Identification of Soybean Mutant Lines Based on Dual-Branch CNN Model Fusion Framework Utilizing Images from Different Organs
Abstract
:1. Introduction
2. Materials and Methods
2.1. Soybean Samples
2.2. Methods
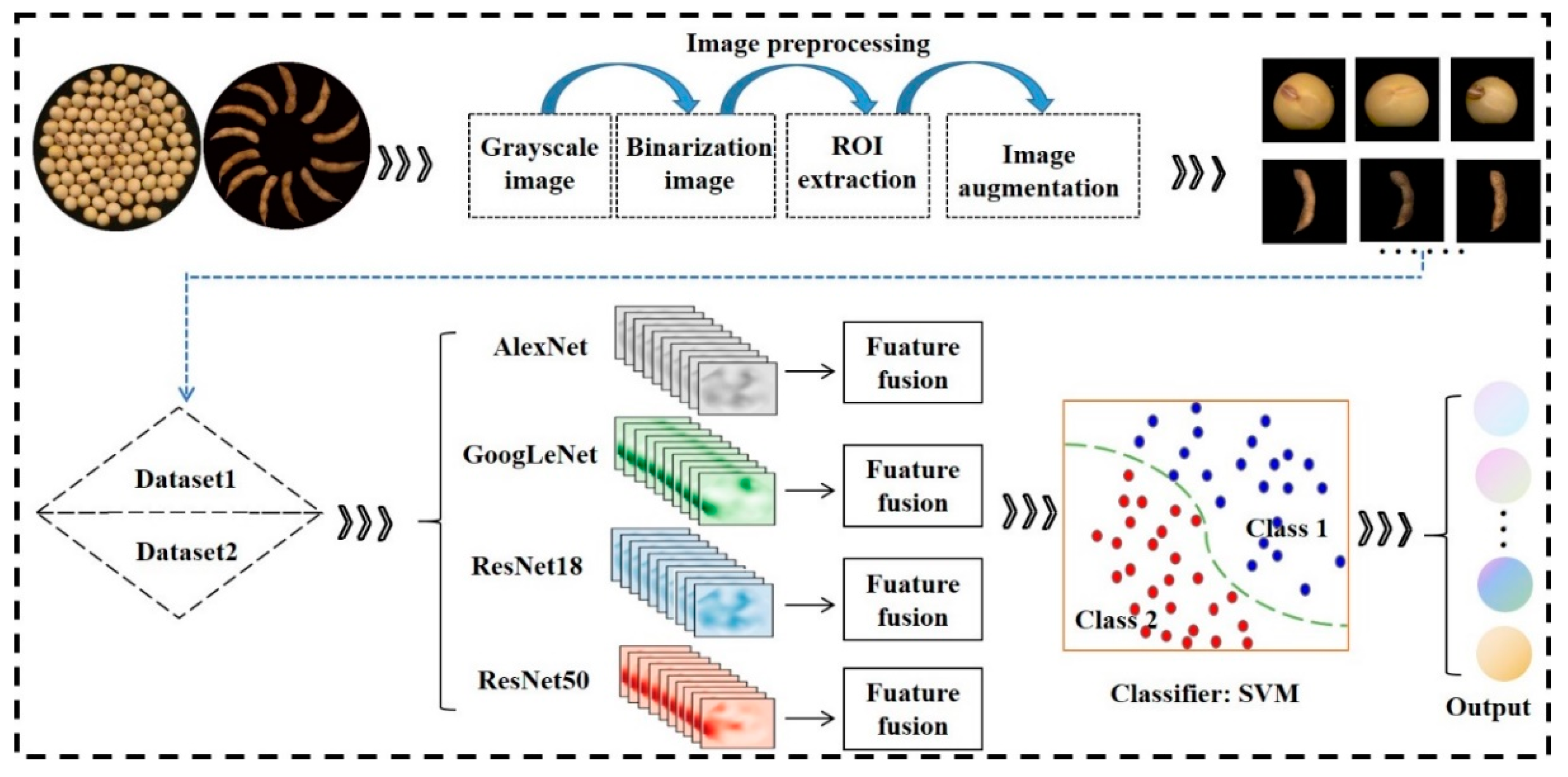
2.2.1. Image Acquisition
2.2.2. Image Segmentation
2.2.3. Image Augmentation
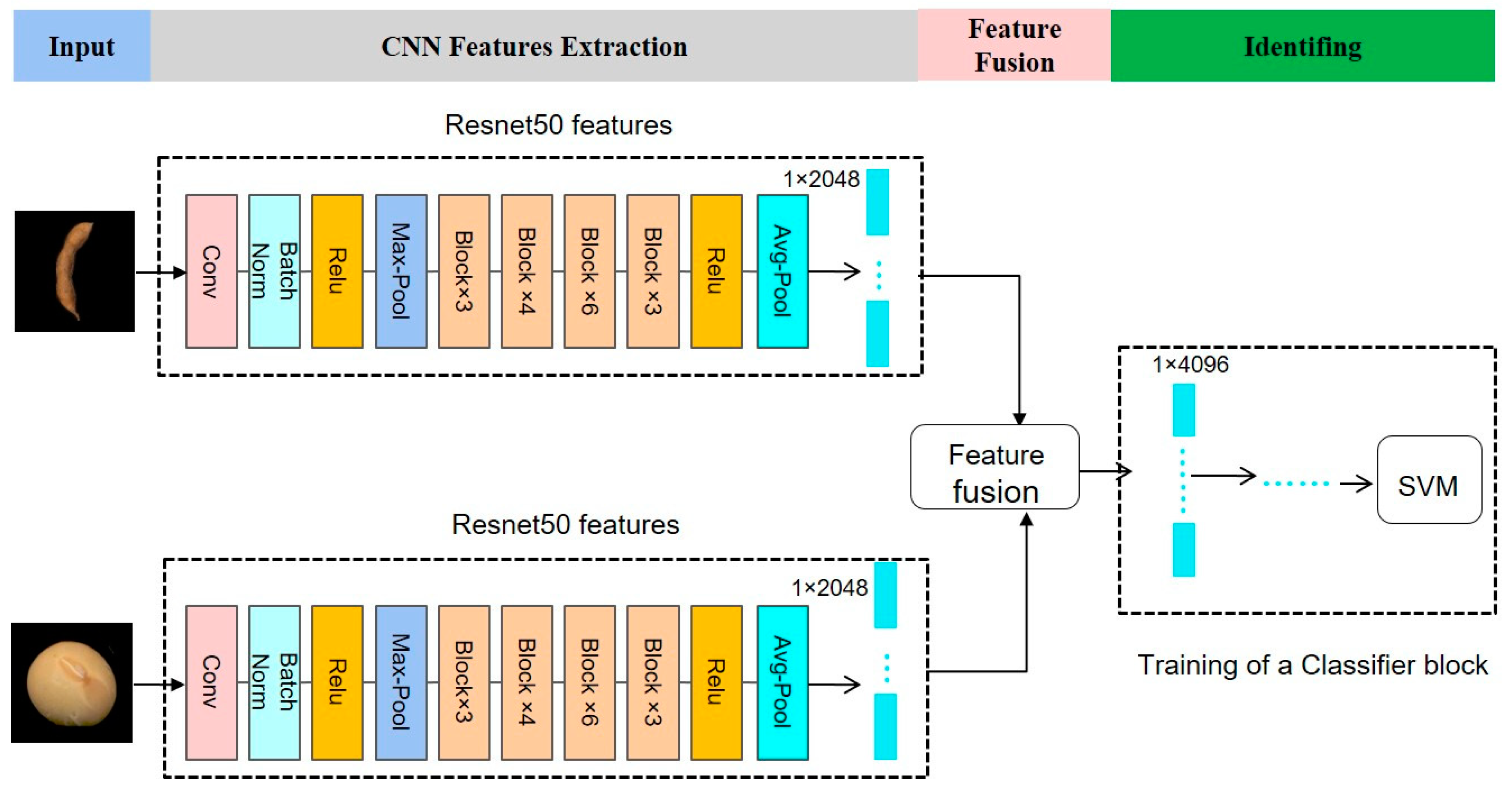
2.2.4. Dual-Convolution Neural Network Model Fusion Frameworks
2.2.5. Workflow Diagram
3. Results and Analysis
3.1. Comparison of Different Single Model Training Results
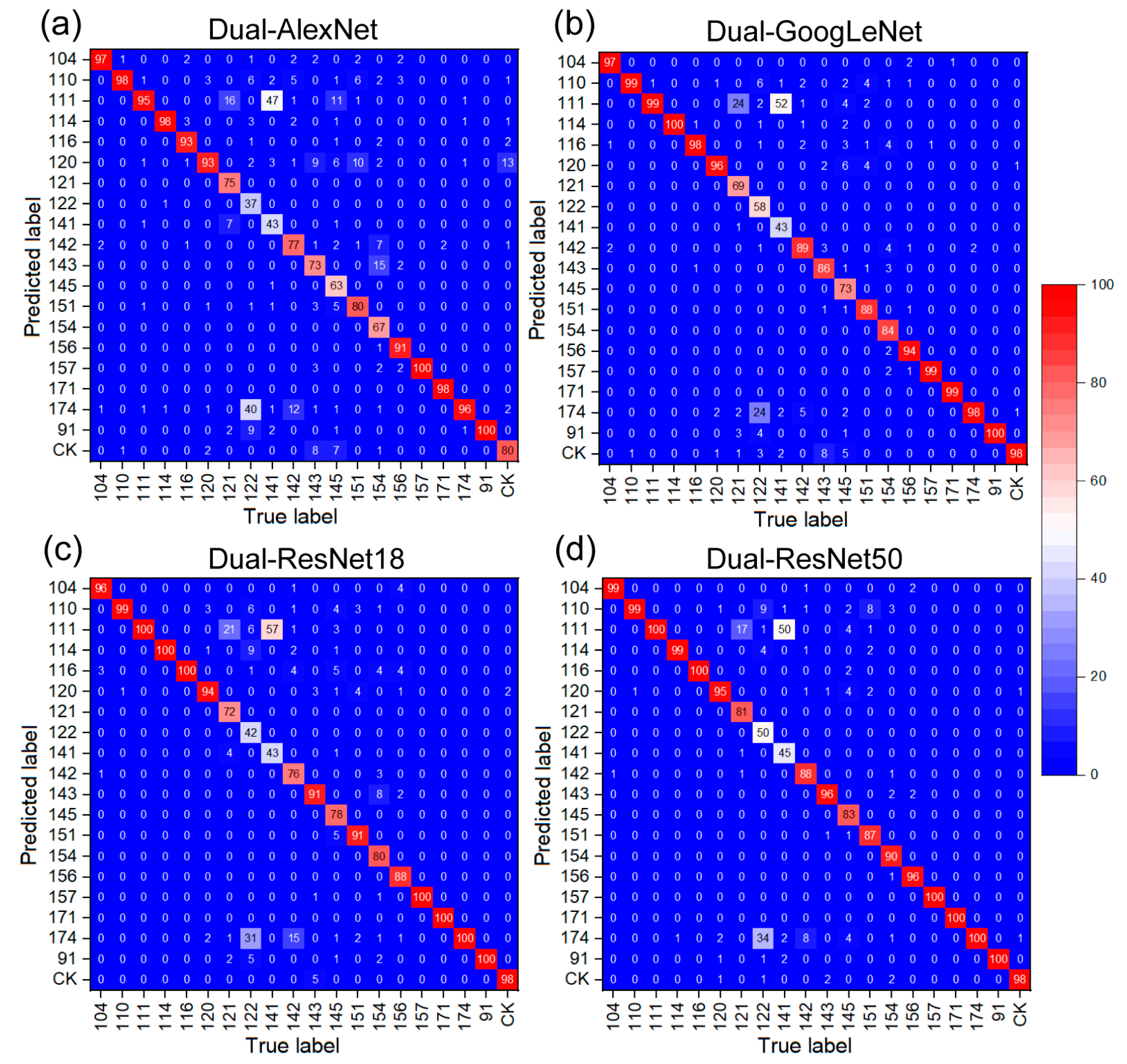
3.2. Dual Network Selection and Evaluation
3.3. Feature Visualization Analysis
3.4. Clustering Results among Soybean Mutant Lines
4. Discussion
4.1. Superiority of Dual-Branch CNN over Single Classical CNN
4.2. Utilization of Clustering Tree
4.3. Significance of Joint Identification of Soybean Mutant Lines
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Wijewardana, C.; Reddy, K.R.; Bellaloui, N. Soybean seed physiology, quality, and chemical composition under soil moisture stress. Food Chem. 2019, 278, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Arslan, H.; KarakuŞ, M.; HatipoĞLu, H.; Arslan, D.; Bayraktar, Ö.V. Assessment of Performances of Yield and Factors Affecting the Yield in Some Soybean Varieties/Lines Grown under Semi-Arid Climate Conditions. Appl. Ecol. Environ. Res. 2018, 16, 4289–4298. [Google Scholar] [CrossRef]
- Liu, S.; Zhang, M.; Feng, F.; Tian, Z. Toward a “Green Revolution” for Soybean. Mol. Plant 2020, 13, 688–697. [Google Scholar] [CrossRef] [PubMed]
- Lammerts van Bueren, E.T.; Myers, J.R. Organic Crop Breeding: Integrating Organic Agricultural Approaches and Traditional and Modern Plant Breeding Methods. In Organic Crop Breeding; John Wiley & Sons, Ltd.: West Sussex, UK, 2012; pp. 1–13. [Google Scholar] [CrossRef]
