Supplementation of the Plant Conditioner ELICE Vakcina® Product with β-Aminobutyric Acid and Salicylic Acid May Lead to Trans-Priming Signaling in Barley (Hordeum vulgare)
Abstract
:1. Introduction
2. Results
2.1. Treatment of Barley Seedlings by Priming Active Agents
2.2. De Novo Assembly, Mapping, and Functional Annotation of Illumina RNA-seq Reads
2.3. Pairwise DEGs and Fisher’s Exact Test
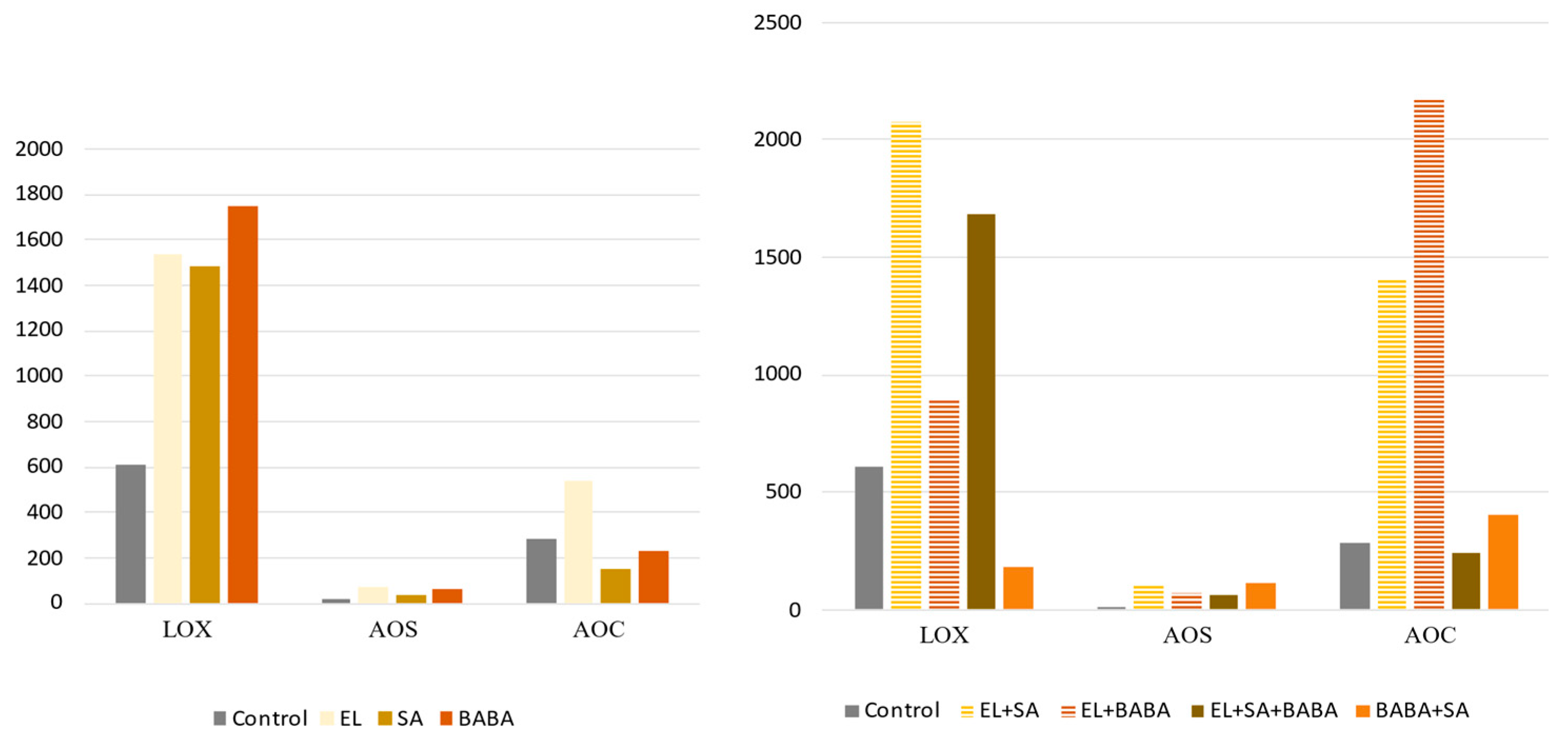
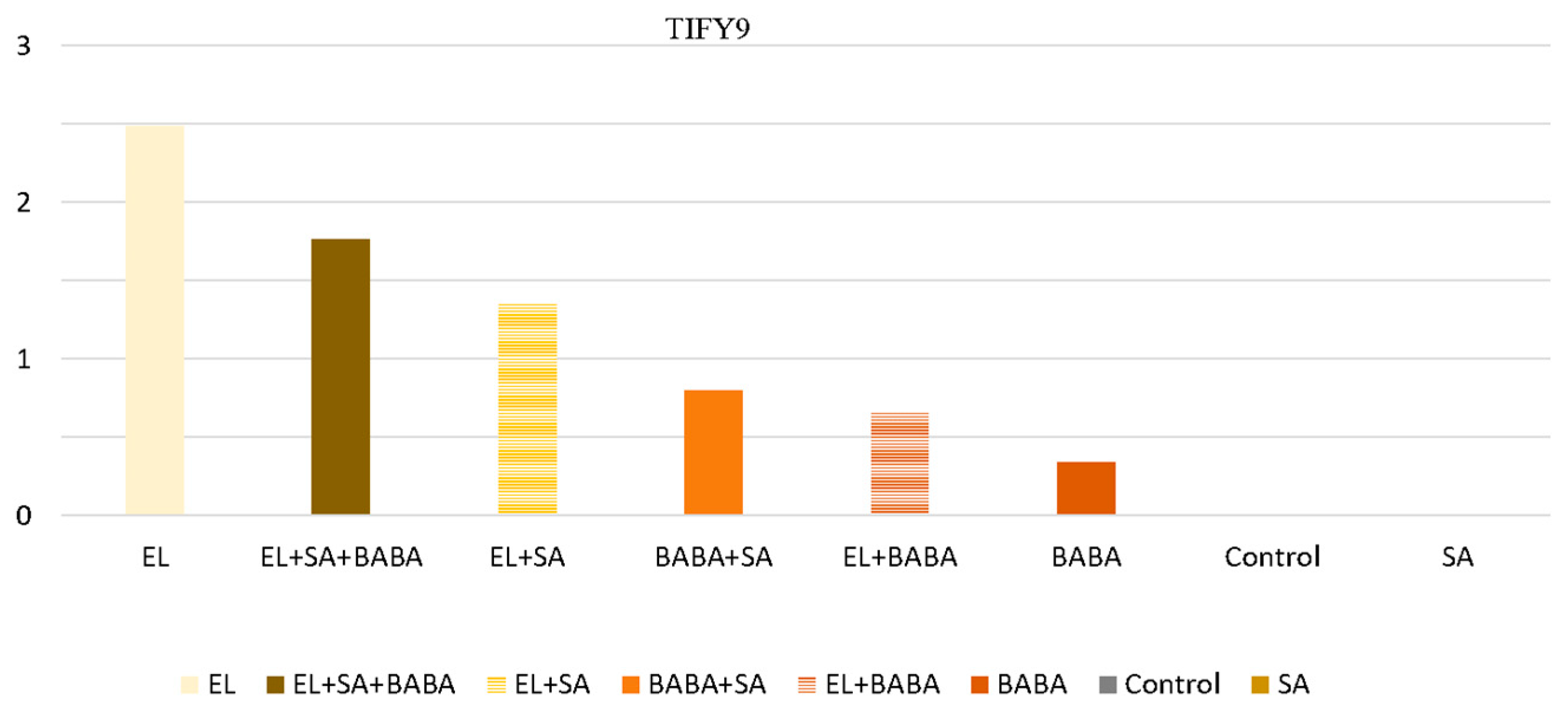
2.4. Analysis of the Genes of the JA- and SA-Pathway
3. Discussion
3.1. Synergistic/Antagonistic Activation of JA Pathway
3.2. Synergistic/Antagonistic Activation of SA Pathway
4. Materials and Methods
4.1. Treatment of Barley Seedlings by Priming Active Agents
4.2. RNA Isolation and Sequencing
4.3. Data Preprocessing, De Novo Assembly, Gene-Level Quantification, and DEG Determination
4.4. Functional Annotation of Transcripts
4.5. Enrichment Analysis (Fisher’s Exact Test)
4.6. Calculation of RPM Index
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bulgari, R.; Trivellini, A.; Ferrante, A. Effects of two doses of organic extract-based biostimulant on greenhouse lettuce grown under increasing NaCl concentrations. Front. Plant Sci. 2019, 9, 1870. [Google Scholar] [CrossRef] [PubMed]
- Husen, H. Plant Performance under Environmental Stress; Hormones, Biostimulants and Sustainable Plant Growth Management; Springer: Cham, Switzerland, 2021; Volume XIV, 606p. [Google Scholar]
- Beckers, G.J.; Conrath, U. Priming for stress resistance: From the lab to the field. Curr. Opin. Plant Biol. 2007, 10, 425–431. [Google Scholar] [CrossRef] [PubMed]
- Johnson, R.; Puthur, J.T. Seed priming as a cost effective technique for developing plants with cross tolerance to salinity stress. Plant Physiol. Biochem. 2021, 162, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Hilker, M.; Schwachtje, J.; Baier, M.; Balazadeh, S.; Bäurle, I.; Geiselhardt, S.; Hincha, D.K.; Kunze, R.; Mueller-Roeber, B.; Rillig, M.C. Priming and memory of stress responses in organisms lacking a nervous system. Biol. Rev. 2016, 91, 1118–1133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baier, M.; Bittner, A.; Prescher, A.; van Buer, J. Preparing plants for improved cold tolerance by priming. Plant Cell Environ. 2019, 42, 782–800. [Google Scholar] [CrossRef]
- Aswathi, K.R.; Sen, A.; Puthur, J.T. Comparative Study of Cis-and Trans-Priming Effect of PEG and BABA in Cowpea Seedlings on Exposure to PEG-Induced Osmotic Stress. Seeds 2023, 2, 85–100. [Google Scholar] [CrossRef]
- Walters, D.R.; Havis, N.D.; Paterson, L.; Taylor, J.; Walsh, D.J.; Sablou, C. Control of foliar pathogens of spring barley using a combination of resistance elicitors. Front. Plant Sci. 2014, 5, 241. [Google Scholar] [CrossRef]
- Schwessinger, B.; Ronald, P.C. Plant innate immunity: Perception of conserved microbial signatures. Annu. Rev. Plant Biol. 2012, 63, 451–482. [Google Scholar] [CrossRef] [Green Version]
- Hirayama, T.; Mochida, K. Plant hormonomics: A key tool for deep physiological phenotyping to improve crop productivity. Plant Cell Physiol. 2022, 63, 1826–1839. [Google Scholar] [CrossRef]
