The Microbial Connection to Sustainable Agriculture
Abstract
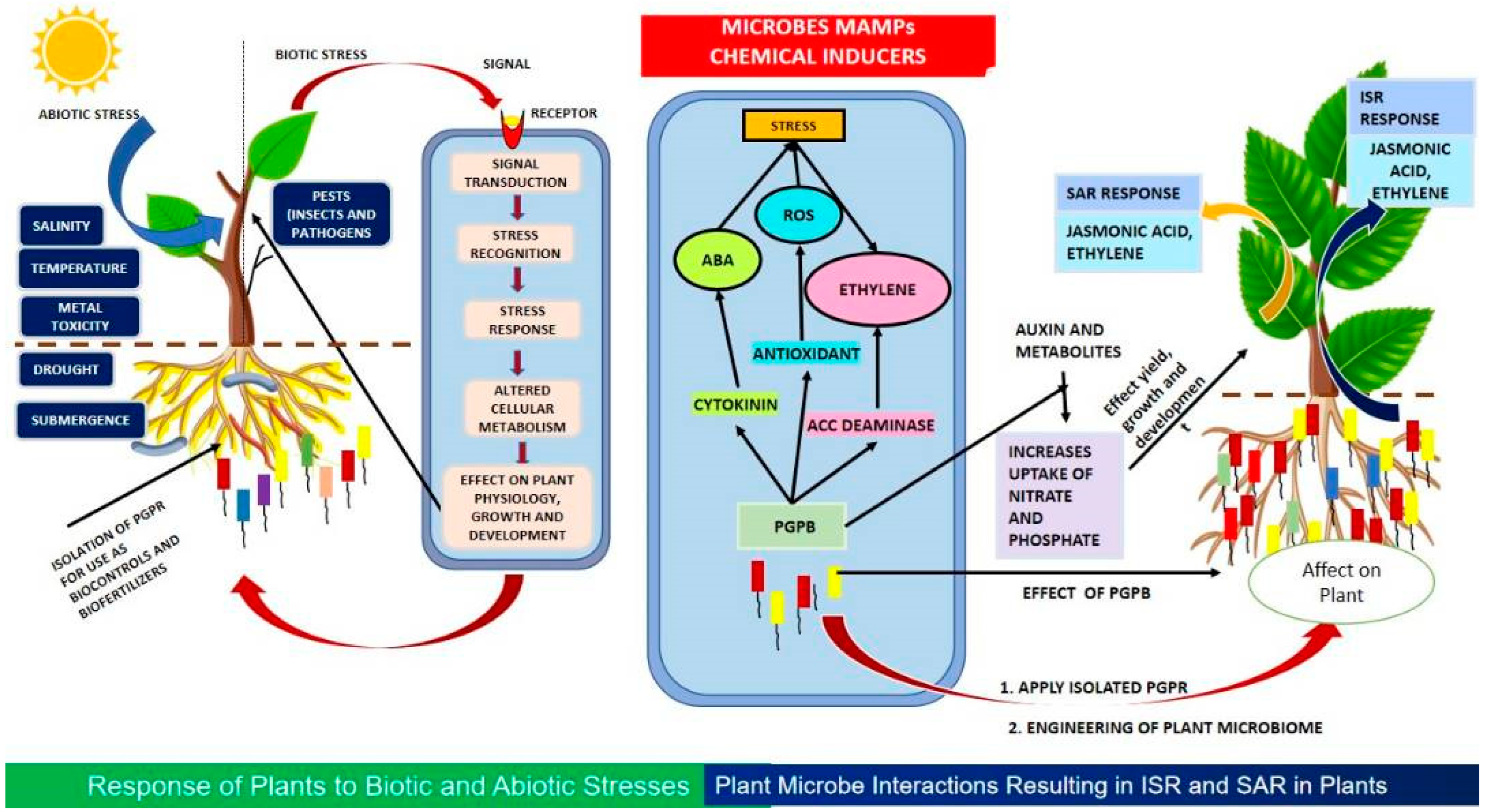
:1. Introduction
2. Connected Plant Microbiome
3. Microbiomes: Above- and Belowground Connection
Factors Affecting the Microbiome
4. Role of Core and Hub Microbiome
5. Beneficial Microbes for Sustainable Agriculture
5.1. Beneficial Microbes and Their Mode of Action
5.2. Plant Growth Promoting Bacteria and Biotic Stressors
5.3. Plant Growth-Promoting Bacteria and Abiotic Stressors
5.4. The Application of PGPB in Sustainable Agriculture
6. Microbiome Engineering
6.1. Rhizosphere Microbiome
6.2. Rhizosphere Engineering
6.3. Shaping the Microbiome
6.4. Rules That Govern Microbiome Engineering
6.5. Shaping the Microbiome through Plant-Mediated Strategies
7. Translation of Knowledge to Field Application
7.1. Engineering a Host-Mediated Microbiome for Sustainable Agriculture
7.2. Engineering Plant Microbiomes for Sustainable Agriculture
7.3. Management Practices That Optimize the Microbiome
7.4. Optimized of Microbiomes through Genetical Engineering
8. Methods of Microbiome Studies
8.1. CRISPR/Cas9
8.2. Genome-Wide Association Studies (GWAS)
8.3. Microbiome Sequencing Platforms
8.4. Metatranscriptomics, Metaproteomics, and Metabolomics for Understanding the Microbiomes
8.5. Culturomics to Support the Molecular Techniques
9. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Malhi, G.S.; Kaur, M.; Kaushik, P. Impact of Climate Change on Agriculture and Its Mitigation Strategies: A Review. Sustainability 2021, 13, 1318. [Google Scholar] [CrossRef]
- Tilman, D. Global environmental impacts of agricultural expansion: The need for sustainable and efficient practices. Proc. Natl. Acad. Sci. USA 1999, 96, 5995–6000. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomiero, T. Soil Degradation, Land Scarcity and Food Security: Reviewing a Complex Challenge. Sustainability 2016, 8, 281. [Google Scholar] [CrossRef] [Green Version]
- Nadarajah, K. Soil Health: The Contribution of Microflora and Microfauna. In Mycorrhizosphere Pedogenesis; Springer: Singapore, 2019; pp. 383–400. [Google Scholar] [CrossRef]
- Koskey, G.; Mburu, S.W.; Awino, R.; Njeru, E.M.; Maingi, J.M. Potential Use of Beneficial Microorganisms for Soil Amelioration, Phytopathogen Biocontrol, and Sustainable Crop Production in Smallholder Agroecosystems. Front. Sustain. Food Syst. 2021, 5, 130. [Google Scholar] [CrossRef]
- Lopes, M.J.D.S.; Dias-Filho, M.B.; Gurgel, E.S.C. Successful Plant Growth-Promoting Microbes: Inoculation Methods and Abiotic Factors. Front. Sustain. Food Syst. 2021, 5, 606454. [Google Scholar] [CrossRef]
- Harman, G.; Khadka, R.; Doni, F.; Uphoff, N. Benefits to Plant Health and Productivity from Enhancing Plant Microbial Symbionts. Front. Plant Sci. 2021, 11, 2001. [Google Scholar] [CrossRef]
- Nishad, R.; Ahmed, T.; Rahman, V.J.; Kareem, A. Modulation of Plant Defense System in Response to Microbial Interactions. Front. Microbiol. 2020, 11, 1298. [Google Scholar] [CrossRef]
- Farrar, K.; Bryant, D.; Cope-Selby, N. Understanding and engineering beneficial plant–microbe interactions: Plant growth promotion in energy crops. Plant Biotechnol. J. 2014, 12, 1193–1206. [Google Scholar] [CrossRef] [Green Version]
- Bhattacharyya, P.N.; Jha, D.K. Plant growth-promoting rhizobacteria (PGPR): Emergence in agriculture. World J. Microbiol. Biotechnol. 2012, 28, 1327–1350. [Google Scholar] [CrossRef]
- Gupta, R.; Anand, G.; Gaur, R.; Yadav, D. Plant–microbiome interactions for sustainable agriculture: A review. Physiol. Mol. Biol. Plants 2021, 27, 165–179. [Google Scholar] [CrossRef]
- Fendrihan, S.; Pop, C.-E. Biotechnological potential of plant associated microorganisms. Rom. Biotechnol. Lett. 2021, 26, 2700–2706. [Google Scholar] [CrossRef]
- Sergaki, C.; Lagunas, B.; Lidbury, I.; Gifford, M.L.; Schäfer, P. Challenges and Approaches in Microbiome Research: From Fundamental to Applied. Front. Plant Sci. 2018, 9, 1205. [Google Scholar] [CrossRef]
- Gupta, A.; Singh, U.B.; Sahu, P.K.; Paul, S.; Kumar, A.; Malviya, D.; Singh, S.; Kuppusamy, P.; Singh, P.; Paul, D.; et al. Linking Soil Microbial Diversity to Modern Agriculture Practices: A Review. Int. J. Environ. Res. Public Health 2022, 19, 3141. [Google Scholar] [CrossRef]
- Anguita-Maeso, M.; Navas-Cortés, J.A.; Landa, B.B. Insights into the Methodological, Biotic and Abiotic Factors Influencing the Characterization of Xylem-Inhabiting Microbial Communities of Olive Trees. Plants 2023, 12, 912. [Google Scholar] [CrossRef]
- Trivedi, P.; Leach, J.E.; Tringe, S.G.; Sa, T.; Singh, B.K. Plant–microbiome interactions: From community assembly to plant health. Nat. Rev. Microbiol. 2020, 18, 607–621. [Google Scholar] [CrossRef]
- Fitzpatrick, C.R.; Schneider, A.C. Unique bacterial assembly, composition, and interactions in a parasitic plant and its host. J. Exp. Bot. 2020, 71, 2198–2209. [Google Scholar] [CrossRef]
- Kaul, S.; Choudhary, M.; Gupta, S.; Dhar, M.K. Engineering Host Microbiome for Crop Improvement and Sustainable Agriculture. Front. Microbiol. 2021, 12, 1125. [Google Scholar] [CrossRef]
- Afridi, M.S.; Ali, S.; Salam, A.; Terra, W.C.; Hafeez, A.; Sumaira; Ali, B.; AlTami, M.S.; Ameen, F.; Ercisli, S.; et al. Plant Microbiome Engineering: Hopes or Hypes. Biology 2022, 11, 1782. [Google Scholar] [CrossRef]
- Cullen, C.M.; Aneja, K.K.; Beyhan, S.; Cho, C.E.; Woloszynek, S.; Convertino, M.; McCoy, S.J.; Zhang, Y.; Anderson, M.Z.; Alvarez-Ponce, D.; et al. Emerging Priorities for Microbiome Research. Front. Microbiol. 2020, 11, 136. [Google Scholar] [CrossRef] [Green Version]
- Brunel, C.; Pouteau, R.; Dawson, W.; Pester, M.; Ramirez, K.S.; van Kleunen, M. Towards Unraveling Macroecological Patterns in Rhizosphere Microbiomes. Trends Plant Sci. 2020, 25, 1017–1029. [Google Scholar] [CrossRef]
- Compant, S.; Samad, A.; Faist, H.; Sessitsch, A. A review on the plant microbiome: Ecology, functions, and emerging trends in microbial application. J. Adv. Res. 2019, 19, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Santoyo, G.; Moreno-Hagelsieb, G.; del Carmen Orozco-Mosqueda, M.; Glick, B.R. Plant growth-promoting bacterial endophytes. Microbiol. Res. 2016, 183, 92–99. [Google Scholar] [CrossRef] [PubMed]
- Vandenkoornhuyse, P.; Quaiser, A.; Duhamel, M.; Le Van, A.; Dufresne, A. The importance of the microbiome of the plant holobiont. New Phytol. 2015, 206, 1196–1206. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Whalley, W.R.; Miller, A.J.; White, P.J.; Zhang, F.; Shen, J. Sustainable Cropping Requires Adaptation to a Heterogeneous Rhizosphere. Trends Plant Sci. 2020, 25, 1194–1202. [Google Scholar] [CrossRef]