- Cavassim, J.E.; Bespalhok, J.C.; Alliprandi, L.F.; De Oliveir, R.A.; Daros, E.; Guerra, E.P. AMMI Analysis to Determine Relative Maturity Groups for the Classification of Soybean Genotypes. J. Agron. 2013, 12, 168–178. [Google Scholar] [CrossRef] [Green Version]
- Minervini, M.; Fischbach, A.; Scharr, H.; Tsaftaris, S.A. Finely-grained annotated datasets for image-based plant phenotyping. Pattern Recognit. Lett. 2016, 81, 80–89. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.P.J.; Susmita, C.; Agarwal, D.K.; Pal, G.; Rai, A.K.; Simal-Gandara, J. Assessment of Genetic Purity in Rice Using Polymorphic SSR Markers and Its Economic Analysis with Grow-Out-Test. Food Anal. Methods 2021, 14, 856–864. [Google Scholar] [CrossRef]
- Zheng, Y.C.; Li, S.; Huang, J.Z.; Fan, L.J.; Shu, Q.Y. Identification and Characterization of gamma-Ray-Induced Mutations in Rice Cytoplasmic Genomes by Whole-Genome Sequencing. Cytogenet. Genome Res. 2020, 160, 100–109. [Google Scholar] [CrossRef]
- Wang, Y.H.; Su, W.H. Convolutional Neural Networks in Computer Vision for Grain Crop Phenotyping: A Review. Agronomy 2022, 12, 2659. [Google Scholar] [CrossRef]
- Ni, J.; Gao, J.; Deng, L.; Han, Z. Monitoring the Change Process of Banana Freshness by GoogLeNet. IEEE Access 2020, 8, 228369–228376. [Google Scholar] [CrossRef]
- Xu, W.; Zhu, Z.; Ge, F.; Han, Z.; Li, J. Analysis of Behavior Trajectory Based on Deep Learning in Ammonia Environment for Fish. Sensors 2020, 20, 4425. [Google Scholar] [CrossRef]
- Li, J.; Xu, C.; Jiang, L.; Xiao, Y.; Deng, L.; Han, Z. Detection and Analysis of Behavior Trajectory for Sea Cucumbers Based on Deep Learning. IEEE Access 2020, 8, 18832–18840. [Google Scholar] [CrossRef]