- Vidhyasekaran, P. Plant Hormone Signaling Systems in Plant Innate Immunity; Signaling and Communication in Plants; Springer: Dordrecht, The Netherlands, 2015; 458p. [Google Scholar]
- Baccelli, I.; Mauch-Mani, B. Beta-aminobutyric acid priming of plant defense: The role of ABA and other hormones. Plant Mol. Biol. 2016, 91, 703–711. [Google Scholar] [CrossRef]
- Zimmerli, L.; Jakab, G.; Métraux, J.-P.; Mauch-Mani, B. Potentiation of pathogen-specific defense mechanisms in Arabidopsis by β-aminobutyric acid. Proc. Natl. Acad. Sci. USA 2000, 97, 12920–12925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, H.S.; Fleming, C.; Selby, C.; Rao, J.; Martin, T. Plant biostimulants: A review on the processing of macroalgae and use of extracts for crop management to reduce abiotic and biotic stresses. J. Appl. Phycol. 2014, 26, 465–490. [Google Scholar] [CrossRef]
- Hegedűs, G.; Nagy, Á.; Decsi, K.; Kutasy, B.; Virág, E. Transcriptome datasets of β-Aminobutyric acid (BABA)-primed mono-and dicotyledonous plants, Hordeum vulgare and Arabidopsis thaliana. Data Brief 2022, 41, 107983. [Google Scholar] [CrossRef]
- Cohen, Y.R. β-aminobutyric acid-induced resistance against plant pathogens. Plant Dis. 2002, 86, 448–457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kutasy, B.; Decsi, K.; Hegedűs, G.; Virág, E. Dataset of conditioning effect of herbal extract-based plant biostimulants in pea (Pisum sativum). Data Brief 2023, 46, 108800. [Google Scholar] [CrossRef]
- Decsi, K.; Hegedűs, G.; Kutasy, B.; Virág, E. RNA-seq datasets of field rapeseed (Brassica napus) cultures conditioned by Elice16Indures® biostimulator. Data Brief 2022, 45, 108602. [Google Scholar] [CrossRef] [PubMed]
- Decsi, K.; Kutasy, B.; Hegedűs, G.; Alföldi, Z.P.; Kálmán, N.; Nagy, Á.; Virág, E. Natural immunity stimulation using ELICE16INDURES® plant conditioner in field culture of soybean. Heliyon 2023, 9, e12907. [Google Scholar] [CrossRef] [PubMed]
- Hegedűs, G.; Kutasy, B.; Kiniczky, M.; Decsi, K.; Juhász, Á.; Nagy, Á.; Pallos, J.P.; Virág, E. Liposomal formulation of botanical extracts may enhance yield triggering PR genes and phenylpropanoid pathway in Barley (Hordeum vulgare). Plants 2022, 11, 2969. [Google Scholar] [CrossRef]
- Turner, J.G.; Ellis, C.; Devoto, A. The jasmonate signal pathway. Plant Cell 2002, 14, S153–S164. [Google Scholar] [CrossRef] [Green Version]
- Rojo, E.; Solano, R.; Sánchez-Serrano, J.J. Interactions between signaling compounds involved in plant defense. J. Plant Growth Regul. 2003, 22, 82–98. [Google Scholar] [CrossRef]
- Huang, J.; Gu, M.; Lai, Z.; Fan, B.; Shi, K.; Zhou, Y.-H.; Yu, J.-Q.; Chen, Z. Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol. 2010, 153, 1526–1538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.S.; Hwang, B.K. An important role of the pepper phenylalanine ammonia-lyase gene (PAL1) in salicylic acid-dependent signalling of the defence response to microbial pathogens. J. Exp. Bot. 2014, 65, 2295–2306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Mostafa, S.; Zeng, W.; Jin, B. Function and mechanism of jasmonic acid in plant responses to abiotic and biotic stresses. Int. J. Mol. Sci. 2021, 22, 8568. [Google Scholar] [CrossRef]