- Chiaranunt, P.; White, J.F. Plant Beneficial Bacteria and Their Potential Applications in Vertical Farming Systems. Plants 2023, 12, 400. [Google Scholar] [CrossRef]
- Li, P.; Liu, J.; Saleem, M.; Li, G.; Luan, L.; Wu, M.; Li, Z. Reduced chemodiversity suppresses rhizosphere microbiome functioning in the mono-cropped agroecosystems. Microbiome 2022, 10, 108. [Google Scholar] [CrossRef]
- Jin, Y.; Zhu, H.; Luo, S.; Yang, W.; Zhang, L.; Li, S.; Jin, Q.; Cao, Q.; Sun, S.; Xiao, M. Role of Maize Root Exudates in Promotion of Colonization of Bacillus velezensis Strain S3-1 in Rhizosphere Soil and Root Tissue. Curr. Microbiol. 2019, 76, 855–862. [Google Scholar] [CrossRef]
- Chen, Y.; Bonkowski, M.; Shen, Y.; Griffiths, B.; Jiang, Y.; Wang, X.; Sun, B. Root ethylene mediates rhizosphere microbial community reconstruction when chemically detecting cyanide produced by neighbouring plants. Microbiome 2020, 8, 4. [Google Scholar] [CrossRef] [Green Version]
- Carrión, V.J.; Perez-Jaramillo, J.; Cordovez, V.; Tracanna, V.; de Hollander, M.; Ruiz-Buck, D.; Mendes, L.W.; van Ijcken, W.F.J.; Gomez-Exposito, R.; Elsayed, S.S.; et al. Pathogen-induced activation of disease-suppressive functions in the endophytic root microbiome. Science 2019, 366, 606–612. [Google Scholar] [CrossRef]
- Liang, S.; Liu, H.; Wu, S.; Xu, S.; Jin, D.; Faiola, F.; Zhuang, X.; Zhuang, G.; Qu, D.; Fan, H.; et al. Genetic diversity of diazotrophs and total bacteria in the phyllosphere of Pyrus serotina, Prunus armeniaca, Prunus avium, and Vitis vinifera. Can. J. Microbiol. 2019, 65, 642–652. [Google Scholar] [CrossRef] [Green Version]
- Stringlis, I.A.; De Jonge, R.; Pieterse, C.M.J. The Age of Coumarins in Plant–Microbe Interactions. Plant Cell Physiol. 2019, 60, 1405–1419. [Google Scholar] [CrossRef] [Green Version]
- Lombardi, N.; Vitale, S.; Turrà, D.; Reverberi, M.; Fanelli, C.; Vinale, F.; Marra, R.; Ruocco, M.; Pascale, A.; D’errico, G.; et al. Root Exudates of Stressed Plants Stimulate and Attract Trichoderma Soil Fungi. Mol. Plant-Microbe Interact. 2018, 31, 982–994. [Google Scholar] [CrossRef] [Green Version]
- Shaposhnikov, A.I.; Shakhnazarova, V.Y.; Vishnevskaya, N.A.; Borodina, E.V.; Strunnikova, O.K. Aromatic Carboxylic Acids in Barley-Root Exudates and Their Influence on the Growth of Fusarium culmorum and Pseudomonas fluorescens. Appl. Biochem. Microbiol. 2020, 56, 344–351. [Google Scholar] [CrossRef]
- Arif, I.; Batool, M.; Schenk, P.M. Plant Microbiome Engineering: Expected Benefits for Improved Crop Growth and Resilience. Trends Biotechnol. 2020, 38, 1385–1396. [Google Scholar] [CrossRef]
- Velmourougane, K.; Saxena, G.; Prasanna, R. Plant-Microbe Interactions in the Rhizosphere: Mechanisms and Their Ecological Benefits. In Plant-Microbe Interactions in Agro-Ecological Perspectives; Springer: Singapore, 2017; Volume 2, pp. 193–219. [Google Scholar] [CrossRef]
- Chagas, F.O.; Pessotti, R.D.C.; Caraballo-Rodríguez, A.M.; Pupo, M.T. Chemical signaling involved in plant–microbe interactions. Chem. Soc. Rev. 2018, 47, 1652–1704. [Google Scholar] [CrossRef]
- Nadarajah, K. Metagenomics for Improving Soil Fertility. In Soil Nitrogen Ecology. Soil Biology; Springer: Cham, Switzerland, 2021; Volume 62, pp. 267–282. [Google Scholar] [CrossRef]
- Navgire, G.S.; Goel, N.; Sawhney, G.; Sharma, M.; Kaushik, P.; Mohanta, Y.K.; Mohanta, T.K.; Al-Harrasi, A. Analysis and Interpretation of metagenomics data: An approach. Biol. Proced. Online 2022, 24, 18. [Google Scholar] [CrossRef]
- Nwachukwu, B.C.; Babalola, O.O. Metagenomics: A Tool for Exploring Key Microbiome with the Potentials for Improving Sustainable Agriculture. Front. Sustain. Food Syst. 2022, 6, 206. [Google Scholar] [CrossRef]
- Dastogeer, K.M.; Tumpa, F.H.; Sultana, A.; Akter, M.A.; Chakraborty, A. Plant microbiome–An account of the factors that shape community composition and diversity. Curr. Plant Biol. 2020, 23, 100161. [Google Scholar] [CrossRef]
- Noman, M.; Ahmed, T.; Ijaz, U.; Shahid, M.; Azizullah; Li, D.; Manzoor, I.; Song, F. Plant–Microbiome Crosstalk: Dawning from Composition and Assembly of Microbial Community to Improvement of Disease Resilience in Plants. Int. J. Mol. Sci. 2021, 22, 6852. [Google Scholar] [CrossRef]
- Saad, M.M.; Eida, A.A.; Hirt, H. Tailoring plant-associated microbial inoculants in agriculture: A roadmap for successful application. J. Exp. Bot. 2020, 71, 3878–3901. [Google Scholar] [CrossRef] [Green Version]
- Sivakumar, N.; Sathishkumar, R.; Selvakumar, G.; Shyamkumar, R.; Arjunekumar, K. Phyllospheric Microbiomes: Diversity, Ecological Significance, and Biotechnological Applications. In Plant Microbiomes for Sustainable Agriculture; Springer: Cham, Switzerland, 2020; Volume 25, pp. 113–172. [Google Scholar] [CrossRef]
- Zheng, Q.; Hu, Y.; Zhang, S.; Noll, L.; Böckle, T.; Dietrich, M.; Herbold, C.W.; Eichorst, S.A.; Woebken, D.; Richter, A.; et al. Soil multifunctionality is affected by the soil environment and by microbial community composition and diversity. Soil Biol. Biochem. 2019, 136, 107521. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Q.; Hu, J.; Wei, H.; Zhang, H.; Zhu, J. Relationship between Plant Roots, Rhizosphere Microorganisms, and Nitrogen and Its Special Focus on Rice. Agriculture 2021, 11, 234. [Google Scholar] [CrossRef]
- Yang, C.; Yue, H.; Ma, Z.; Feng, Z.; Feng, H.; Zhao, L.; Zhang, Y.; Deakin, G.; Xu, X.; Zhu, H.; et al. Influence of plant genotype and soil on the cotton rhizosphere microbiome. Front. Microbiol. 2022, 13, 3674. [Google Scholar] [CrossRef] [PubMed]
- Abdul Rahman, N.S.N.; Abdul Hamid, N.W.; Nadarajah, K. Effects of abiotic stress on soil microbiome. Int. J. Mol. Sci. 2021, 22, 9036. [Google Scholar] [CrossRef]
- Koza, N.A.; Adedayo, A.A.; Babalola, O.O.; Kappo, A.P. Microorganisms in Plant Growth and Development: Roles in Abiotic Stress Tolerance and Secondary Metabolites Secretion. Microorganisms 2022, 10, 1528. [Google Scholar] [CrossRef]
- Blaustein, R.A.; Lorca, G.L.; Meyer, J.L.; Gonzalez, C.F.; Teplitski, M. Defining the Core Citrus Leaf- and Root-Associated Microbiota: Factors Associated with Community Structure and Implications for Managing Huanglongbing (Citrus Greening) Disease. Appl. Environ. Microbiol. 2017, 83, e00210-17. [Google Scholar] [CrossRef] [Green Version]
- Hamonts, K.; Trivedi, P.; Garg, A.; Janitz, C.; Grinyer, J.; Holford, P.; Botha, F.C.; Anderson, I.C.; Singh, B.K. Field study reveals core plant microbiota and relative importance of their drivers. Environ. Microbiol. 2018, 20, 124–140. [Google Scholar] [CrossRef]
- Xu, J.; Zhang, Y.; Zhang, P.; Trivedi, P.; Riera, N.; Wang, Y.; Liu, X.; Fan, G.; Tang, J.; Coletta-Filho, H.D.; et al. The structure and function of the global citrus rhizosphere microbiome. Nat. Commun. 2018, 9, 4894. [Google Scholar] [CrossRef] [Green Version]