- Gao, J.; Zhao, L.; Li, J.; Deng, L.; Ni, J.; Han, Z. Aflatoxin rapid detection based on hyperspectral with 1D-convolution neural network in the pixel level. Food Chem. 2021, 360, 129968. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.; Gao, J. Pixel-level aflatoxin detecting based on deep learning and hyperspectral imaging. Comput. Electron. Agric. 2019, 164, 104888. [Google Scholar] [CrossRef]
- Uçar, E.; Atila, Ü.; Uçar, M.; Akyol, K. Automated detection of Covid-19 disease using deep fused features from chest radiography images. Biomed. Signal Process. Control 2021, 69, 102862. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Li, J.; Li, J.; Xia, Y.; Ding, Z.; Samali, B. Automated damage diagnosis of concrete jack arch beam using optimized deep stacked autoencoders and multi-sensor fusion. Dev. Built Environ. 2023, 14, 100128. [Google Scholar] [CrossRef]
- Yu, Y.; Liang, S.; Samali, B.; Nguyen, T.N.; Zhai, C.; Li, J.; Xie, X. Torsional capacity evaluation of RC beams using an improved bird swarm algorithm optimised 2D convolutional neural network. Eng. Struct. 2022, 273, R713–R715. [Google Scholar] [CrossRef]
- Zhou, L.; Zhang, C.; Taha, M.F.; Wei, X.; He, Y.; Qiu, Z.; Liu, Y. Wheat Kernel Variety Identification Based on a Large Near-Infrared Spectral Dataset and a Novel Deep Learning-Based Feature Selection Method. Front. Plant Sci. 2020, 11, 575810. [Google Scholar] [CrossRef]
- Zhang, J.; Dai, L.; Cheng, F. Corn seed variety classification based on hyperspectral reflectance imaging and deep convolutional neural network. J. Food Meas. Charact. 2020, 15, 484–494. [Google Scholar] [CrossRef]
- Yang, H.; Ni, J.; Gao, J.; Han, Z.; Luan, T. A novel method for peanut variety identification and classification by Improved VGG16. Sci. Rep. 2021, 11, 15756. [Google Scholar] [CrossRef]