- Li, X.-P.; Gilmore, A.M.; Caffarri, S.; Bassi, R.; Golan, T.; Kramer, D.; Niyogi, K.K. Regulation of photosynthetic light harvesting involves intrathylakoid lumen pH sensing by the PsbS protein. J. Biol. Chem. 2004, 279, 22866–22874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, L.; Li, G. FAR1-related sequence (FRS) and FRS-related factor (FRF) family proteins in Arabidopsis growth and development. Front. Plant Sci. 2018, 9, 692. [Google Scholar] [CrossRef] [Green Version]
- Tang, W.; Ji, Q.; Huang, Y.; Jiang, Z.; Bao, M.; Wang, H.; Lin, R. FAR-RED ELONGATED HYPOCOTYL3 and FAR-RED IMPAIRED RESPONSE1 transcription factors integrate light and abscisic acid signaling in Arabidopsis. Plant Physiol. 2013, 163, 857–866. [Google Scholar] [CrossRef] [Green Version]
- Skubacz, A.; Daszkowska-Golec, A.; Szarejko, I. The role and regulation of ABI5 (ABA-Insensitive 5) in plant development, abiotic stress responses and phytohormone crosstalk. Front. Plant Sci. 2016, 7, 1884. [Google Scholar] [CrossRef] [Green Version]
- van der Linde, K.; Mueller, A.N.; Hemetsberger, C.; Kashani, F.; van der Hoorn, R.A.; Doehlemann, G. The maize cystatin CC9 interacts with apoplastic cysteine proteases. Plant Signal. Behav. 2012, 7, 1397–1401. [Google Scholar] [CrossRef] [Green Version]
- Ilyas, M.; Hörger, A.C.; Bozkurt, T.O.; Van Den Burg, H.A.; Kaschani, F.; Kaiser, M.; Belhaj, K.; Smoker, M.; Joosten, M.H.; Kamoun, S. Functional divergence of two secreted immune proteases of tomato. Curr. Biol. 2015, 25, 2300–2306. [Google Scholar] [CrossRef] [Green Version]
- Kaschani, F.; Van der Hoorn, R.A. A model of the C14-EPIC complex indicates hotspots for a protease-inhibitor arms race in the oomycete-potato interaction. Plant Signal. Behav. 2011, 6, 109–112. [Google Scholar] [CrossRef] [Green Version]
- Misas-Villamil, J.C.; van der Hoorn, R.A.; Doehlemann, G. Papain-like cysteine proteases as hubs in plant immunity. New Phytol. 2016, 212, 902–907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plattner, S.; Gruber, C.; Stadlmann, J.; Widmann, S.; Gruber, C.W.; Altmann, F.; Bohlmann, H. Isolation and characterization of a thionin proprotein-processing enzyme from barley. J. Biol. Chem. 2015, 290, 18056–18067. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, R.-F.; Li, T.-T.; Liu, W.-C. Jasmonic acid impairs Arabidopsis seedling salt stress tolerance through MYC2-mediated repression of CAT2 expression. Front. Plant Sci. 2021, 12, 730228. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Song, R.F.; Yuan, H.M.; Li, T.T.; Wang, L.F.; Lu, K.K.; Guo, J.X.; Liu, W.C. Overexpressing the N-terminus of CATALASE2 enhances plant jasmonic acid biosynthesis and resistance to necrotrophic pathogen Botrytis cinerea B05. 10. Mol. Plant Pathol. 2021, 22, 1226–1238. [Google Scholar] [CrossRef]