- Simonin, M.; Dasilva, C.; Terzi, V.; Ngonkeu, E.L.M.; Diouf, D.; Kane, A.; Béna, G.; Moulin, L. Influence of plant genotype and soil on the wheat rhizosphere microbiome: Evidences for a core microbiome across eight African and European soils. FEMS Microbiol. Ecol. 2020, 96, fiaa067. [Google Scholar] [CrossRef]
- Singh, B.K.; Trivedi, P.; Egidi, E.; Macdonald, C.A.; Delgado-Baquerizo, M. Crop microbiome and sustainable agriculture. Nat. Rev. Microbiol. 2020, 18, 601–602. [Google Scholar] [CrossRef]
- de Souza, R.S.C.; Okura, V.K.; Armanhi, J.S.L.; Jorrín, B.; Lozano, N.; da Silva, M.J.; González-Guerrero, M.; de Araújo, L.M.; Verza, N.C.; Bagheri, H.C.; et al. Unlocking the bacterial and fungal communities assemblages of sugarcane microbiome. Sci. Rep. 2016, 6, 28774. [Google Scholar] [CrossRef] [Green Version]
- Dai, Y.; Yang, F.; Zhang, L.; Xu, Z.; Fan, X.; Tian, Y.; Wang, T. Wheat-associated microbiota and their correlation with stripe rust reaction. J. Appl. Microbiol. 2020, 128, 544–555. [Google Scholar] [CrossRef]
- Lemanceau, P.; Blouin, M.; Muller, D.; Moënne-Loccoz, Y. Let the Core Microbiota Be Functional. Trends Plant Sci. 2017, 22, 583–595. [Google Scholar] [CrossRef]
- Banerjee, S.; Schlaeppi, K.; Van Der Heijden, M.G.A. Keystone taxa as drivers of microbiome structure and functioning. Nat. Rev. Microbiol. 2018, 16, 567–576. [Google Scholar] [CrossRef]
- Roman-Reyna, V.; Pinili, D.; Borja, F.N.; Quibod, I.L.; Groen, S.C.; Mulyaningsih, E.S.; Rachmat, A.; Slamet-Loedin, I.H.; Alexandrov, N.; Mauleon, R.; et al. The rice leaf microbiome has a conserved community structure controlled by complex host-microbe interactions. bioRxiv 2019, 615278. [Google Scholar] [CrossRef]
- Niu, Y.; Bainard, L.; Bandara, M.S.; Hamel, C.; Gan, Y. Soil residual water and nutrients explain about 30% of the rotational effect in 4-year pulse-intensified rotation systems. Can. J. Plant Sci. 2017, 97, 852–864. [Google Scholar] [CrossRef] [Green Version]
- Egamberdieva, D.; Wirth, S.J.; Alqarawi, A.A.; Abd_Allah, E.F.; Hashem, A. Phytohormones and Beneficial Microbes: Essential Components for Plants to Balance Stress and Fitness. Front. Microbiol. 2017, 8, 2104. [Google Scholar] [CrossRef]
- Vannier, N.; Agler, M.; Hacquard, S. Microbiota-mediated disease resistance in plants. PLoS Pathog. 2019, 15, e1007740. [Google Scholar] [CrossRef] [Green Version]
- Ebrahimi-Zarandi, M.; Riseh, R.S.; Tarkka, M.T. Actinobacteria as Effective Biocontrol Agents against Plant Pathogens, an Overview on Their Role in Eliciting Plant Defense. Microorganisms 2022, 10, 1739. [Google Scholar] [CrossRef]
- Inbaraj, M.P. Plant-Microbe Interactions in Alleviating Abiotic Stress—A Mini Review. Front. Agron. 2021, 3, 667903. [Google Scholar] [CrossRef]
- Munir, N.; Hanif, M.; Abideen, Z.; Sohail, M.; El-Keblawy, A.; Radicetti, E.; Mancinelli, R.; Haider, G. Mechanisms and Strategies of Plant Microbiome Interactions to Mitigate Abiotic Stresses. Agronomy 2022, 12, 2069. [Google Scholar] [CrossRef]
- Poria, V.; Dębiec-Andrzejewska, K.; Fiodor, A.; Lyzohub, M.; Ajijah, N.; Singh, S.; Pranaw, K. Plant Growth-Promoting Bacteria (PGPB) integrated phytotechnology: A sustainable approach for remediation of marginal lands. Front. Plant Sci. 2022, 13, 999866. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.-M.; Guo, S.-J.; Tian, W.; Chen, Y.; Han, H.; Chen, E.; Li, B.-L.; Li, Y.-Y.; Chen, Z.-J. Effects of Plant Growth-Promoting Bacteria (PGPB) Inoculation on the Growth, Antioxidant Activity, Cu Uptake, and Bacterial Community Structure of Rape (Brassica napus L.) Grown in Cu-Contaminated Agricultural Soil. Front. Microbiol. 2019, 10, 1455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamid, N.W.A.; Nadarajah, K. Microbe Related Chemical Signalling and Its Application in Agriculture. Int. J. Mol. Sci. 2022, 23, 8998. [Google Scholar] [CrossRef]
- El-Saadony, M.T.; Saad, A.M.; Soliman, S.M.; Salem, H.M.; Ahmed, A.I.; Mahmood, M.; El-Tahan, A.M.; Ebrahim, A.A.M.; El-Mageed, T.A.A.; Negm, S.H.; et al. Plant growth-promoting microorganisms as biocontrol agents of plant diseases: Mechanisms, challenges and future perspectives. Front. Plant Sci. 2022, 13, 923880. [Google Scholar] [CrossRef]
- Olanrewaju, O.S.; Glick, B.R.; Babalola, O.O. Mechanisms of action of plant growth promoting bacteria. World J. Microbiol. Biotechnol. 2017, 33, 197. [Google Scholar] [CrossRef] [Green Version]
- Dong, O.X.; Ronald, P.C. Genetic Engineering for Disease Resistance in Plants: Recent Progress and Future Perspectives. Plant Physiol. 2019, 180, 26. [Google Scholar] [CrossRef] [Green Version]
- Shah, A.; Nazari, M.; Antar, M.; Msimbira, L.A.; Naamala, J.; Lyu, D.; Rabileh, M.; Zajonc, J.; Smith, D.L. PGPR in Agriculture: A Sustainable Approach to Increasing Climate Change Resilience. Front. Sustain. Food Syst. 2021, 5, 667546. [Google Scholar] [CrossRef]
- Singh, J.; Saini, D.K.; Kashyap, R.; Kumar, S.; Chopra, Y.; Sandhu, K.S.; Goraya, M.; Aggarwal, R. Omics technologies for agricultural microbiology research. In Trends of Applied Microbiology for Sustainable Economy; Elsevier: Amsterdam, The Netherlands, 2022; pp. 343–394. [Google Scholar] [CrossRef]
- Nadarajah, K.; Rahman, N.S.N.A. Plant–Microbe Interaction: Aboveground to Belowground, from the Good to the Bad. Int. J. Mol. Sci. 2021, 22, 10388. [Google Scholar] [CrossRef]
- Verma, V.; Ravindran, P.; Kumar, P.P. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016, 16, 86. [Google Scholar] [CrossRef] [Green Version]
- Cohen, A.C.; Bottini, R.; Pontin, M.; Berli, F.J.; Moreno, D.; Boccanlandro, H.; Travaglia, C.N.; Piccoli, P.N. Azospirillum brasilense ameliorates the response of Arabidopsis thaliana to drought mainly via enhancement of ABA levels. Physiol. Plant. 2015, 153, 79–90. [Google Scholar] [CrossRef]
- Kumawat, K.C.; Sharma, B.; Nagpal, S.; Kumar, A.; Tiwari, S.; Nair, R.M. Plant growth-promoting rhizobacteria: Salt stress alleviators to improve crop productivity for sustainable agriculture development. Front. Plant Sci. 2022, 13, 1101862. [Google Scholar] [CrossRef]
- Morcillo, R.J.L.; Manzanera, M. The Effects of Plant-Associated Bacterial Exopolysaccharides on Plant Abiotic Stress Tolerance. Metabolites 2021, 11, 337. [Google Scholar] [CrossRef]
- Vacheron, J.; Desbrosses, G.; Bouffaud, M.-L.; Touraine, B.; Moënne-Loccoz, Y.; Muller, D.; Legendre, L.; Wisniewski-Dyé, F.; Prigent-Combaret, C. Plant growth-promoting rhizobacteria and root system functioning. Front. Plant Sci. 2013, 4, 356. [Google Scholar] [CrossRef] [Green Version]