- Zhu, S.; Zhang, J.; Chao, M.; Xu, X.; Song, P.; Zhang, J.; Huang, Z. A Rapid and Highly Efficient Method for the Identification of Soybean Seed Varieties: Hyperspectral Images Combined with Transfer Learning. Molecules 2019, 25, 152. [Google Scholar] [CrossRef] [Green Version]
- Zhu, S.; Zhou, L.; Zhang, C.; Bao, Y.; Wu, B.; Chu, H.; Yu, Y.; He, Y.; Feng, L. Identification of Soybean Varieties Using Hyperspectral Imaging Coupled with Convolutional Neural Network. Sensors 2019, 19, 4065. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Zhang, L.; Sun, H.; Rao, Z.; Ji, H. Identification of soybean varieties based on hyperspectral imaging technology and one-dimensional convolutional neural network. J. Food Process Eng. 2021, 44, 5225. [Google Scholar] [CrossRef]
- Huang, Z.; Wang, R.; Cao, Y.; Zheng, S.; Teng, Y.; Wang, F.; Wang, L.; Du, J. Deep learning based soybean seed classification. Comput. Electron. Agric. 2022, 202, 107393. [Google Scholar] [CrossRef]
- Song, X.F.; Wei, H.C.; Cheng, W.; Yang, S.X.; Zhao, Y.X.; Li, X.; Luo, D.; Zhang, H.; Feng, X.Z. Development of INDEL Markers for Genetic Mapping Based on Whole Genome Resequencing in Soybean. G3-Genes Genomes Genet. 2015, 5, 2793–2799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nieto-Hidalgo, M.; Gallego, A.-J.; Gil, P.; Pertusa, A. Two-Stage Convolutional Neural Network for Ship and Spill Detection Using SLAR Images. IEEE Trans. Geosci. Remote Sens. 2018, 56, 5217–5230. [Google Scholar] [CrossRef] [Green Version]
- Mikołajczyk, A.; Grochowski, M. Data augmentation for improving deep learning in image classification problem. In Proceedings of the 2018 International Interdisciplinary PhD Workshop (IIPhDW), Swinoujscie, Poland, 9–12 May 2018; pp. 117–122. [Google Scholar]
- Yoo, J.; Kang, S. Class-Adaptive Data Augmentation for Image Classification. IEEE Access 2023, 11, 26393–26402. [Google Scholar] [CrossRef]
