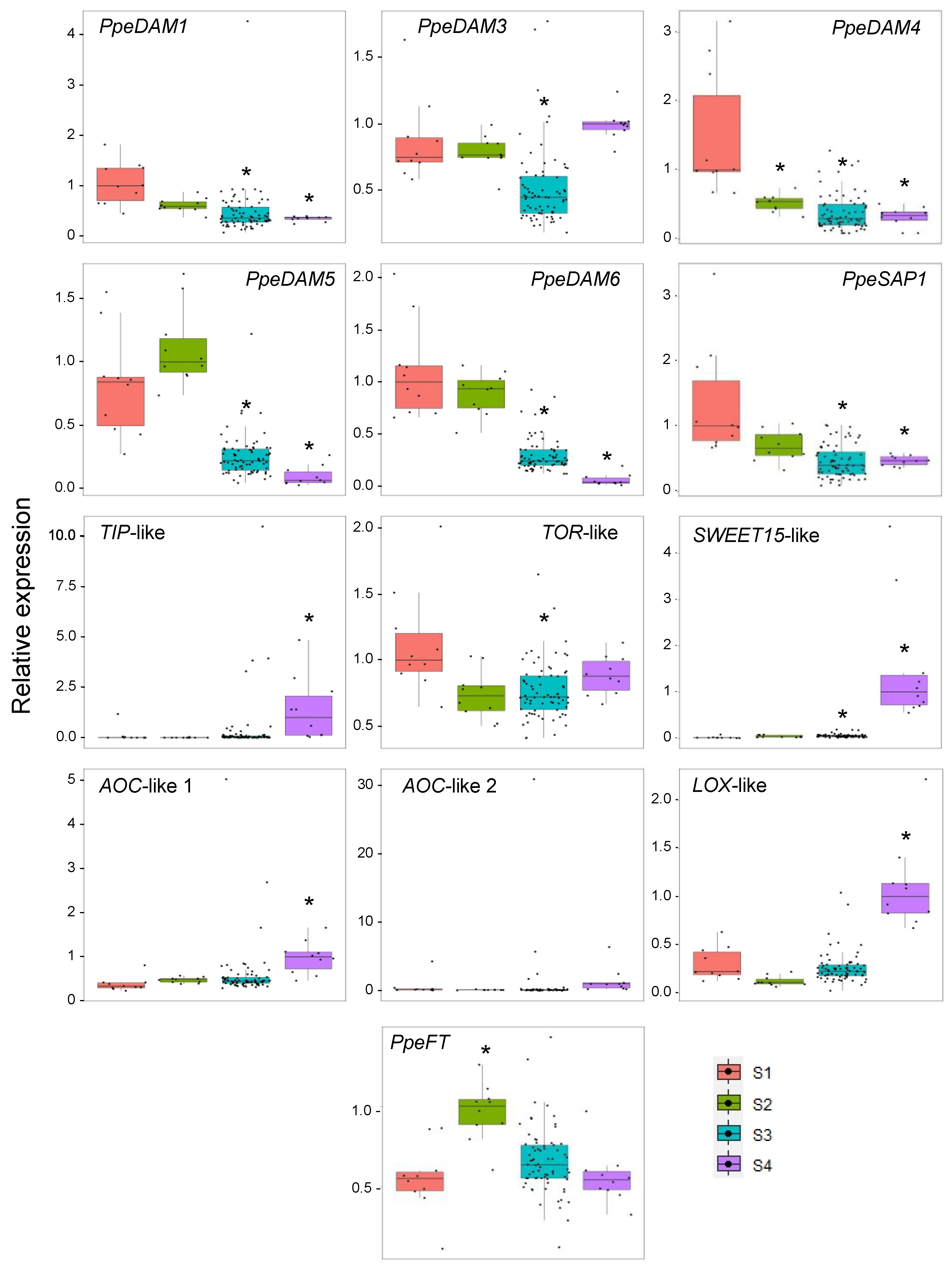
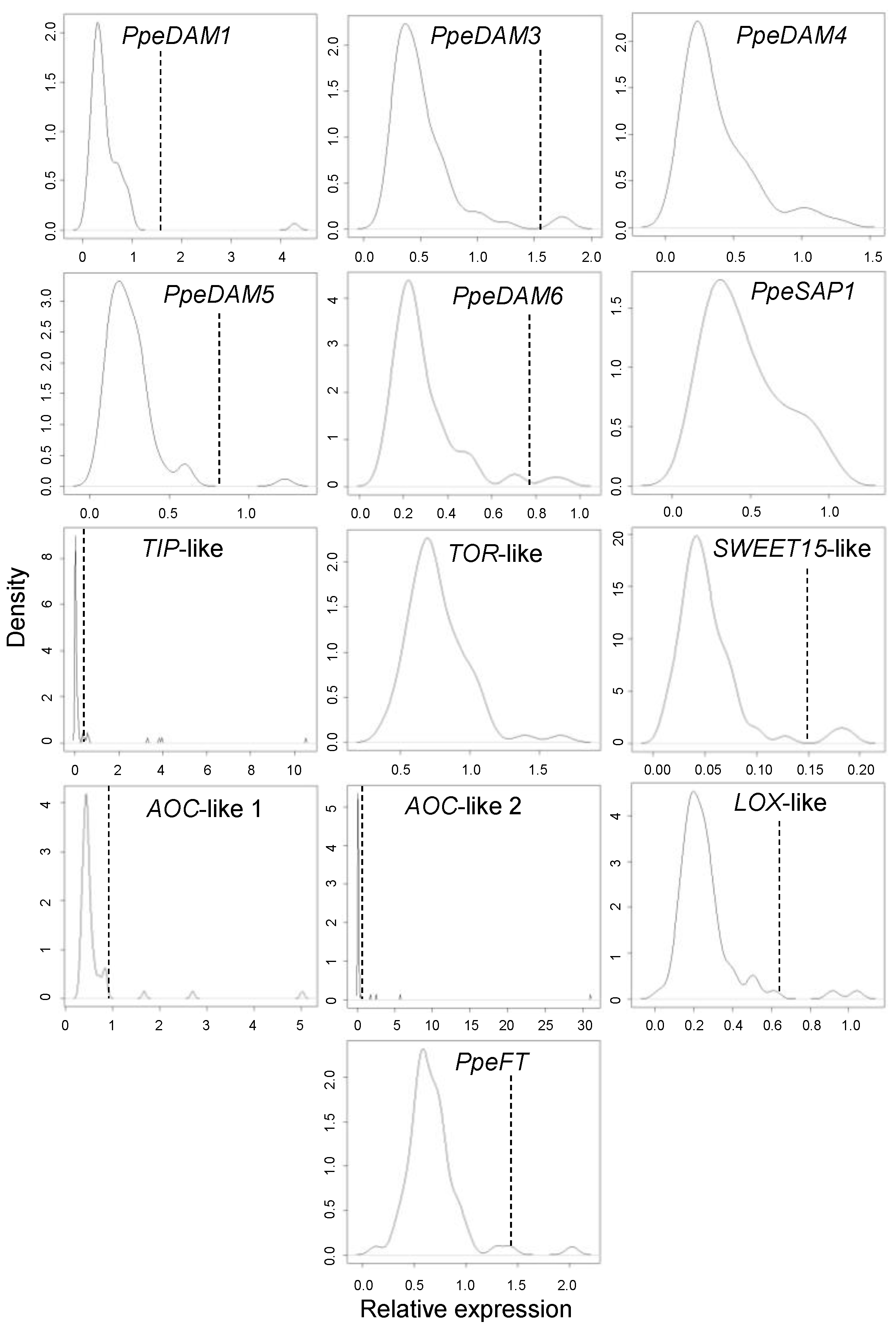
2.2. Single-Bud Analysis of Dormancy-Associated Gene Expression
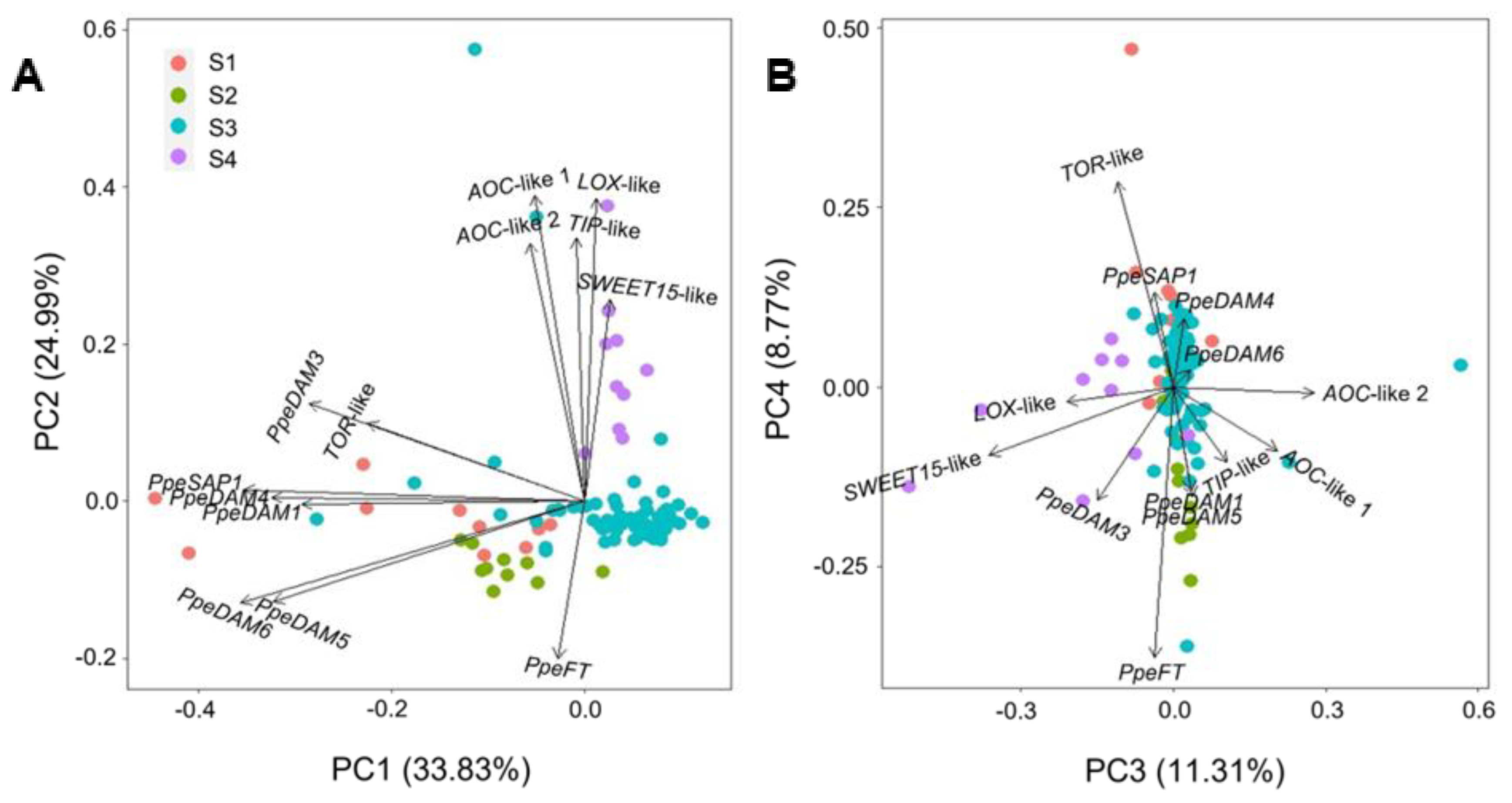
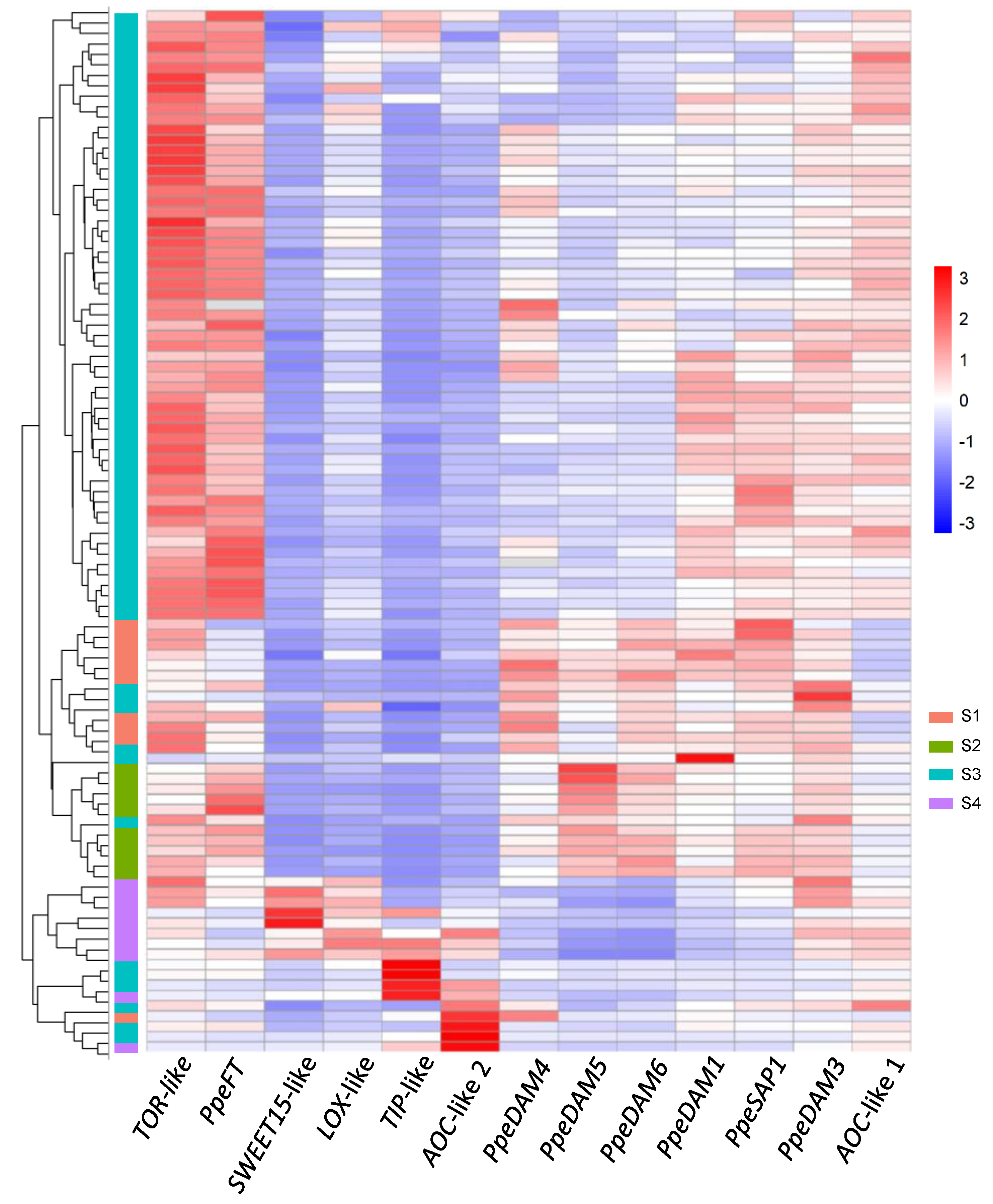
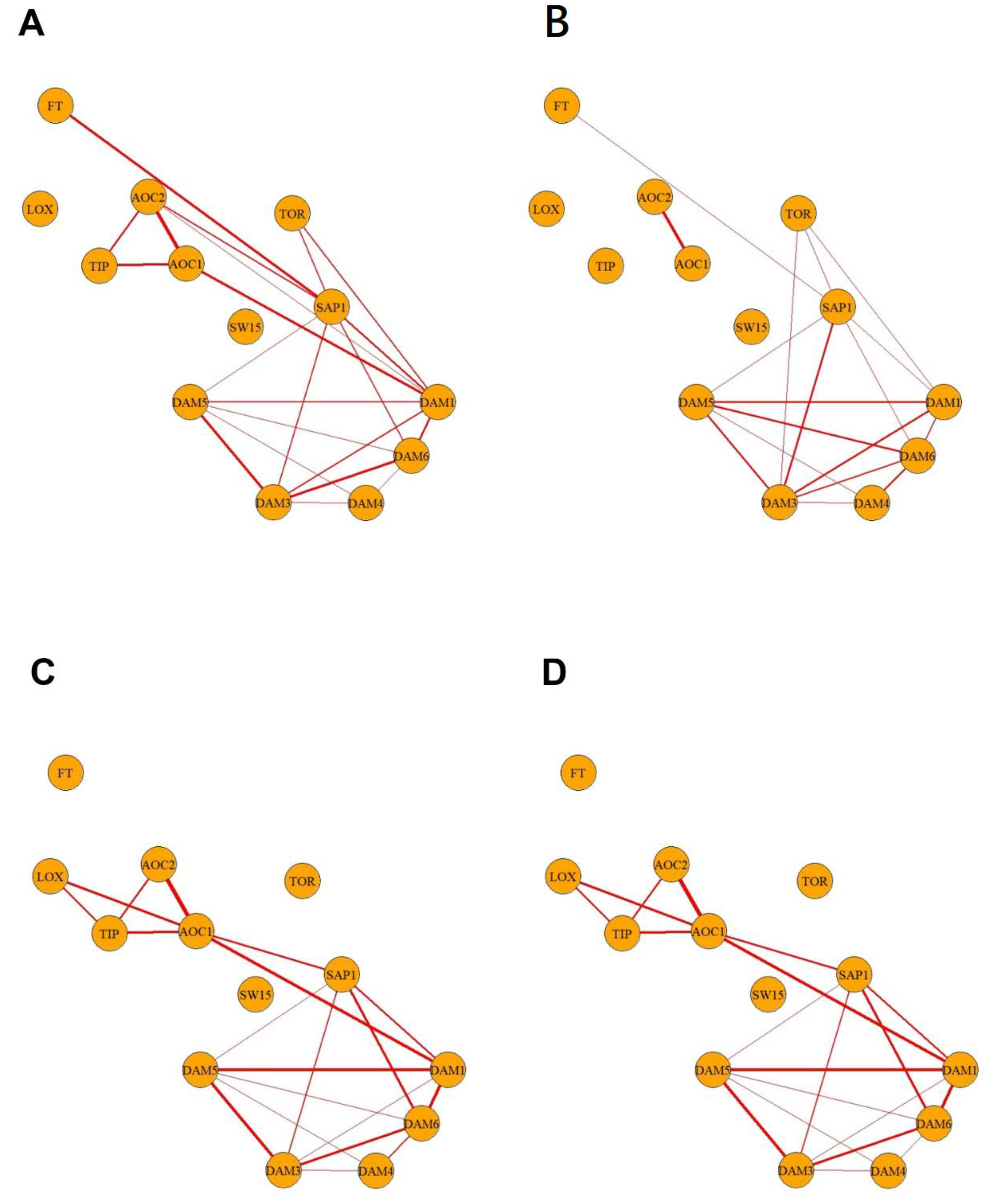
2.3. Single-Bud Variability in Dormancy-Transition Sample S3
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Dormancy Assessment
4.3. Single Bud RNA Isolation and qRT-PCR
4.4. Statistical and Bioinformatic Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Arora, R.; Rowland, L.J.; Tanino, K. Induction and Release of Bud Dormancy in Woody Perennials: A Science Comes of Age. HortScience 2003, 38, 911–921. [Google Scholar] [CrossRef]
- Coville, F.V. The Influence of Cold in Stimulating the Growth of Plants. Proc. Natl. Acad. Sci. USA 1920, 6, 434–435. [Google Scholar] [CrossRef]
- Weinberger, J.H. Chilling Requirements of Peach Varieties. Proc. Am. Soc. Hortic. Sci. 1950, 56, 122–128. [Google Scholar]
- Richardson, E.A. A Model for Estimating the Completion of Rest for “Redheaven” and “Elberta” Peach Trees. HortScience 1974, 9, 331–332. [Google Scholar] [CrossRef]
- Fishman, S.; Erez, A.; Couvillon, G.A. The Temperature Dependence of Dormancy Breaking in Plants: Computer Simulation of Processes Studied under Controlled Temperatures. J. Theor. Biol. 1987, 126, 309–321. [Google Scholar] [CrossRef]
- Anderson, J.L.; Richardson, E.A.; Kesner, C.D. Validation of chill unit and flower bud phenology models for “Montmorency” sour cherry. Acta Hortic. 1986, 184, 71–78. [Google Scholar] [CrossRef]
- Lloret, A.; Quesada-Traver, C.; Ríos, G. Models for a Molecular Calendar of Bud-Break in Fruit Trees. Sci. Hortic. 2022, 297, 110972. [Google Scholar] [CrossRef]
- Campoy, J.A.; Ruiz, D.; Egea, J. Dormancy in Temperate Fruit Trees in a Global Warming Context: A Review. Sci. Hortic. 2011, 130, 357–372. [Google Scholar] [CrossRef]
- Guo, L.; Dai, J.; Ranjitkar, S.; Yu, H.; Xu, J.; Luedeling, E. Chilling and Heat Requirements for Flowering in Temperate Fruit Trees. Int. J. Biometeorol. 2014, 58, 1195–1206. [Google Scholar] [CrossRef]