- Bala, S.; Garg, D.; Thirumalesh, B.V.; Sharma, M.; Sridhar, K.; Inbaraj, B.S.; Tripathi, M. Recent Strategies for Bioremediation of Emerging Pollutants: A Review for a Green and Sustainable Environment. Toxics 2022, 10, 484. [Google Scholar] [CrossRef]
- Gopalakrishnan, S.; Sathya, A.; Vijayabharathi, R.; Varshney, R.K.; Gowda, C.L.; Krishnamurthy, L. Plant growth promoting rhizobia: Challenges and opportunities. 3 Biotech 2015, 5, 355. [Google Scholar] [CrossRef] [Green Version]
- Tariq, M.R.; Shaheen, F.; Mustafa, S.; Ali, S.; Fatima, A.; Shafiq, M.; Safdar, W.; Sheas, M.N.; Hameed, A.; Nasir, M.A. Phosphate solubilizing microorganisms isolated from medicinal plants improve growth of mint. PeerJ 2022, 10, e13782. [Google Scholar] [CrossRef]
- Nadarajah, K.; Kumar, I.S. Molecular Microbial Biodiversity Assessment in the Mycorrhizosphere. In Mycorrhizosphere and Pedogenesis; Springer: Singapore, 2019; pp. 401–420. [Google Scholar] [CrossRef]
- Pérez-Izquierdo, L.; Zabal-Aguirre, M.; González-Martínez, S.C.; Buée, M.; Verdú, M.; Rincón, A.; Goberna, M. Plant intraspecific variation modulates nutrient cycling through its below ground rhizospheric microbiome. J. Ecol. 2019, 107, 1594–1605. [Google Scholar] [CrossRef]
- Lumibao, C.Y.; Bernik, B.M.; Formel, S.K.; Kandalepas, D.; Mighell, K.L.; Pardue, J.; Van Bael, S.A.; Blum, M.J. Rhizosphere microbial communities reflect genotypic and trait variation in a salt marsh ecosystem engineer. Am. J. Bot. 2020, 107, 941–949. [Google Scholar] [CrossRef]
- Dubey, A.; Malla, M.A.; Khan, F.; Chowdhary, K.; Yadav, S.; Kumar, A.; Sharma, S.; Khare, P.K.; Khan, M.L. Soil microbiome: A key player for conservation of soil health under changing climate. Biodivers. Conserv. 2019, 28, 2405–2429. [Google Scholar] [CrossRef]
- Dubey, A.; Kumar, A.; Abd_Allah, E.F.; Hashem, A.; Khan, M.L. Growing more with less: Breeding and developing drought resilient soybean to improve food security. Ecol. Indic. 2019, 105, 425–437. [Google Scholar] [CrossRef]
- Lakshmanan, V.; Selvaraj, G.; Bais, H.P. Functional Soil Microbiome: Belowground Solutions to an Aboveground Problem. Plant Physiol. 2014, 166, 689–700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mendes, R.; Garbeva, P.; Raaijmakers, J.M. The rhizosphere microbiome: Significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 2013, 37, 634–663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Kloepper, J.W.; Ryu, C.-M. Rhizosphere bacteria help plants tolerate abiotic stress. Trends Plant Sci. 2009, 14, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Lundberg, D.S.; Yourstone, S.; Mieczkowski, P.; Jones, C.D.; Dangl, J.L. Practical innovations for high-throughput amplicon sequencing. Nat. Methods 2013, 10, 999–1002. [Google Scholar] [CrossRef]
- Igiehon, N.; Babalola, O. Rhizosphere Microbiome Modulators: Contributions of Nitrogen Fixing Bacteria towards Sustainable Agriculture. Int. J. Environ. Res. Public Health 2018, 15, 574. [Google Scholar] [CrossRef] [Green Version]
- Khalifa, A.Y.Z.; Alsyeeh, A.-M.; Almalki, M.A.; Saleh, F.A. Characterization of the plant growth promoting bacterium, Enterobacter cloacae MSR1, isolated from roots of non-nodulating Medicago sativa. Saudi J. Biol. Sci. 2016, 23, 79–86. [Google Scholar] [CrossRef]
- Mishra, J.; Arora, N.K. Bioformulations for Plant Growth Promotion and Combating Phytopathogens: A Sustainable Approach. In Bioformulations: For Sustainable Agriculture; Springer: New Delhi, India, 2016; pp. 3–33. [Google Scholar] [CrossRef]
- García-Fraile, P.; Menéndez, E.; Rivas, R. Role of bacterial biofertilizers in agriculture and forestry. AIMS Bioeng. 2015, 2, 183–205. [Google Scholar] [CrossRef]
- Khan, N.; Bano, A.; Rahman, M.A.; Guo, J.; Kang, Z.; Babar, M.A. Comparative physiological and metabolic analysis reveals a complex mechanism involved in drought tolerance in Chickpea (Cicer arietinum L.) induced by PGPR and PGRs. Sci. Rep. 2019, 9, 2097. [Google Scholar] [CrossRef] [Green Version]
- Hashem, A.; Kumar, A.; Al-Dbass, A.M.; Alqarawi, A.A.; Al-Arjani, A.-B.F.; Singh, G.; Farooq, M.; Abd_Allah, E.F. Arbuscular mycorrhizal fungi and biochar improves drought tolerance in chickpea. Saudi J. Biol. Sci. 2019, 26, 614–624. [Google Scholar] [CrossRef]
- Wang, Q.; Coleman, J.J. CRISPR/Cas9-mediated endogenous gene tagging in Fusarium oxysporum. Fungal Genet. Biol. 2019, 126, 17–24. [Google Scholar] [CrossRef]
- Moreira, Z.P.M.; Chen, M.Y.; Ortuno, D.L.Y.; Haney, C.H. Engineering plant microbiomes by integrating eco-evolutionary principles into current strategies. Curr. Opin. Plant Biol. 2023, 71, 102316. [Google Scholar] [CrossRef]
- Hayat, R.; Ahmed, I.; Sheirdil, R.A. An Overview of Plant Growth Promoting Rhizobacteria (PGPR) for Sustainable Agriculture. In Crop Production for Agricultural Improvement; Springer: Dordrecht, The Netherlands, 2012; pp. 557–579. [Google Scholar] [CrossRef]
- Kour, D.; Rana, K.L.; Yadav, N.; Yadav, A.N.; Kumar, A.; Meena, V.S.; Singh, B.; Chauhan, V.S.; Dhaliwal, H.S.; Saxena, A.K. Rhizospheric Microbiomes: Biodiversity, Mechanisms of Plant Growth Promotion, and Biotechnological Applications for Sustainable Agriculture. In Plant Growth Promoting Rhizobacteria for Agricultural Sustainability; Springer: Singapore, 2019; pp. 19–65. [Google Scholar] [CrossRef]
- Caulier, S.; Nannan, C.; Gillis, A.; Licciardi, F.; Bragard, C.; Mahillon, J. Overview of the Antimicrobial Compounds Produced by Members of the Bacillus subtilis Group. Front. Microbiol. 2019, 10, 302. [Google Scholar] [CrossRef] [Green Version]
- Hashem, A.; Abd_Allah, E.F.; Alqarawi, A.A.; Radhakrishnan, R.; Kumar, A. Plant defense approach of Bacillus subtilis (BERA 71) against Macrophomina phaseolina (Tassi) Goid in mung bean. J. Plant Interact. 2017, 12, 390–401. [Google Scholar] [CrossRef] [Green Version]
- Tsurumaru, H.; Okubo, T.; Okazaki, K.; Hashimoto, M.; Kakizaki, K.; Hanzawa, E.; Takahashi, H.; Asanome, N.; Tanaka, F.; Sekiyama, Y.; et al. Metagenomic Analysis of the Bacterial Community Associated with the Taproot of Sugar Beet. Microbes Environ. 2015, 30, 63–69. [Google Scholar] [CrossRef] [Green Version]
- Massart, S.; Martinez-Medina, M.; Jijakli, M.H. Biological control in the microbiome era: Challenges and opportunities. Biol. Control. 2015, 89, 98–108. [Google Scholar] [CrossRef]
- Elias, F.; Woyessa, D.; Muleta, D. Phosphate Solubilization Potential of Rhizosphere Fungi Isolated from Plants in Jimma Zone, Southwest Ethiopia. Int. J. Microbiol. 2016, 2016, 5472601. [Google Scholar] [CrossRef] [Green Version]
- Gupta, A.; Sharma, A.; Pathak, R.; Kumar, A.; Sharma, S. Solid State Fermentation of Non-Edible Oil Seed Cakes for Production of Proteases and Cellulases and Degradation of Anti Nutritional Factors. J. Food Biotechnol. Res. 2018, 2, 1–6. [Google Scholar]