- Baker, A.; Lin, C.-C.; Lett, C.; Karpinska, B.; Wright, M.H.; Foyer, C.H. Catalase: A critical node in the regulation of cell fate. Free Radic. Biol. Med. 2023, 199, 56–66. [Google Scholar] [CrossRef]
- Yuan, H.-M.; Liu, W.-C.; Lu, Y.-T. CATALASE2 coordinates SA-mediated repression of both auxin accumulation and JA biosynthesis in plant defenses. Cell Host Microbe 2017, 21, 143–155. [Google Scholar] [CrossRef] [Green Version]
- Tola, A.J.; Jaballi, A.; Germain, H.; Missihoun, T.D. Recent development on plant aldehyde dehydrogenase enzymes and their functions in plant development and stress signaling. Genes 2020, 12, 51. [Google Scholar] [CrossRef]
- Qian, Y.; Zhang, T.; Yu, Y.; Gou, L.; Yang, J.; Xu, J.; Pi, E. Regulatory mechanisms of bHLH transcription factors in plant adaptive responses to various abiotic stresses. Front. Plant Sci. 2021, 12, 677611. [Google Scholar] [CrossRef]
- Ji, X.; Nie, X.; Liu, Y.; Zheng, L.; Zhao, H.; Zhang, B.; Huo, L.; Wang, Y. A bHLH gene from Tamarix hispida improves abiotic stress tolerance by enhancing osmotic potential and decreasing reactive oxygen species accumulation. Tree Physiol. 2016, 36, 193–207. [Google Scholar] [CrossRef] [Green Version]
- Lu, X.; Zhang, H.; Hu, J.; Nie, G.; Khan, I.; Feng, G.; Zhang, X.; Wang, X.; Huang, L. Genome-wide identification and characterization of bHLH family genes from orchardgrass and the functional characterization of DgbHLH46 and DgbHLH128 in drought and salt tolerance. Funct. Integr. Genom. 2022, 22, 1331–1344. [Google Scholar] [CrossRef]
- Aleman, F.; Yazaki, J.; Lee, M.; Takahashi, Y.; Kim, A.Y.; Li, Z.; Kinoshita, T.; Ecker, J.R.; Schroeder, J.I. An ABA-increased interaction of the PYL6 ABA receptor with MYC2 transcription factor: A putative link of ABA and JA signaling. Sci. Rep. 2016, 6, 28941. [Google Scholar] [CrossRef]
- Sharma, A.D.; Rakhra, G.; Vyas, D. Data set of in-silico analysis and 3D modelling of boiling stable stress-responsive protein from drought tolerant wheat. Data Brief 2019, 27, 104657. [Google Scholar] [CrossRef] [PubMed]
- Rakhra, G.; Kaur, T.; Vyas, D.; Sharma, A.D.; Singh, J.; Ram, G. Molecular cloning, characterization, heterologous expression and in-silico analysis of disordered boiling soluble stress-responsive wBsSRP protein from drought tolerant wheat cv. PBW 175. Plant Physiol. Biochem. 2017, 112, 29–44. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.; Xu, L.; Singh, A.; Wang, H.; Du, L.; Poovaiah, B. Involvement of calmodulin and calmodulin-like proteins in plant responses to abiotic stresses. Front. Plant Sci. 2015, 6, 600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ranty, B.; Aldon, D.; Cotelle, V.; Galaud, J.-P.; Thuleau, P.; Mazars, C. Calcium sensors as key hubs in plant responses to biotic and abiotic stresses. Front. Plant Sci. 2016, 7, 327. [Google Scholar] [CrossRef] [Green Version]
- Zhu, X.; Perez, M.; Aldon, D.; Galaud, J.-P. Respective contribution of CML8 and CML9, two arabidopsis calmodulin-like proteins, to plant stress responses. Plant Signal. Behav. 2017, 12, e1322246. [Google Scholar] [CrossRef] [Green Version]