- Jiménez, S.; Lawton-Rauh, A.L.; Reighard, G.L.; Abbott, A.G.; Bielenberg, D.G. Phylogenetic Analysis and Molecular Evolution of the Dormancy Associated MADS-Box Genes from Peach. BMC Plant Biol. 2009, 9, 81. [Google Scholar] [CrossRef]
- Quesada-Traver, C.; Lloret, A.; Carretero-Paulet, L.; Badenes, M.L.; Ríos, G. Evolutionary Origin and Functional Specialization of Dormancy-Associated MADS Box (DAM) Proteins in Perennial Crops. BMC Plant Biol. 2022, 22, 473. [Google Scholar] [CrossRef]
- Bielenberg, D.G.; Wang, Y.; Li, Z.; Zhebentyayeva, T.; Fan, S.; Reighard, G.L.; Scorza, R.; Abbott, A.G. Sequencing and Annotation of the Evergrowing Locus in Peach [Prunus persica (L.) Batsch] Reveals a Cluster of Six MADS-Box Transcription Factors as Candidate Genes for Regulation of Terminal Bud Formation. Tree Genet. Genomes 2008, 4, 495–507. [Google Scholar] [CrossRef]
- Moser, M.; Asquini, E.; Miolli, G.V.; Weigl, K.; Hanke, M.-V.; Flachowsky, H.; Si-Ammour, A. The MADS-Box Gene MdDAM1 Controls Growth Cessation and Bud Dormancy in Apple. Front. Plant Sci. 2020, 11, 1003. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, R.; Yamane, H.; Ooka, T.; Jotatsu, H.; Kitamura, Y.; Akagi, T.; Tao, R. Functional and Expressional Analyses of PmDAM Genes Associated with Endodormancy in Japanese Apricot. Plant Physiol. 2011, 157, 485–497. [Google Scholar] [CrossRef] [PubMed]
- Yamane, H.; Wada, M.; Honda, C.; Matsuura, T.; Ikeda, Y.; Hirayama, T.; Osako, Y.; Gao-Takai, M.; Kojima, M.; Sakakibara, H.; et al. Overexpression of Prunus DAM6 Inhibits Growth, Represses Bud Break Competency of Dormant Buds and Delays Bud Outgrowth in Apple Plants. PLoS ONE 2019, 14, e0214788. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Tomes, S.; Karunairetnam, S.; Tustin, S.D.; Hellens, R.P.; Allan, A.C.; Macknight, R.C.; Varkonyi-Gasic, E. SVP-like MADS Box Genes Control Dormancy and Budbreak in Apple. Front. Plant Sci. 2017, 8, 477. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.; Bielenberg, D.G.; Zhebentyayeva, T.N.; Reighard, G.L.; Okie, W.R.; Holland, D.; Abbott, A.G. Mapping Quantitative Trait Loci Associated with Chilling Requirement, Heat Requirement and Bloom Date in Peach (Prunus persica). New Phytol. 2010, 185, 917–930. [Google Scholar] [CrossRef]