- Momeny, M.; Jahanbakhshi, A.; Jafarnezhad, K.; Zhang, Y.D. Accurate classification of cherry fruit using deep CNN based on hybrid pooling approach. Postharvest Biol. Technol. 2020, 166, 111204. [Google Scholar] [CrossRef]
- Liu, S.P.; Tian, G.H.; Xu, Y. A novel scene classification model combining ResNet based transfer learning and data augmentation with a filter. Neurocomputing 2019, 338, 191–206. [Google Scholar] [CrossRef]
- Suh, S.; Lukowicz, P.; Lee, Y.O. Discriminative feature generation for classification of imbalanced data. Pattern Recognit. 2022, 122, 108302. [Google Scholar] [CrossRef]
- Jing, X.Y.; Zhang, X.; Zhu, X.; Wu, F.; You, X.; Gao, Y.; Shan, S.; Yang, J.Y. Multiset Feature Learning for Highly Imbalanced Data Classification. IEEE Trans. Pattern Anal. Mach. Intell. 2021, 43, 139–156. [Google Scholar] [CrossRef]
- Zhang, T.; Fan, S.; Hu, J.; Guo, X.; Li, Q.; Zhang, Y.; Wulamu, A. A Feature Fusion Method with Guided Training for Classification Tasks. Comput. Intell. Neurosci. 2021, 2021, 6647220. [Google Scholar] [CrossRef] [PubMed]
- Afifi, S.; GholamHosseini, H.; Sinha, R. FPGA Implementations of SVM Classifiers: A Review. SN Comput. Sci. 2020, 1, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet classification with deep convolutional neural networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef] [Green Version]
- Szegedy, C.; Wei, L.; Yangqing, J.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual Explanations from Deep Networks via Gradient-Based Localization. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar]
- Yan, Q.; Gong, D.; Zhang, Y. Two-Stream Convolutional Networks for Blind Image Quality Assessment. IEEE Trans. Image Process. 2019, 28, 2200–2211. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; He, W.; Zhang, Y.; Yao, S.; Cui, Z. Effect of dual-convolutional neural network model fusion for Aluminum profile surface defects classification and recognition. Math. Biosci. Eng. 2022, 19, 997–1025. [Google Scholar] [CrossRef] [PubMed]