- Magnan, F.; Ranty, B.; Charpenteau, M.; Sotta, B.; Galaud, J.P.; Aldon, D. Mutations in AtCML9, a calmodulin-like protein from Arabidopsis thaliana, alter plant responses to abiotic stress and abscisic acid. Plant J. 2008, 56, 575–589. [Google Scholar] [CrossRef]
- Delk, N.A.; Johnson, K.A.; Chowdhury, N.I.; Braam, J. CML24, regulated in expression by diverse stimuli, encodes a potential Ca2+ sensor that functions in responses to abscisic acid, daylength, and ion stress. Plant Physiol. 2005, 139, 240–253. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.; Guo, T.; Wang, H.; Li, J.; Sun, Z.; Liu, Y.; Xing, Y.; Xu, B.; Yang, B.; Li, J. Characterization and expression profile analysis of the 3-phosphoglycerate dehydrogenase family in rice. Agron. J. 2021, 113, 1039–1049. [Google Scholar] [CrossRef]
- Xu, S.; Zhang, M.; Ye, J.; Hu, D.; Zhang, Y.; Li, Z.; Liu, J.; Sun, Y.; Wang, S.; Yuan, X. Brittle culm 25, which encodes an UDP-xylose synthase, affects cell wall properties in rice. Crop J. 2023, 11, 733–743. [Google Scholar] [CrossRef]
- Ruan, N.; Dang, Z.; Wang, M.; Cao, L.; Wang, Y.; Liu, S.; Tang, Y.; Huang, Y.; Zhang, Q.; Xu, Q. FRAGILE CULM 18 encodes a UDP-glucuronic acid decarboxylase required for xylan biosynthesis and plant growth in rice. J. Exp. Bot. 2022, 73, 2320–2335. [Google Scholar] [CrossRef] [PubMed]
- Shen, T.; Li, K.; Yan, R.; Xu, F.; Ni, L.; Jiang, M. The Udp-Glucuronic Acid Decarboxylase Osuxs3 Regulates Na+ Ions Osmotic Stress Tolerance by Interacting with Oscats in Rice. Available SSRN 4220806 2022, preprint. Available online: https://ssrn.com/abstract=4220806 (accessed on 16 September 2022).
- Napieraj, N.; Janicka, M.; Reda, M. Interactions of Polyamines and Phytohormones in Plant Response to Abiotic Stress. Plants 2023, 12, 1159. [Google Scholar] [CrossRef] [PubMed]
- Morsy, M.R.; Almutairi, A.M.; Gibbons, J.; Yun, S.J.; de Los Reyes, B.G. The OsLti6 genes encoding low-molecular-weight membrane proteins are differentially expressed in rice cultivars with contrasting sensitivity to low temperature. Gene 2005, 344, 171–180. [Google Scholar] [CrossRef]
- Razzaque, S.; Haque, T.; Elias, S.M.; Rahman, M.; Biswas, S.; Schwartz, S.; Ismail, A.M.; Walia, H.; Juenger, T.E.; Seraj, Z.I. Reproductive stage physiological and transcriptional responses to salinity stress in reciprocal populations derived from tolerant (Horkuch) and susceptible (IR29) rice. Sci. Rep. 2017, 7, 46138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, W.; Gao, E.; Shaban, M.; Wang, Y.; Wang, H.; Nie, X.; Zhu, L. GhUMC1, a blue copper-binding protein, regulates lignin synthesis and cotton immune response. Biochem. Biophys. Res. Commun. 2018, 504, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Ariza, J.; Campo, S.; Rufat, M.; Estopà, M.; Messeguer, J.; Segundo, B.S.; Coca, M. Sucrose-mediated priming of plant defense responses and broad-spectrum disease resistance by overexpression of the maize pathogenesis-related PRms protein in rice plants. Mol. Plant-Microbe Interact. 2007, 20, 832–842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conrath, U.; Pieterse, C.M.; Mauch-Mani, B. Priming in plant–pathogen interactions. Trends Plant Sci. 2002, 7, 210–216. [Google Scholar] [CrossRef] [Green Version]