- Celton, J.-M.; Martinez, S.; Jammes, M.-J.; Bechti, A.; Salvi, S.; Legave, J.-M.; Costes, E. Deciphering the Genetic Determinism of Bud Phenology in Apple Progenies: A New Insight into Chilling and Heat Requirement Effects on Flowering Dates and Positional Candidate Genes. New Phytol. 2011, 192, 378–392. [Google Scholar] [CrossRef]
- Sánchez-Pérez, R.; Dicenta, F.; Martínez-Gómez, P. Inheritance of Chilling and Heat Requirements for Flowering in Almond and QTL Analysis. Tree Genet. Genomes 2012, 8, 379–389. [Google Scholar] [CrossRef]
- Castède, S.; Campoy, J.A.; Dantec, L.L.; Quero-García, J.; Barreneche, T.; Wenden, B.; Dirlewanger, E. Mapping of Candidate Genes Involved in Bud Dormancy and Flowering Time in Sweet Cherry (Prunus avium). PLoS ONE 2015, 10, e0143250. [Google Scholar] [CrossRef]
- Gabay, G.; Dahan, Y.; Izhaki, Y.; Isaacson, T.; Elkind, Y.; Ben-Ari, G.; Flaishman, M.A. Identification of QTLs Associated with Spring Vegetative Budbreak Time after Dormancy Release in Pear (Pyrus communis L.). Plant Breed. 2017, 136, 749–758. [Google Scholar] [CrossRef]
- Kitamura, Y.; Habu, T.; Yamane, H.; Nishiyama, S.; Kajita, K.; Sobue, T.; Kawai, T.; Numaguchi, K.; Nakazaki, T.; Kitajima, A.; et al. Identification of QTLs Controlling Chilling and Heat Requirements for Dormancy Release and Bud Break in Japanese Apricot (Prunus mume). Tree Genet. Genomes 2018, 14, 33. [Google Scholar] [CrossRef]
- Li, Z.; Reighard, G.L.; Abbott, A.G.; Bielenberg, D.G. Dormancy-Associated MADS Genes from the EVG Locus of Peach [Prunus persica (L.) Batsch] Have Distinct Seasonal and Photoperiodic Expression Patterns. J. Exp. Bot. 2009, 60, 3521–3530. [Google Scholar] [CrossRef] [PubMed]
- Leida, C.; Terol, J.; Martí, G.; Agustí, M.; Llácer, G.; Badenes, M.L.; Ríos, G. Identification of Genes Associated with Bud Dormancy Release in Prunus Persica by Suppression Subtractive Hybridization. Tree Physiol. 2010, 30, 655–666. [Google Scholar] [CrossRef]
- Ubi, B.E.; Sakamoto, D.; Ban, Y.; Shimada, T.; Ito, A.; Nakajima, I.; Takemura, Y.; Tamura, F.; Saito, T.; Moriguchi, T. Molecular Cloning of Dormancy-Associated MADS-Box Gene Homologs and Their Characterization during Seasonal Endodormancy Transitional Phases of Japanese Pear. J. Am. Soc. Hortic. Sci. 2010, 135, 174–182. [Google Scholar] [CrossRef]