- Yuan, F.; Yang, Y.; Yuan, T. A dissimilarity measure for mixed nominal and ordinal attribute data in k-Modes algorithm. Appl. Intell. 2020, 50, 1498–1509. [Google Scholar] [CrossRef]
- Banerjee, J.; Singh, Y.; Shrivastava, M. Mutagenesis in soybean: A review. Pharma Innov. J. 2021, 10, 322–327. [Google Scholar]
- Deng, L.; Han, Z. Image features and DUS testing traits for peanut pod variety identification and pedigree analysis. J. Sci. Food Agric. 2019, 99, 2572–2578. [Google Scholar] [CrossRef]





| Category | Irradiation Intensity (Gy) | Pedigree Source | Category | Irradiation Intensity (Gy) | Pedigree Source |
|---|---|---|---|---|---|
| CK | \ | Hedou12 | 141 | 250 | 14-9-2 |
| 91 | 150 | 1-1-2-1 | 142 | 250 | 3-1-6 |
| 104 | 150 | 5-1-1-7 | 143 | 250 | 14-2-2 |
| 110 | 150 | 8-7-2-2 | 145 | 150 | 3-1 |
| 111 | 150 | 10-3-1 | 151 | 250 | 14-1-11 |
| 114 | 250 | 3-1-2 | 154 | 250 | 14-1-14 |
| 116 | 250 | 3-6-2 | 156 | 250 | 14-3-1 |
| 120 | 250 | 11-2-1-2 | 157 | 250 | 14-8-1 |
| 121 | 250 | 14-2-1 | 171 | 250 | 14-11 |
| 122 | 250 | 14-2-13-2-1 | 174 | 350 | 15-3-9 |
| Category | Pod | Seed | Category | Pod | Seed |
|---|---|---|---|---|---|
| CK | 155 | 377 | 141 | 220 | 752 |
| 91 | 148 | 820 | 142 | 199 | 674 |
| 104 | 355 | 375 | 143 | 193 | 694 |
| 110 | 138 | 620 | 145 | 225 | 427 |
| 111 | 156 | 518 | 151 | 170 | 652 |
| 114 | 241 | 577 | 154 | 225 | 444 |
| 116 | 264 | 411 | 156 | 209 | 663 |
| 120 | 145 | 565 | 157 | 263 | 479 |
| 121 | 222 | 457 | 171 | 269 | 488 |
| 122 | 228 | 436 | 174 | 154 | 818 |
| Model | Feature Extraction Layer | Extracted Feature Vector (Pod) | Extracted Feature Vector (Seed) | Fused Feature Vector |
|---|---|---|---|---|