- Zamioudis, C.; Pieterse, C.M.J. Modulation of Host Immunity by Beneficial Microbes. Mol. Plant Microbe Interact. 2012, 25, 139–150. [Google Scholar] [CrossRef] [Green Version]
- De Vleesschauwer, D.; Höfte, M. Rhizobacteria-Induced Systemic Resistance. Adv. Bot. Res. 2009, 51, 223–281. [Google Scholar] [CrossRef]
- Chowdhury, F.T.; Islam, M.R.; Islam, M.R.; Khan, H. Diversity of Plant Endophytic Volatile Organic Compound (VOC) and Their Potential Applications. In Endophytes and Secondary Metabolites; Springer International Publishing: Cham, Switzerland, 2018; pp. 1–27. [Google Scholar]
- Farag, M.A.; Zhang, H.; Ryu, C.-M. Dynamic Chemical Communication between Plants and Bacteria through Airborne Signals: Induced Resistance by Bacterial Volatiles. J. Chem. Ecol. 2013, 39, 1007–1018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rajesh, P.; Rai, V.R. Quorum quenching activity in cell-free lysate of endophytic bacteria isolated from Pterocarpus santalinus Linn., and its effect on quorum sensing regulated biofilm in Pseudomonas aeruginosa PAO1. Microbiol. Res. 2014, 169, 561–569. [Google Scholar] [CrossRef] [PubMed]
- Cordovez, V.; Dini-Andreote, F.; Carrión, V.J.; Raaijmakers, J.M. Ecology and Evolution of Plant Microbiomes. Annu. Rev. Microbiol. 2019, 73, 69–88. [Google Scholar] [CrossRef] [PubMed]
- Fierer, N.; Ferrenberg, S.; Flores, G.E.; González, A.; Kueneman, J.; Legg, T.; Lynch, R.C.; McDonald, D.; Mihaljevic, J.R.; O’Neill, S.P.; et al. From Animalcules to an Ecosystem: Application of Ecological Concepts to the Human Microbiome. Annu. Rev. Ecol. Evol. Syst. 2012, 43, 137–155. [Google Scholar] [CrossRef] [Green Version]
- Miller, E.T.; Svanbäck, R.; Bohannan, B.J. Microbiomes as Metacommunities: Understanding Host-Associated Microbes through Metacommunity Ecology. Trends Ecol. Evol. 2018, 33, 926–935. [Google Scholar] [CrossRef]
- Yuan, H.; Li, T.; Li, H.; Wang, C.; Li, L.; Lin, X.; Lin, S. Diversity Distribution, Driving Factors and Assembly Mechanisms of Free-Living and Particle-Associated Bacterial Communities at a Subtropical Marginal Sea. Microorganisms 2021, 9, 2445. [Google Scholar] [CrossRef]
- Wei, Z.; Gu, Y.; Friman, V.-P.; Kowalchuk, G.A.; Xu, Y.; Shen, Q.; Jousset, A. Initial soil microbiome composition and functioning predetermine future plant health. Sci. Adv. 2019, 5, eaaw0759. [Google Scholar] [CrossRef] [Green Version]
- Van Geel, M.; Aavik, T.; Ceulemans, T.; Träger, S.; Mergeay, J.; Peeters, G.; van Acker, K.; Zobel, M.; Koorem, K.; Honnay, O. The role of genetic diversity and arbuscular mycorrhizal fungal diversity in population recovery of the semi-natural grassland plant species Succisa pratensis. BMC Ecol. Evol. 2021, 21, 200. [Google Scholar] [CrossRef]
- Ramírez-Flores, M.R.; Perez-Limon, S.; Li, M.; Barrales-Gamez, B.; Albinsky, D.; Paszkowski, U.; Olalde-Portugal, V.; Sawers, R.J. The genetic architecture of host response reveals the importance of arbuscular mycorrhizae to maize cultivation. Elife 2020, 9, e61701. [Google Scholar] [CrossRef]
- Trivedi, P.; Schenk, P.M.; Wallenstein, M.D.; Singh, B.K. Tiny Microbes, Big Yields: Enhancing food crop production with biological solutions. Microb. Biotechnol. 2017, 10, 999. [Google Scholar] [CrossRef] [Green Version]
- Yin, C.; Hulbert, S.H.; Schroeder, K.L.; Mavrodi, O.; Mavrodi, D.; Dhingra, A.; Schillinger, W.F.; Paulitz, T.C. Role of Bacterial Communities in the Natural Suppression of Rhizoctonia solani Bare Patch Disease of Wheat (Triticum aestivum L.). Appl. Environ. Microbiol. 2013, 79, 7428. [Google Scholar] [CrossRef] [Green Version]
- Lebedev, V.; Lebedeva, T.; Tikhonova, E.; Shestibratov, K. Assessing Impacts of Transgenic Plants on Soil Using Functional Indicators: Twenty Years of Research and Perspectives. Plants 2022, 11, 2439. [Google Scholar] [CrossRef]
- Gévaudant, F.; Duby, G.; von Stedingk, E.; Zhao, R.; Morsomme, P.; Boutry, M. Expression of a Constitutively Activated Plasma Membrane H+-ATPase Alters Plant Development and Increases Salt Tolerance. Plant Physiol. 2007, 144, 1763–1776. [Google Scholar] [CrossRef] [Green Version]
- Cameron, D.D.; Neal, A.L.; Van Wees, S.C.; Ton, J. Mycorrhiza-induced resistance: More than the sum of its parts? Trends Plant Sci. 2013, 18, 545. [Google Scholar] [CrossRef] [Green Version]
- Ellouze, W.; Hamel, C.; Vujanovic, V.; Gan, Y.; Bouzid, S.; St-Arnaud, M. Chickpea genotypes shape the soil microbiome and affect the establishment of the subsequent durum wheat crop in the semiarid North American Great Plains. Soil Biol. Biochem. 2013, 63, 129–141. [Google Scholar] [CrossRef]
- Gu, Y.; Dong, K.; Geisen, S.; Yang, W.; Yan, Y.; Gu, D.; Liu, N.; Borisjuk, N.; Luo, Y.; Friman, V.-P. The effect of microbial inoculant origin on the rhizosphere bacterial community composition and plant growth-promotion. Plant Soil 2020, 452, 105–117. [Google Scholar] [CrossRef]
- Morella, N.M.; Weng, F.C.-H.; Joubert, P.M.; Metcalf, C.J.E.; Lindow, S.; Koskella, B. Successive passaging of a plant-associated microbiome reveals robust habitat and host genotype-dependent selection. Proc. Natl. Acad. Sci. USA 2020, 117, 1148–1159. [Google Scholar] [CrossRef]
- Arias-Sánchez, F.I.; Vessman, B.; Mitri, S. Artificially selecting microbial communities: If we can breed dogs, why not microbiomes? PLoS Biol. 2019, 17, e3000356. [Google Scholar] [CrossRef] [Green Version]
- Thakur, N.; Nigam, M.; Mann, N.A.; Gupta, S.; Hussain, C.M.; Shukla, S.K.; Shah, A.A.; Casini, R.; Elansary, H.O.; Khan, S.A. Host-mediated gene engineering and microbiome-based technology optimization for sustainable agriculture and environment. Funct. Integr. Genom. 2023, 23, 57. [Google Scholar] [CrossRef]
- Trivedi, P.; Batista, B.D.; Bazany, K.E.; Singh, B.K. Plant–microbiome interactions under a changing world: Responses, consequences and perspectives. New Phytol. 2022, 234, 1951–1959. [Google Scholar] [CrossRef]
- Kazan, K.; Lyons, R. The link between flowering time and stress tolerance. J. Exp. Bot. 2016, 67, 47–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Müller, D.B.; Vogel, C.; Bai, Y.; Vorholt, J.A. The Plant Microbiota: Systems-Level Insights and Perspectives. Annu. Rev. Genet. 2016, 50, 211–234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kavamura, V.N.; Robinson, R.J.; Hughes, D.; Clark, I.; Rossmann, M.; de Melo, I.S.; Hirsch, P.R.; Mendes, R.; Mauchline, T.H. Wheat dwarfing influences selection of the rhizosphere microbiome. Sci. Rep. 2020, 10, 1452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taye, Z.M.; Helgason, B.L.; Bell, J.K.; Norris, C.E.; Vail, S.; Robinson, S.J.; Parkin, I.A.P.; Arcand, M.; Mamet, S.; Links, M.G.; et al. Core and Differentially Abundant Bacterial Taxa in the Rhizosphere of Field Grown Brassica napus Genotypes: Implications for Canola Breeding. Front. Microbiol. 2020, 10, 3007. [Google Scholar] [CrossRef] [Green Version]