- Falavigna, V.d.S.; Porto, D.D.; Buffon, V.; Margis-Pinheiro, M.; Pasquali, G.; Revers, L.F. Differential Transcriptional Profiles of Dormancy-Related Genes in Apple Buds. Plant Mol. Biol. Rep. 2014, 32, 796–813. [Google Scholar] [CrossRef]
- Falavigna, V.d.S.; Guitton, B.; Costes, E.; Andrés, F. I Want to (Bud) Break Free: The Potential Role of DAM and SVP-Like Genes in Regulating Dormancy Cycle in Temperate Fruit Trees. Front. Plant Sci. 2019, 9, 1990. [Google Scholar] [CrossRef]
- Hsiang, T.-F.; Chen, W.; Yamane, H. The MADS-Box Gene Family Involved in the Regulatory Mechanism of Dormancy and Flowering in Rosaceae Fruit Trees. In Annual Plant Reviews Online; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2021; pp. 649–686. [Google Scholar]
- Hao, X.; Chao, W.; Yang, Y.; Horvath, D. Coordinated Expression of FLOWERING LOCUS T and DORMANCY ASSOCIATED MADS-BOX-Like Genes in Leafy Spurge. PLoS ONE 2015, 10, e0126030. [Google Scholar] [CrossRef]
- Niu, Q.; Li, J.; Cai, D.; Qian, M.; Jia, H.; Bai, S.; Hussain, S.; Liu, G.; Teng, Y.; Zheng, X. Dormancy-Associated MADS-Box Genes and MicroRNAs Jointly Control Dormancy Transition in Pear (Pyrus pyrifolia White Pear Group) Flower Bud. J. Exp. Bot. 2016, 67, 239–257. [Google Scholar] [CrossRef]
- Valverde, F.; Mouradov, A.; Soppe, W.; Ravenscroft, D.; Samach, A.; Coupland, G. Photoreceptor Regulation of CONSTANS Protein in Photoperiodic Flowering. Science 2004, 303, 1003–1006. [Google Scholar] [CrossRef]
- Hsu, C.-Y.; Adams, J.P.; Kim, H.; No, K.; Ma, C.; Strauss, S.H.; Drnevich, J.; Vandervelde, L.; Ellis, J.D.; Rice, B.M.; et al. FLOWERING LOCUS T Duplication Coordinates Reproductive and Vegetative Growth in Perennial Poplar. Proc. Natl. Acad. Sci. USA 2011, 108, 10756–10761. [Google Scholar] [CrossRef] [PubMed]
- Tuan, P.A.; Bai, S.; Saito, T.; Ito, A.; Moriguchi, T. Dormancy-Associated MADS-Box (DAM) and the Abscisic Acid Pathway Regulate Pear Endodormancy Through a Feedback Mechanism. Plant Cell Physiol. 2017, 58, 1378–1390. [Google Scholar] [CrossRef] [PubMed]