| Dual-AlexNet | fc8 | 1 × 4096 | 1 × 4096 | 1 × 8192 |
| relu7 | 1 × 4096 | 1 × 4096 | 1 × 8192 | |
| prob | 1 × 20 | 1 × 20 | 1 × 40 | |
| Dual-GoogLeNet | inception_5b-output | 1 × 50,176 | 1 × 50,176 | 1 × 100,352 |
| pool5-7x7_s1 | 1 × 1024 | 1 × 1024 | 1 × 2048 | |
| prob | 1 × 20 | 1 × 20 | 1 × 40 | |
| Dual-ResNet18 | res5b_relu | 1 × 25,088 | 1 × 25,088 | 1 × 50,176 |
| pool5 | 1 × 512 | 1 × 512 | 1 × 1024 | |
| prob | 1 × 20 | 1 × 20 | 1 × 40 | |
| Dual-ResNet50 | activation_48_relu | 1 × 50,176 | 1 × 50,176 | 1 × 100,352 |
| avg_pool | 1 × 2048 | 1 × 2048 | 1 × 4096 | |
| fc1000_softmax | 1 × 20 | 1 × 20 | 1 × 40 |
| Model | Depth Layer | Size/MB | Batch Size | Learning Rate | Validation Frequency | Input Size |
|---|---|---|---|---|---|---|
| AlexNet | 25 | 227 | 32 | 0.0003 | 64 | 227 × 227 × 3 |
| GoogLeNet | 144 | 27 | 32 | 0.0003 | 64 | 224 × 224 × 3 |
| ResNet18 | 71 | 44 | 32 | 0.0003 | 64 | 224 × 224 × 3 |
| ResNet50 | 177 | 96 | 32 | 0.0003 | 64 | 224 × 224 × 3 |
| Accuracy (%) | Replicates | Dual-AlexNet | Dual-GoogLeNet | Dual-ResNet18 | Dual-ResNet50 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| fc8 | relu7 | prob | inception_5b-output | pool5-7x7_s1 | prob | res5b_relu | pool5 | prob | activation_48_relu | avg_pool | fc1000_softmax | ||
| Validation | 1 | 94.10 | 94.20 | 88.50 | 95.95 | 96.75 | 91.75 | 95.80 | 95.60 | 90.55 | 97.50 | 97.90 | 93.75 |
| 2 | 94.10 | 94.85 | 87.25 | 94.85 | 95.50 | 91.30 | 96.15 | 96.70 | 90.85 | 96.20 | 97.10 | 93.65 | |
| 3 | 94.85 | 94.80 | 88.85 | 95.80 | 96.50 | 91.85 | 94.25 | 95.75 | 88.80 | 96.70 | 97.80 | 93.15 | |
| Average | 94.35 ± 0.35 | 94.62 ± 0.30 | 88.20 ± 0.69 | 95.53 ± 0.49 | 96.25 ± 0.54 | 91.63 ± 0.24 | 95.40 ± 0.83 | 96.02 ± 0.49 | 90.07 ± 0.90 | 96.80 ± 0.54 | 97.60 ± 0.36 | 93.52 ± 0.26 | |
| Test | 1 | 82.70 | 81.75 | 66.10 | 86.80 | 88.35 | 78.10 | 85.70 | 87.40 | 74.05 | 89.90 | 90.30 | 81.05 |