- Wohor, O.Z.; Rispail, N.; Ojiewo, C.O.; Rubiales, D. Pea Breeding for Resistance to Rhizospheric Pathogens. Plants 2022, 11, 2664. [Google Scholar] [CrossRef]
- Busby, P.E.; Soman, C.; Wagner, M.; Friesen, M.; Kremer, J.; Bennett, A.; Morsy, M.; Eisen, J.A.; Leach, J.E.; Dangl, J.L. Research priorities for harnessing plant microbiomes in sustainable agriculture. PLoS Biol. 2017, 15, e2001793. [Google Scholar] [CrossRef]
- Wille, L.; Messmer, M.M.; Studer, B.; Hohmann, P. Insights to plant-microbe interactions provide opportunities to improve resistance breeding against root diseases in grain legumes. Plant Cell Environ. 2019, 42, 20–40. [Google Scholar] [CrossRef] [Green Version]
- Deng, S.; Caddell, D.F.; Xu, G.; Dahlen, L.; Washington, L.; Yang, J.; Coleman-Derr, D. Genome wide association study reveals plant loci controlling heritability of the rhizosphere microbiome. ISME J. 2021, 15, 3181–3194. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, X.; Sun, S.; Jin, C.; Su, J.; Wei, J.; Luo, X.; Wen, J.; Wei, T.; Sahu, S.K.; et al. GWAS, MWAS and mGWAS provide insights into precision agriculture based on genotype-dependent microbial effects in foxtail millet. Nat. Commun. 2022, 13, 5913. [Google Scholar] [CrossRef]
- Jochum, M.D.; McWilliams, K.L.; Pierson, E.A.; Jo, Y.-K. Host-mediated microbiome engineering (HMME) of drought tolerance in the wheat rhizosphere. PLoS ONE 2019, 14, e0225933. [Google Scholar] [CrossRef] [Green Version]
- Awany, D.; Allali, I.; Dalvie, S.; Hemmings, S.; Mwaikono, K.S.; Thomford, N.E.; Gomez, A.; Mulder, N.; Chimusa, E.R. Host and Microbiome Genome-Wide Association Studies: Current State and Challenges. Front. Genet. 2019, 9, 637. [Google Scholar] [CrossRef]
- Li, Y.; Liang, J.; Deng, B.; Jiang, Y.; Zhu, J.; Chen, L.; Li, M.; Li, J. Applications and Prospects of CRISPR/Cas9-Mediated Base Editing in Plant Breeding. Curr. Issues Mol. Biol. 2023, 45, 918–935. [Google Scholar] [CrossRef]
- Geddes, B.A.; Paramasivan, P.; Joffrin, A.; Thompson, A.L.; Christensen, K.; Jorrin, B.; Brett, P.; Conway, S.J.; Oldroyd, G.E.D.; Poole, P.S. Engineering transkingdom signalling in plants to control gene expression in rhizosphere bacteria. Nat. Commun. 2019, 10, 3430. [Google Scholar] [CrossRef] [Green Version]
- Lebeis, S.L.; Paredes, S.H.; Lundberg, D.S.; Breakfield, N.; Gehring, J.; McDonald, M.; Malfatti, S.; Del Rio, T.G.; Jones, C.D.; Tringe, S.G.; et al. Plant Microbiome. Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science 2015, 349, 860–864. [Google Scholar] [CrossRef] [Green Version]
- Palojärvi, A.; Kellock, M.; Parikka, P.; Jauhiainen, L.; Alakukku, L. Tillage System and Crop Sequence Affect Soil Disease Suppressiveness and Carbon Status in Boreal Climate. Front. Microbiol. 2020, 11, 534786. [Google Scholar] [CrossRef]
- De Corato, U. Soil microbiota manipulation and its role in suppressing soil-borne plant pathogens in organic farming systems under the light of microbiome-assisted strategies. Chem. Biol. Technol. Agric. 2020, 7, 17. [Google Scholar] [CrossRef]
- Hartman, K.; Van Der Heijden, M.G.A.; Wittwer, R.A.; Banerjee, S.; Walser, J.-C.; Schlaeppi, K. Correction to: Cropping practices manipulate abundance patterns of root and soil microbiome members paving the way to smart farming. Microbiome 2020, 8, 14. [Google Scholar] [CrossRef]
- de Souza, R.S.C.; Armanhi, J.S.L.; Damasceno, N.D.B.; Imperial, J.; Arruda, P. Genome Sequences of a Plant Beneficial Synthetic Bacterial Community Reveal Genetic Features for Successful Plant Colonization. Front. Microbiol. 2019, 10, 1779. [Google Scholar] [CrossRef] [Green Version]
- Leonard, S.P.; Powell, J.E.; Perutka, J.; Geng, P.; Heckmann, L.C.; Horak, R.D.; Davies, B.W.; Ellington, A.D.; Barrick, J.E.; Moran, N.A. Engineered symbionts activate honey bee immunity and limit pathogens. Science 2020, 367, 573. [Google Scholar] [CrossRef]
- Goold, H.D.; Wright, P.; Hailstones, D. Emerging Opportunities for Synthetic Biology in Agriculture. Genes 2018, 9, 341. [Google Scholar] [CrossRef] [Green Version]
- Kemal, R.; Islamiah, P.W.N.; Lusiany, T. Synthetic Biology for BioControl: A Mini-Review. In Proceedings of the 6th International Conference on Global Resource Conservation, Malang, Indonesia, 1–2 December 2015; Volume 6, pp. 92–94. [Google Scholar]
- Ryu, M.-H.; Zhang, J.; Toth, T.; Khokhani, D.; Geddes, B.A.; Mus, F.; Garcia-Costas, A.; Peters, J.W.; Poole, P.S.; Ané, J.-M.; et al. Control of nitrogen fixation in bacteria that associate with cereals. Nat. Microbiol. 2019, 5, 314–330. [Google Scholar] [CrossRef] [PubMed]
- Goyal, R.K.; Schmidt, M.A.; Hynes, M.F. Molecular Biology in the Improvement of Biological Nitrogen Fixation by Rhizobia and Extending the Scope to Cereals. Microorganisms 2021, 9, 125. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Diksha; Sindhu, S.S.; Kumar, R. Biofertilizers: An ecofriendly technology for nutrient recycling and environmental sustainability. Curr. Res. Microb. Sci. 2021, 3, 100094. [Google Scholar] [CrossRef] [PubMed]
- Mahmud, K.; Makaju, S.; Ibrahim, R.; Missaoui, A. Current Progress in Nitrogen Fixing Plants and Microbiome Research. Plants 2020, 9, 97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prabhukarthikeyan, S.R.; Parameswaran, C.; Keerthana, U.; Teli, B.; Jagannadham, P.T.K.; Cayalvizhi, B.; Panneerselvam, P.; Senapati, A.; Nagendran, K.; Kumari, S.; et al. Understanding the Plant-microbe Interactions in CRISPR/Cas9 Era: Indeed a Sprinting Start in Marathon. Curr. Genom. 2020, 21, 429–443. [Google Scholar] [CrossRef] [PubMed]
- Glandorf, D.C.M. Re-evaluation of biosafety questions on genetically modified biocontrol bacteria. Eur. J. Plant Pathol. 2019, 154, 43–51. [Google Scholar] [CrossRef]
- Nakamura, M.; Okamura, Y.; Iwai, H. Plasmid-based and -free methods using CRISPR/Cas9 system for replacement of targeted genes in Colletotrichum sansevieriae. Sci. Rep. 2019, 9, 18947. [Google Scholar] [CrossRef] [Green Version]
- Ferrara, M.; Haidukowski, M.; Logrieco, A.F.; Leslie, J.F.; Mulè, G. A CRISPR-Cas9 System for Genome Editing of Fusarium proliferatum. Sci. Rep. 2019, 9, 19836. [Google Scholar] [CrossRef] [Green Version]
- Fang, C.; Chen, X. Potential biocontrol efficacy of Trichoderma atroviride with cellulase expression regulator ace1 gene knock-out. 3 Biotech 2018, 8, 302. [Google Scholar] [CrossRef]