- Lloret, A.; Quesada-Traver, C.; Conejero, A.; Arbona, V.; Gómez-Mena, C.; Petri, C.; Sánchez-Navarro, J.A.; Zuriaga, E.; Leida, C.; Badenes, M.L.; et al. Regulatory Circuits Involving Bud Dormancy Factor PpeDAM6. Hortic. Res. 2021, 8, 261. [Google Scholar] [CrossRef]
- Lloret, A.; Badenes, M.L.; Ríos, G. Modulation of Dormancy and Growth Responses in Reproductive Buds of Temperate Trees. Front. Plant Sci. 2018, 9, 1368. [Google Scholar] [CrossRef] [PubMed]
- Fennell, A. Genomics and Functional Genomics of Winter Low Temperature Tolerance in Temperate Fruit Crops. Crit. Rev. Plant Sci. 2014, 33, 125–140. [Google Scholar] [CrossRef]
- Lloret, A.; Conejero, A.; Leida, C.; Petri, C.; Gil-Muñoz, F.; Burgos, L.; Badenes, M.L.; Ríos, G. Dual Regulation of Water Retention and Cell Growth by a Stress-Associated Protein (SAP) Gene in Prunus. Sci. Rep. 2017, 7, 332. [Google Scholar] [CrossRef]
- Hsiang, T.-F.; Lin, Y.-J.; Yamane, H.; Tao, R. Characterization of Japanese Apricot (Prunus mume) Floral Bud Development Using a Modified BBCH Scale and Analysis of the Relationship between BBCH Stages and Floral Primordium Development and the Dormancy Phase Transition. Horticulturae 2021, 7, 142. [Google Scholar] [CrossRef]
- Herrera, S.; Lora, J.; Fadón, E.; Hedhly, A.; Alonso, J.M.; Hormaza, J.I.; Rodrigo, J. Male Meiosis as a Biomarker for Endo- to Ecodormancy Transition in Apricot. Front. Plant Sci. 2022, 13, 842333. [Google Scholar] [CrossRef]
- Liu, B.; Mo, W.-J.; Zhang, D.; De Storme, N.; Geelen, D. Cold Influences Male Reproductive Development in Plants: A Hazard to Fertility, but a Window for Evolution. Plant Cell Physiol. 2019, 60, 7–18. [Google Scholar] [CrossRef]
- Ríos, G.; Tadeo, F.R.; Leida, C.; Badenes, M.L. Prediction of Components of the Sporopollenin Synthesis Pathway in Peach by Genomic and Expression Analyses. BMC Genom. 2013, 14, 40. [Google Scholar] [CrossRef]
- Leida, C.; Conesa, A.; Llácer, G.; Badenes, M.L.; Ríos, G. Histone Modifications and Expression of DAM6 Gene in Peach Are Modulated during Bud Dormancy Release in a Cultivar-Dependent Manner. New Phytol. 2012, 193, 67–80. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Islam, M.T.; Sapkota, S.; Ravindran, P.; Kumar, P.P.; Artlip, T.S.; Sherif, S.M. Ethylene-Mediated Modulation of Bud Phenology, Cold Hardiness, and Hormone Biosynthesis in Peach (Prunus persica). Plants 2021, 10, 1266. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; An, L.; Nguyen, T.H.; Liang, H.; Wang, R.; Liu, X.; Li, T.; Qi, Y.; Yu, F. The Cloning and Functional Characterization of Peach CONSTANS and FLOWERING LOCUS T Homologous Genes PpCO and PpFT. PLoS ONE 2015, 10, e0124108. [Google Scholar] [CrossRef]
- Leida, C.; Romeu, J.F.; García-Brunton, J.; Ríos, G.; Badenes, M.L. Gene Expression Analysis of Chilling Requirements for Flower Bud Break in Peach. Plant Breed. 2012, 131, 329–334. [Google Scholar] [CrossRef]
- de la Fuente, L.; Conesa, A.; Lloret, A.; Badenes, M.L.; Ríos, G. Genome-Wide Changes in Histone H3 Lysine 27 Trimethylation Associated with Bud Dormancy Release in Peach. Tree Genet. Genomes 2015, 11, 45. [Google Scholar] [CrossRef]
- Horvath, D.P.; Sung, S.; Kim, D.; Chao, W.; Anderson, J. Characterization, Expression and Function of DORMANCY ASSOCIATED MADS-BOX Genes from Leafy Spurge. Plant Mol. Biol. 2010, 73, 169–179. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Chen, P.-Y.; Zhong, S.; Dardick, C.; Callahan, A.; An, Y.-Q.; van Knocker, S.; Yang, Y.; Zhong, G.-Y.; Abbott, A.; et al. Thermal-Responsive Genetic and Epigenetic Regulation of DAM Cluster Controlling Dormancy and Chilling Requirement in Peach Floral Buds. Hortic. Res. 2020, 7, 114. [Google Scholar] [CrossRef]
- Saito, T.; Bai, S.; Imai, T.; Ito, A.; Nakajima, I.; Moriguchi, T. Histone Modification and Signalling Cascade of the Dormancy-Associated MADS-Box Gene, PpMADS13-1, in Japanese Pear (Pyrus pyrifolia) during Endodormancy. Plant Cell Environ. 2015, 38, 1157–1166. [Google Scholar] [CrossRef]
- Vimont, N.; Quah, F.X.; Schöepfer, D.G.; Roudier, F.; Dirlewanger, E.; Wigge, P.A.; Wenden, B.; Cortijo, S. ChIP-Seq and RNA-Seq for Complex and Low-Abundance Tree Buds Reveal Chromatin and Expression Co-Dynamics during Sweet Cherry Bud Dormancy. Tree Genet. Genomes 2019, 16, 9. [Google Scholar] [CrossRef]