| 2 | 83.00 | 83.25 | 67.10 | 85.25 | 87.00 | 75.60 | 87.45 | 88.40 | 78.95 | 87.90 | 89.95 | 80.50 | |
| 3 | 83.55 | 83.25 | 68.40 | 87.10 | 88.70 | 77.45 | 84.75 | 87.45 | 75.05 | 88.70 | 90.40 | 81.45 | |
| Average | 83.08 ± 0.35 | 82.75 ± 0.71 | 67.20 ± 0.94 | 86.38 ± 0.81 | 88.02 ± 0.73 | 77.05 ± 1.06 | 85.97 ± 1.12 | 87.75 ± 0.46 | 76.02 ± 2.11 | 88.83 ± 0.82 | 90.22 ± 0.19 | 81.00 ± 0.39 | |
| Method | Precision (%) | Recall (%) | F1-Score (%) |
|---|---|---|---|
| Dual-GoogLeNet | 88.35 | 91.18 | 88.12 |
| Dual-AlexNet | 82.7 | 85.98 | 82.18 |
| Dual-ResNet18 | 87.4 | 90.55 | 87.01 |
| Dual-ResNet50 (proposed) | 90.3 | 92.99 | 90.08 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, G.; Fei, L.; Deng, L.; Yang, H.; Han, M.; Han, Z.; Zhao, L. Identification of Soybean Mutant Lines Based on Dual-Branch CNN Model Fusion Framework Utilizing Images from Different Organs. Plants 2023, 12, 2315. https://doi.org/10.3390/plants12122315
Wu G, Fei L, Deng L, Yang H, Han M, Han Z, Zhao L. Identification of Soybean Mutant Lines Based on Dual-Branch CNN Model Fusion Framework Utilizing Images from Different Organs. Plants. 2023; 12(12):2315. https://doi.org/10.3390/plants12122315
Chicago/Turabian StyleWu, Guangxia, Lin Fei, Limiao Deng, Haoyan Yang, Meng Han, Zhongzhi Han, and Longgang Zhao. 2023. "Identification of Soybean Mutant Lines Based on Dual-Branch CNN Model Fusion Framework Utilizing Images from Different Organs" Plants 12, no. 12: 2315. https://doi.org/10.3390/plants12122315
APA StyleWu, G., Fei, L., Deng, L., Yang, H., Han, M., Han, Z., & Zhao, L. (2023). Identification of Soybean Mutant Lines Based on Dual-Branch CNN Model Fusion Framework Utilizing Images from Different Organs. Plants, 12(12), 2315. https://doi.org/10.3390/plants12122315