- Xie, Y.; Ravet, K.; Pearce, S. Extensive structural variation in the Bowman-Birk inhibitor family in common wheat (Triticum aestivum L.). BMC Genom. 2021, 22, 218. [Google Scholar] [CrossRef]
- Malefo, M.; Mathibela, E.; Crampton, B.; Makgopa, M. Investigating the role of Bowman-Birk serine protease inhibitor in Arabidopsis plants under drought stress. Plant Physiol. Biochem. 2020, 149, 286–293. [Google Scholar] [CrossRef]
- Mestre, P.; Arista, G.; Piron, M.C.; Rustenholz, C.; Ritzenthaler, C.; Merdinoglu, D.; Chich, J.F. Identification of a Vitis vinifera endo-β-1, 3-glucanase with antimicrobial activity against Plasmopara viticola. Mol. Plant Pathol. 2017, 18, 708–719. [Google Scholar] [CrossRef]
- Wang, X.; Yuan, Z.; Shi, Y.; Cai, F.; Zhao, J.; Wang, J.; Wang, Y. Bacillus amyloliquefaciens HG01 induces resistance in loquats against anthracnose rot caused by Colletotrichum acutatum. Postharvest Biol. Technol. 2020, 160, 111034. [Google Scholar] [CrossRef]
- Hata, E.M.; Yusof, M.T.; Zulperi, D. Induction of systemic resistance against bacterial leaf streak disease and growth promotion in rice plant by Streptomyces shenzhenesis TKSC3 and Streptomyces sp. SS8. Plant Pathol. J. 2021, 37, 173. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; Zhang, Q.; Wang, Q.; Wang, X.; Liu, J.; Li, M.; Huang, L.; Kang, Z. Target of tae-miR408, a chemocyanin-like protein gene (TaCLP1), plays positive roles in wheat response to high-salinity, heavy cupric stress and stripe rust. Plant Mol. Biol. 2013, 83, 433–443. [Google Scholar] [CrossRef] [PubMed]
- Tholl, D.; Sohrabi, R.; Huh, J.-H.; Lee, S. The biochemistry of homoterpenes–common constituents of floral and herbivore-induced plant volatile bouquets. Phytochemistry 2011, 72, 1635–1646. [Google Scholar] [CrossRef]
- Song, J.; Zou, X.; Liu, P.; Cardoso, J.A.; Schultze-Kraft, R.; Liu, G.; Luo, L.; Chen, Z. Differential expressions and enzymatic properties of malate dehydrogenases in response to nutrient and metal stresses in Stylosanthes guianensis. Plant Physiol. Biochem. 2022, 170, 325–337. [Google Scholar] [CrossRef]
- Balk, J.; Pilon, M. Ancient and essential: The assembly of iron–sulfur clusters in plants. Trends Plant Sci. 2011, 16, 218–226. [Google Scholar] [CrossRef]
- Lamb, C.; Dixon, R.A. The oxidative burst in plant disease resistance. Annu. Rev. Plant Biol. 1997, 48, 251–275. [Google Scholar] [CrossRef]
- Skopelitis, D.S.; Paranychianakis, N.V.; Paschalidis, K.A.; Pliakonis, E.D.; Delis, I.D.; Yakoumakis, D.I.; Kouvarakis, A.; Papadakis, A.K.; Stephanou, E.G.; Roubelakis-Angelakis, K.A. Abiotic stress generates ROS that signal expression of anionic glutamate dehydrogenases to form glutamate for proline synthesis in tobacco and grapevine. Plant Cell 2006, 18, 2767–2781. [Google Scholar] [CrossRef] [Green Version]