- Chen, W.; Tamada, Y.; Yamane, H.; Matsushita, M.; Osako, Y.; Gao-Takai, M.; Luo, Z.; Tao, R. H3K4me3 Plays a Key Role in Establishing Permissive Chromatin States during Bud Dormancy and Bud Break in Apple. Plant J. 2022, 111, 1015–1031. [Google Scholar] [CrossRef]
- Ríos, G.; Leida, C.; Conejero, A.; Badenes, M.L. Epigenetic Regulation of Bud Dormancy Events in Perennial Plants. Front. Plant Sci. 2014, 5, 247. [Google Scholar]
- Conde, D.; Perales, M.; Sreedasyam, A.; Tuskan, G.A.; Lloret, A.; Badenes, M.L.; González-Melendi, P.; Ríos, G.; Allona, I. Engineering Tree Seasonal Cycles of Growth Through Chromatin Modification. Front. Plant Sci. 2019, 10, 412. [Google Scholar] [CrossRef]
- Acosta, I.F.; Przybyl, M. Jasmonate Signaling during Arabidopsis Stamen Maturation. Plant Cell Physiol. 2019, 60, 2648–2659. [Google Scholar] [CrossRef]
- Julian, C.; Herrero, M.; Rodrigo, J. Anther Meiosis Time Is Related to Winter Cold Temperatures in Apricot (Prunus armeniaca L.). Environ. Exp. Bot. 2014, 100, 20–25. [Google Scholar] [CrossRef]
- Huang, B.; Fan, Y.; Cui, L.; Li, C.; Guo, C. Cold Stress Response Mechanisms in Anther Development. Int. J. Mol. Sci. 2023, 24, 30. [Google Scholar] [CrossRef] [PubMed]
- Vergara, R.; Noriega, X.; Pérez, F.J. VvDAM-SVPs Genes Are Regulated by FLOWERING LOCUS T (VvFT) and Not by ABA/Low Temperature-Induced VvCBFs Transcription Factors in Grapevine Buds. Planta 2021, 253, 31. [Google Scholar] [CrossRef]
- Baggiolini, M. Les stades repères du pêcher. Rev. Rom. D’agriculture D’arboriculture 1952, 8, 29. [Google Scholar]
- Wang, T.; Hao, R.; Pan, H.; Cheng, T.; Zhang, Q. Selection of Suitable Reference Genes for Quantitative Real-time Polymerase Chain Reaction in Prunus mume during Flowering Stages and under Different Abiotic Stress Conditions. J. Am. Soc. Hort. Sci. 2014, 139, 113–122. [Google Scholar] [CrossRef]
- Quesada-Traver, C.; Guerrero, B.I.; Badenes, M.L.; Rodrigo, J.; Ríos, G.; Lloret, A. Structure and Expression of Bud Dormancy-Associated MADS-Box Genes (DAM) in European Plum. Front. Plant Sci. 2020, 11, 1288. [Google Scholar] [CrossRef]

| Name | Transcript | Protein ID | Description | Process | Peach Reference |
|---|---|---|---|---|---|
| PpeDAM1 | Prupe.1G531100 | ABJ96361 | MADS-box transcription factor | Bud dormancy regulation | [23] |