- Xin, W.; Wang, J.; Li, J.; Zhao, H.; Liu, H.; Zheng, H.; Yang, L.; Wang, C.; Yang, F.; Chen, J.; et al. Candidate Gene Analysis for Nitrogen Absorption and Utilization in Japonica Rice at the Seedling Stage Based on a Genome-Wide Association Study. Front. Plant Sci. 2021, 12, 670861. [Google Scholar] [CrossRef]
- Mohapatra, S.; Mishra, S.S.; Bhalla, P.; Thatoi, H. Engineering grass biomass for sustainable and enhanced bioethanol production. Planta 2019, 250, 395–412. [Google Scholar] [CrossRef] [Green Version]
- Luo, M.L.; Leenay, R.T.; Beisel, C.L. Current and future prospects for CRISPR-based tools in bacteria. Biotechnol. Bioeng. 2016, 113, 930–943. [Google Scholar] [CrossRef] [Green Version]
- Wheatley, R.M.; MacLean, R.C. CRISPR-Cas systems restrict horizontal gene transfer in Pseudomonas aeruginosa. ISME J. 2021, 15, 1420–1433. [Google Scholar] [CrossRef]
- Kim, S.-M.; Reinke, R.F. A novel resistance gene for bacterial blight in rice, Xa43(t) identified by GWAS, confirmed by QTL mapping using a bi-parental population. PLoS ONE 2019, 14, e0211775. [Google Scholar] [CrossRef] [Green Version]
- Xiao, Y.; Liu, H.; Wu, L.; Warburton, M.; Yan, J. Genome-wide Association Studies in Maize: Praise and Stargaze. Mol. Plant 2017, 10, 359–374. [Google Scholar] [CrossRef] [Green Version]
- Raboin, L.-M.; Ballini, E.; Tharreau, D.; Ramanantsoanirina, A.; Frouin, J.; Courtois, B.; Ahmadi, N. Association mapping of resistance to rice blast in upland field conditions. Rice 2016, 9, 59. [Google Scholar] [CrossRef] [Green Version]
- Zhu, D.; Kang, H.; Li, Z.; Liu, M.; Zhu, X.; Wang, Y.; Wang, D.; Wang, Z.; Liu, W.; Wang, G.-L. A Genome-Wide Association Study of Field Resistance to Magnaporthe oryzae in Rice. Rice 2016, 9, 44. [Google Scholar] [CrossRef] [Green Version]
- Mgonja, E.M.; Balimponya, E.G.; Kang, H.; Bellizzi, M.; Park, C.H.; Li, Y.; Mabagala, R.; Sneller, C.; Correll, J.; Opiyo, S.; et al. Genome-Wide Association Mapping of Rice Resistance Genes against Magnaporthe oryzae Isolates from Four African Countries. Phytopathology 2016, 106, 1359–1365. [Google Scholar] [CrossRef] [Green Version]
- Bartoli, C.; Roux, F. Genome-Wide Association Studies in Plant Pathosystems: Toward an Ecological Genomics Approach. Front. Plant Sci. 2017, 8, 763. [Google Scholar] [CrossRef] [Green Version]
- Oladzad, A.; González, A.; Macchiavelli, R.; De Jensen, C.E.; Beaver, J.; Porch, T.; McClean, P. Genetic Factors Associated with Nodulation and Nitrogen Derived from Atmosphere in a Middle American Common Bean Panel. Front. Plant Sci. 2020, 11, 576078. [Google Scholar] [CrossRef]
- Horton, M.W.; Bodenhausen, N.; Beilsmith, K.; Meng, D.; Muegge, B.D.; Subramanian, S.; Vetter, M.M.; Vilhjálmsson, B.J.; Nordborg, M.; Gordon, J.I.; et al. Genome-wide association study of Arabidopsis thaliana leaf microbial community. Nat. Commun. 2014, 5, 6320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bartram, A.K.; Lynch, M.D.J.; Stearns, J.C.; Moreno-Hagelsieb, G.; Neufeld, J.D. Generation of Multimillion-Sequence 16S rRNA Gene Libraries from Complex Microbial Communities by Assembling Paired-End Illumina Reads. Appl. Environ. Microbiol. 2011, 77, 3846–3852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M.; et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huttenhower, C.; Gevers, D.; Knight, R.; Abubucker, S.; Badger, J.H.; Chinwalla, A.T.; Creasy, H.H.; Earl, A.M.; FitzGerald, M.G.; Fulton, R.S.; et al. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef] [Green Version]
- Krishna, B.M.; Khan, M.A.; Khan, S.T. Next-Generation Sequencing (NGS) Platforms: An Exciting Era of Genome Sequence Analysis. In Microbial Genomics in Sustainable Agroecosystems; Springer: Singapore, 2019; pp. 89–109. [Google Scholar] [CrossRef]
- Zhang, C.; Li, X.; Yin, L.; Liu, C.; Zou, H.; Wu, Z.; Zhang, Z. Analysis of the complete genome sequence of Brevibacterium frigoritolerans ZB201705 isolated from drought- and salt-stressed rhizosphere soil of maize. Ann. Microbiol. 2019, 69, 1489–1496. [Google Scholar] [CrossRef]
- Duan, J.; Jiang, W.; Cheng, Z.; Heikkila, J.J.; Glick, B.R. The Complete Genome Sequence of the Plant Growth-Promoting Bacterium pseudomonas sp. UW4. PLoS ONE 2013, 8, e58640. [Google Scholar] [CrossRef] [Green Version]
- Johnston-Monje, D.; Mejia, J.L. Botanical microbiomes on the cheap: Inexpensive molecular fingerprinting methods to study plant-associated communities of bacteria and fungi. Appl. Plant Sci. 2020, 8, e11334. [Google Scholar] [CrossRef] [Green Version]
- Afzal, I.; Shinwari, Z.K.; Sikandar, S.; Shahzad, S. Plant beneficial endophytic bacteria: Mechanisms, diversity, host range and genetic determinants. Microbiol. Res. 2019, 221, 36–49. [Google Scholar] [CrossRef]
- Orozco-Mosqueda, M.D.C.; Santoyo, G. Plant-microbial endophytes interactions: Scrutinizing their beneficial mechanisms from genomic explorations. Curr. Plant Biol. 2021, 25, 100189. [Google Scholar] [CrossRef]
- Turner, T.R.; Ramakrishnan, K.; Walshaw, J.; Heavens, D.; Alston, M.; Swarbreck, D.; Osbourn, A.; Grant, A.; Poole, P.S. Comparative metatranscriptomics reveals kingdom level changes in the rhizosphere microbiome of plants. ISME J. 2013, 7, 2248–2258. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Yao, Z.; Sun, Y.; Wang, E.; Tian, C.; Sun, Y.; Liu, J.; Sun, C.; Tian, L. Current Studies of the Effects of Drought Stress on Root Exudates and Rhizosphere Microbiomes of Crop Plant Species. Int. J. Mol. Sci. 2022, 23, 2374. [Google Scholar] [CrossRef]
- Xie, J.; Dawwam, G.E.; Sehim, A.E.; Li, X.; Wu, J.; Chen, S.; Zhang, D. Drought Stress Triggers Shifts in the Root Microbial Community and Alters Functional Categories in the Microbial Gene Pool. Front. Microbiol. 2021, 12, 744897. [Google Scholar] [CrossRef]
- Srivastava, A.K.; Das, A.K.; Jagannadham, P.T.K.; Bora, P.; Ansari, F.A.; Bhate, R. Bioprospecting Microbiome for Soil and Plant Health Management Amidst Huanglongbing Threat in Citrus: A Review. Front. Plant Sci. 2022, 13, 858842. [Google Scholar] [CrossRef]
- Knief, C.; Delmotte, N.; Chaffron, S.; Stark, M.; Innerebner, G.; Wassmann, R.; von Mering, C.; Vorholt, J.A. Metaproteogenomic analysis of microbial communities in the phyllosphere and rhizosphere of rice. ISME J. 2011, 6, 1378–1390. [Google Scholar] [CrossRef] [Green Version]
- Huang, A.C.; Jiang, T.; Liu, Y.-X.; Bai, Y.-C.; Reed, J.; Qu, B.; Goossens, A.; Nützmann, H.-W.; Bai, Y.; Osbourn, A. A specialized metabolic network selectively modulates Arabidopsis root microbiota. Science 2019, 364, aau6389. [Google Scholar] [CrossRef]
- Hong, Y.; Zhou, Q.; Hao, Y.; Huang, A.C. Crafting the plant root metabolome for improved microbe-assisted stress resilience. New Phytol. 2022, 234, 1945–1950. [Google Scholar] [CrossRef]