- Laudert, D.; Weiler, E.W. Allene oxide synthase: A major control point in Arabidopsis thaliana octadecanoid signalling. Plant J. 1998, 15, 675–684. [Google Scholar] [CrossRef]
- Yalpani, N.; Raskin, I. Salicylic acid: A systemic signal in induced plant disease resistance. Trends Microbiol. 1993, 1, 88–92. [Google Scholar] [CrossRef]
- Kutasy, B.; Decsi, K.; Kiniczky, M.; Hegedűs, G.; Virág, E. Time-course gene expression profiling data of Triticum aestivum treated by supercritical CO2 garlic extract encapsulated in nanoscale liposomes. Data Brief 2022, 42, 108287. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q. Trinity: Reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 2011, 29, 644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 251. [Google Scholar] [CrossRef] [Green Version]
- Tarazona, S.; García, F.; Ferrer, A.; Dopazo, J.; Conesa, A. NOIseq: A RNA-seq differential expression method robust for sequencing depth biases. EMBnet. J. 2011, 17, 18–19. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Forslund, K.; Coelho, L.P.; Szklarczyk, D.; Jensen, L.J.; Von Mering, C.; Bork, P. Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol. Biol. Evol. 2017, 34, 2115–2122. [Google Scholar] [CrossRef] [Green Version]
- Al-Shahrour, F.; Díaz-Uriarte, R.; Dopazo, J. FatiGO: A web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics 2004, 20, 578–580. [Google Scholar] [CrossRef] [Green Version]
- Bedre, R. Gene Expression Units Explained: Rpm, Rpkm, Fpkm, Tpm, Deseq, Tmm, Scnorm, Getmm, and Combat-Seq. 2017. Available online: https://www.reneshbedre.com/blog/expression_units.html (accessed on 16 April 2023).
- Atallah, J.; Plachetzki, D.C.; Jasper, W.C.; Johnson, B.R. The utility of shallow RNA-seq for documenting differential gene expression in genes with high and low levels of expression. PLoS ONE 2013, 8, e84160. [Google Scholar] [CrossRef] [Green Version]

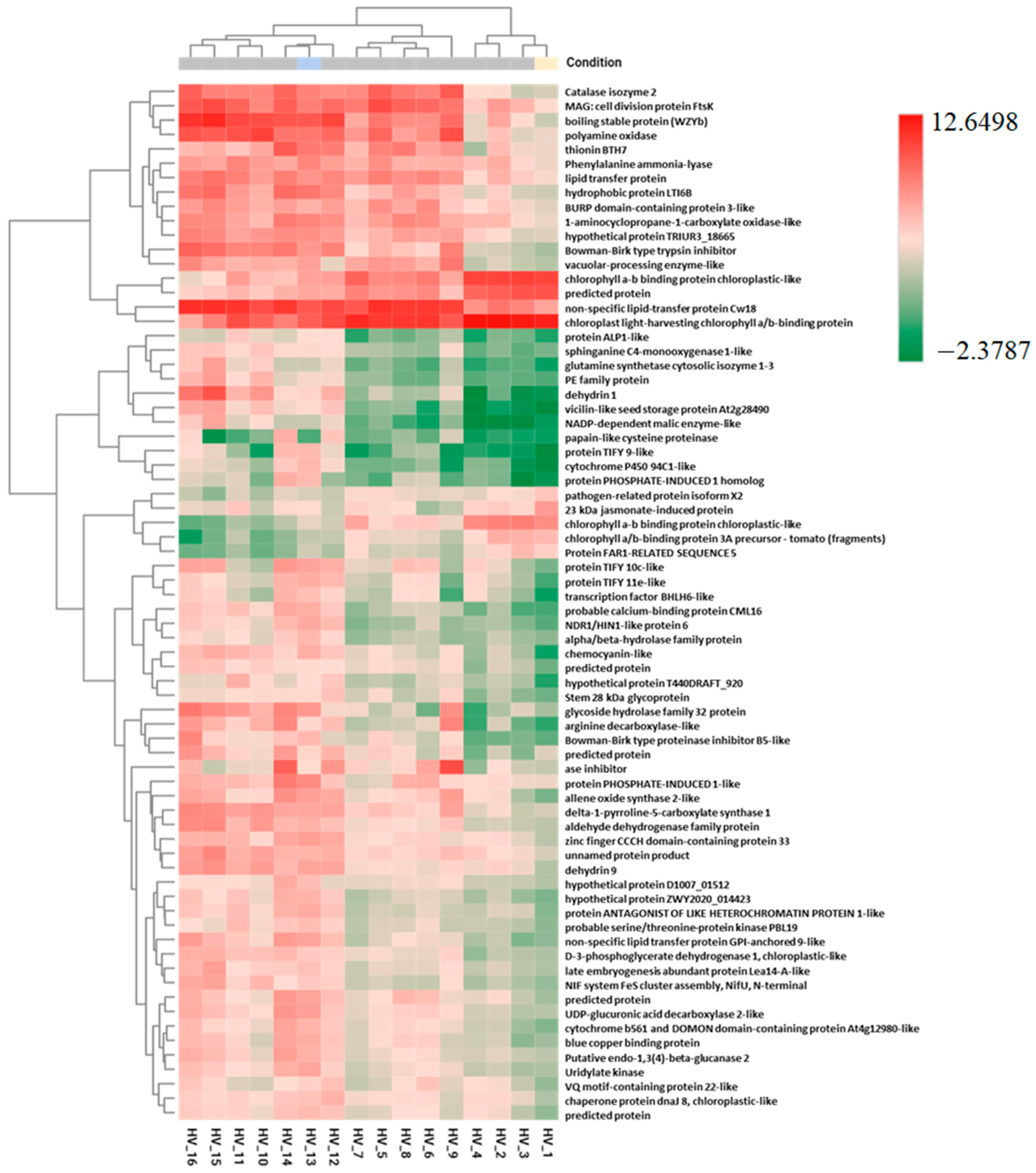
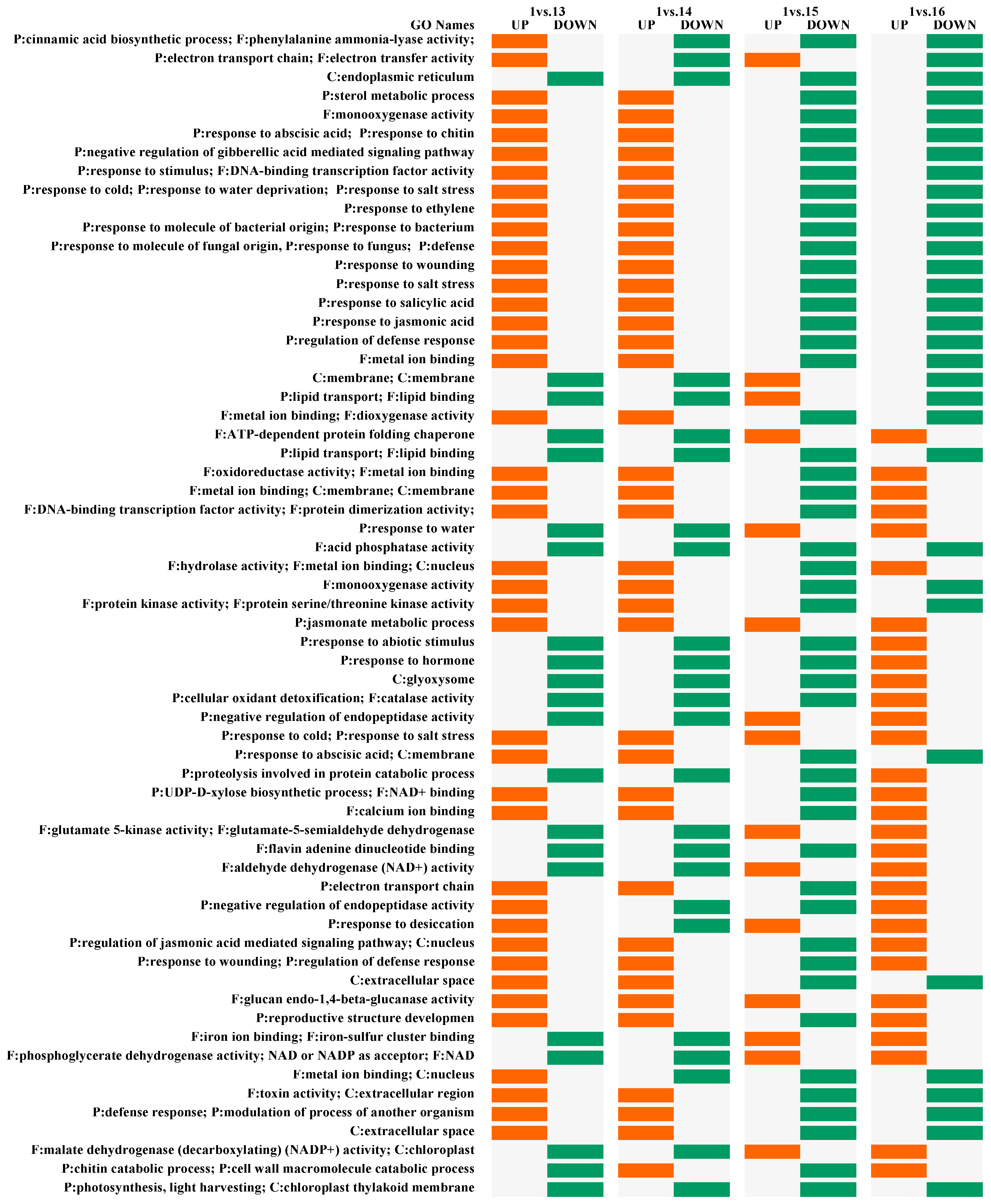


| C | EL | EL + BABA | EL + SA | EL + SA + BABA | Gene name | Function |
| Down | UP | Down | UP | UP | TIFY9 TIFY10 TIF11e | Are involved in JA and other hormone signaling pathways, including auxins, gibberellins (GAs), ABA, SA, and ethylene (ET) [21,22] |
| Down | UP | UP | UP | UP | PAL | PAL gene expression responds to a variety of environmental stresses, including pathogen infection, wounding, nutrient depletion, UV irradiation, and extreme temperatures [23]. It is involved in the biosynthesis of SA, essential signal involved in plant systemic resistance [24]. |
| Down | UP | UP | UP | UP | AOS | It has a key role in the synthesis of JA and biologically active jasmonoyl-isoleucine (JA-Ile) [21]. It plays important roles in the mediation of plant responses and defenses to various biotic (pathogen, insect, and herbivore) and abiotic (drought, cold, salt, heat, and heavy metal toxicity) stresses therefore have received extensive research attention [25]. |
| SA-pathway | JA-pathway | |||||||||
| PAL DEG | PAL RPM | ICS RPM | TIFY DEG | TIFY RPM | AOS DEG | AOS RPM | LOX RPM | AOC RPM | ||