| PpeDAM3 | Prupe.1G531400 | ABJ96364 | MADS-box transcription factor | Bud dormancy regulation | [23] |
| PpeDAM4 | Prupe.1G531500 | ABJ96358 | MADS-box transcription factor | Bud dormancy regulation | [23] |
| PpeDAM5 | Prupe.1G531600 | ABJ96359 | MADS-box transcription factor | Bud dormancy regulation | [23] |
| PpeDAM6 | Prupe.1G531700 | ABJ96360 | MADS-box transcription factor | Bud dormancy regulation | [23] |
| PpeSAP1 | Prupe.2G010400 | XP_007218502 | A20/AN1 Zn finger | Abiotic stress | [37] |
| TIP-like | Prupe.2G229500 | XP_007218847 | Aquaporin | Water movement (*) | [37] |
| TOR-like | Prupe.8G151300 | XP_020425391 | Protein kinase | Cell growth (*) | [37] |
| SWEET15-like | Prupe.1G220700 | XP_007222479 | Sugar transporter | Pollen maturation (*) | [41] |
| AOC-like 1 | Prupe.1G306100 | XP_007223720 | Allene oxide cyclase | Jasmonic acid biosynthesis (*) | [34] |
| AOC-like 2 | Prupe.3G239900 | XP_00721504 | Allene oxide cyclase | Jasmonic acid biosynthesis (*) | [34,43] |
| LOX-like | Prupe.2G005300 | XP_007220253 | Lipoxygenase | Jasmonic acid biosynthesis (*) | [34] |
| PpeFT | Prupe.6G364900 | ACH73165 | Phosphatidylethanolamine-binding protein | Flowering initiation | [44] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Puertes, A.; Polat, H.; Ramón-Núñez, L.A.; González, M.; Ancillo, G.; Zuriaga, E.; Ríos, G. Single-Bud Expression Analysis of Bud Dormancy Factors in Peach. Plants 2023, 12, 2601. https://doi.org/10.3390/plants12142601
Puertes A, Polat H, Ramón-Núñez LA, González M, Ancillo G, Zuriaga E, Ríos G. Single-Bud Expression Analysis of Bud Dormancy Factors in Peach. Plants. 2023; 12(14):2601. https://doi.org/10.3390/plants12142601
Chicago/Turabian StylePuertes, Ana, Helin Polat, Luis Andrés Ramón-Núñez, Matilde González, Gema Ancillo, Elena Zuriaga, and Gabino Ríos. 2023. "Single-Bud Expression Analysis of Bud Dormancy Factors in Peach" Plants 12, no. 14: 2601. https://doi.org/10.3390/plants12142601
APA StylePuertes, A., Polat, H., Ramón-Núñez, L. A., González, M., Ancillo, G., Zuriaga, E., & Ríos, G. (2023). Single-Bud Expression Analysis of Bud Dormancy Factors in Peach. Plants, 12(14), 2601. https://doi.org/10.3390/plants12142601