| Control | 1 | DOWN | UP | DOWN | DOWN | DOWN | DOWN | DOWN | DOWN | DOWN |
| SA | 10 | UP | DOWN | DOWN | DOWN | DOWN | DOWN | DOWN | UP | DOWN |
| BABA | 11 | UP | UP | UP | DOWN | DOWN | DOWN | UP | UP | DOWN |
| BABA + SA | 12 | UP | UP | UP | UP | UP | UP | DOWN | DOWN | DOWN |
| EL | 13 | UP | UP | UP | UP | UP | UP | UP | UP | UP |
| EL + SA | 14 | UP | UP | UP | UP | UP | UP | UP | UP | UP |
| EL + BABA | 15 | UP | UP | UP | DOWN | UP | UP | UP | UP | DOWN |
| EL + BABA + SA | 16 | UP | UP | UP | UP | UP | UP | DOWN | UP | DOWN |
| Treatment | Day 0 | Day 2 |
|---|---|---|
| Control | 1 HV_1) | 9 (HV_9) |
| SA | 2 (HV_2) | 10 (HV_10) |
| BABA | 3 (HV_3) | 11 (HV_11) |
| BABA + SA | 4 (HV_4) | 12 (HV_12) |
| EL | 5 (HV_5) | 13 (HV_13) |
| EL + SA | 6 (HV_6) | 14 (HV_14) |
| EL + BABA | 7 (HV_7) | 15 (HV_15) |
| EL + BABA + SA | 8 (HV_8) | 16 (HV_16) |
| Process | Software | Web Page |
|---|---|---|
| Quality control | FastQc V0.11.9 | https://github.com/s-andrews/FastQC (8 June 2023) |
| Filtering | Trimmomatic v0.39 | https://github.com/usadellab/Trimmomatic/releases (8 June 2023) |
| De novo assembly | Trinity v2.15.1 | https://github.com/trinityrnaseq/trinityrnaseq/wiki (8 June 2023) |
| Functional annotation | EggNOG-Mapper V5 | http://eggnog-mapper.embl.de/ (8 June 2023) |
| RNA-Seq alignment | Bowtie2 v2.4.5 | https://bowtie-bio.sourceforge.net/bowtie2/index.shtml (8 June 2023) |
| Expression quantification | RSEM v1.3.3 | https://deweylab.github.io/RSEM/ (8 June 2023) |
| Distribution of GO term | Blast2GO v6.0 | https://www.blast2go.com/ (8 June 2023) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Virág, E.; Kiniczky, M.; Kutasy, B.; Nagy, Á.; Pallos, J.P.; Laczkó, L.; Freytag, C.; Hegedűs, G. Supplementation of the Plant Conditioner ELICE Vakcina® Product with β-Aminobutyric Acid and Salicylic Acid May Lead to Trans-Priming Signaling in Barley (Hordeum vulgare). Plants 2023, 12, 2308. https://doi.org/10.3390/plants12122308
Virág E, Kiniczky M, Kutasy B, Nagy Á, Pallos JP, Laczkó L, Freytag C, Hegedűs G. Supplementation of the Plant Conditioner ELICE Vakcina® Product with β-Aminobutyric Acid and Salicylic Acid May Lead to Trans-Priming Signaling in Barley (Hordeum vulgare). Plants. 2023; 12(12):2308. https://doi.org/10.3390/plants12122308
Chicago/Turabian StyleVirág, Eszter, Márta Kiniczky, Barbara Kutasy, Ágnes Nagy, József Péter Pallos, Levente Laczkó, Csongor Freytag, and Géza Hegedűs. 2023. "Supplementation of the Plant Conditioner ELICE Vakcina® Product with β-Aminobutyric Acid and Salicylic Acid May Lead to Trans-Priming Signaling in Barley (Hordeum vulgare)" Plants 12, no. 12: 2308. https://doi.org/10.3390/plants12122308
APA StyleVirág, E., Kiniczky, M., Kutasy, B., Nagy, Á., Pallos, J. P., Laczkó, L., Freytag, C., & Hegedűs, G. (2023). Supplementation of the Plant Conditioner ELICE Vakcina® Product with β-Aminobutyric Acid and Salicylic Acid May Lead to Trans-Priming Signaling in Barley (Hordeum vulgare). Plants, 12(12), 2308. https://doi.org/10.3390/plants12122308