- Muthuramalingam, P.; Jeyasri, R.; Rakkammal, K.; Satish, L.; Shamili, S.; Karthikeyan, A.; Valliammai, A.; Priya, A.; Selvaraj, A.; Gowri, P.; et al. Multi-Omics and Integrative Approach towards Understanding Salinity Tolerance in Rice: A Review. Biology 2022, 11, 1022. [Google Scholar] [CrossRef]
- Sirangelo, T.M.; Rogers, H.J.; Spadafora, N.D. Multi-Omic Approaches to Investigate Molecular Mechanisms in Peach Post-Harvest Ripening. Agriculture 2022, 12, 553. [Google Scholar] [CrossRef]
- Martínez-Hidalgo, P.; Maymon, M.; Pule-Meulenberg, F.; Hirsch, A.M. Engineering root microbiomes for healthier crops and soils using beneficial, environmentally safe bacteria. Can. J. Microbiol. 2019, 65, 91–104. [Google Scholar] [CrossRef] [Green Version]
- Land, M.; Hauser, L.; Jun, S.-R.; Nookaew, I.; Leuze, M.R.; Ahn, T.-H.; Karpinets, T.; Lund, O.; Kora, G.; Wassenaar, T.; et al. Insights from 20 years of bacterial genome sequencing. Funct. Integr. Genom. 2015, 15, 141. [Google Scholar] [CrossRef] [Green Version]
- Sarhan, M.S.; Hamza, M.A.; Youssef, H.H.; Patz, S.; Becker, M.; ElSawey, H.; Nemr, R.; Daanaa, H.-S.A.; Mourad, E.F.; Morsi, A.T.; et al. Culturomics of the plant prokaryotic microbiome and the dawn of plant-based culture media—A review. J. Adv. Res. 2019, 19, 15–27. [Google Scholar] [CrossRef] [PubMed]
- Fagen, J.R.; Leonard, M.T.; McCullough, C.M.; Edirisinghe, J.N.; Henry, C.S.; Davis, M.J.; Triplett, E.W. Comparative Genomics of Cultured and Uncultured Strains Suggests Genes Essential for Free-Living Growth of Liberibacter. PLoS ONE 2014, 9, e84469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bai, Y.; Müller, D.B.; Srinivas, G.; Garrido-Oter, R.; Potthoff, E.; Rott, M.; Dombrowski, N.; Münch, P.C.; Spaepen, S.; Remus-Emsermann, M.; et al. Functional overlap of the Arabidopsis leaf and root microbiota. Nature 2015, 528, 364–369. [Google Scholar] [CrossRef] [PubMed]
- Tosi, M.; Mitter, E.K.; Gaiero, J.; Dunfield, K.E. It takes three to tango: The importance of microbes, host plant, and soil management to elucidate manipulation strategies for the plant microbiome. Can. J. Microbiol. 2020, 66, 413–433. [Google Scholar] [CrossRef] [PubMed]
- Glick, B.R. The enhancement of plant growth by free-living bacteria. Can. J. Microbiol. 1995, 41, 109–117. [Google Scholar] [CrossRef]
- Kuan, K.B.; Othman, R.; Rahim, K.A.; Shamsuddin, Z.H. Plant Growth-Promoting Rhizobacteria Inoculation to Enhance Vegetative Growth, Nitrogen Fixation and Nitrogen Remobilisation of Maize under Greenhouse Conditions. PLoS ONE 2016, 11, e0152478. [Google Scholar] [CrossRef] [Green Version]
- Vejan, P.; Abdullah, R.; Khadiran, T.; Ismail, S.; Nasrulhaq Boyce, A. Role of Plant Growth Promoting Rhizobacteria in Agricultural Sustainability—A Review. Molecules 2016, 21, 573. [Google Scholar] [CrossRef] [Green Version]
- Sarkar, S.; Kamke, A.; Ward, K.; Hartung, E.; Ran, Q.; Feehan, B.; Galliart, M.; Jumpponen, A.; Johnson, L.; Lee, S.T.M. Culturomics of Andropogon gerardii rhizobiome revealed nitrogen transforming capabilities of stress-tolerant Pseudomonas under drought conditions. bioRxiv 2022, 500515. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, G.; Huang, Y.; Guo, M.; Song, J.; Zhang, T.; Long, Y.; Wang, B.; Liu, H. A Potential Biofertilizer—Siderophilic Bacteria Isolated from the Rhizosphere of Paris polyphylla var. yunnanensis. Front. Microbiol. 2022, 13, 870413. [Google Scholar] [CrossRef]
- Félix, J.D.F.; Sanchez-Juanes, F.; Silva, L.R.; Alves, G. Metagenomic and Culturomic Approaches for Blueberry Biofertilizer Design. Biol. Life Sci. Forum 2021, 3, 49. [Google Scholar] [CrossRef]
- Singh, R.P.; Shelke, G.M.; Kumar, A.; Jha, P.N. Biochemistry and genetics of ACC deaminase: A weapon to “stress ethylene” produced in plants. Front. Microbiol. 2015, 6, 937. [Google Scholar] [CrossRef] [Green Version]
- del Carmen Orozco-Mosqueda, M.; Glick, B.R.; Santoyo, G. ACC deaminase in plant growth-promoting bacteria (PGPB): An efficient mechanism to counter salt stress in crops. Microbiol. Res. 2020, 235, 126439. [Google Scholar] [CrossRef]
- Glick, B.R.; Penrose, D.M.; Li, J. A Model For the Lowering of Plant Ethylene Concentrations by Plant Growth-promoting Bacteria. J. Theor. Biol. 1998, 190, 63–68. [Google Scholar] [CrossRef]
- Chicca, I.; Becarelli, S.; Bernabei, G.; Siracusa, G.; Di Gregorio, S. Innovative Culturomic Approaches and Predictive Functional Metagenomic Analysis: The Isolation of Hydrocarbonoclastic Bacteria with Plant Growth Promoting Capacity. Water 2022, 14, 142. [Google Scholar] [CrossRef]
- Tsukanova, K.A.; Chebotar, V.K.; Meyer, J.J.M.; Bibikova, T.N. Effect of plant growth-promoting Rhizobacteria on plant hormone homeostasis. S. Afr. J. Bot. 2017, 113, 91–102. [Google Scholar] [CrossRef]
- Sahu, K.P.; Patel, A.; Kumar, M.; Sheoran, N.; Mehta, S.; Reddy, B.; Eke, P.; Prabhakaran, N.; Kumar, A. Integrated Metabarcoding and Culturomic-Based Microbiome Profiling of Rice Phyllosphere Reveal Diverse and Functional Bacterial Communities for Blast Disease Suppression. Front. Microbiol. 2021, 12, 780458. [Google Scholar] [CrossRef]
- Haque, M.; Mosharaf, K.; Khatun, M.; Haque, A.; Biswas, S.; Islam, S.; Islam, M.; Shozib, H.B.; Miah, M.U.; Molla, A.H.; et al. Biofilm Producing Rhizobacteria with Multiple Plant Growth-Promoting Traits Promote Growth of Tomato Under Water-Deficit Stress. Front. Microbiol. 2020, 11, 542053. [Google Scholar] [CrossRef]
- Flores-Duarte, N.J.; Pajuelo, E.; Mateos-Naranjo, E.; Navarro-Torre, S.; Rodríguez-Llorente, I.D.; Redondo-Gómez, S.; López, J.A.C. A Culturomics-Based Bacterial Synthetic Community for Improving Resilience towards Arsenic and Heavy Metals in the Nutraceutical Plant Mesembryanthemum crystallinum. Int. J. Mol. Sci. 2023, 24, 7003. [Google Scholar] [CrossRef]
- Bouremani, N.; Cherif-Silini, H.; Silini, A.; Bouket, A.C.; Luptakova, L.; Alenezi, F.N.; Baranov, O.; Belbahri, L. Plant Growth-Promoting Rhizobacteria (PGPR): A Rampart against the Adverse Effects of Drought Stress. Water 2023, 15, 418. [Google Scholar] [CrossRef]
- Rijavec, T.; Lapanje, A. Hydrogen Cyanide in the Rhizosphere: Not Suppressing Plant Pathogens, but Rather Regulating Availability of Phosphate. Front. Microbiol. 2016, 7, 1785. [Google Scholar] [CrossRef] [Green Version]
- Santoyo, G.; Urtis-Flores, C.A.; Loeza-Lara, P.D.; Orozco-Mosqueda, M.D.C.; Glick, B.R. Rhizosphere Colonization Determinants by Plant Growth-Promoting Rhizobacteria (PGPR). Biology 2021, 10, 475. [Google Scholar] [CrossRef] [PubMed]
- Glick, B.R. Plant Growth-Promoting Bacteria: Mechanisms and Applications. Scientifica 2012, 2012, 963401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saeed, Q.; Xiukang, W.; Haider, F.U.; Kučerik, J.; Mumtaz, M.Z.; Holatko, J.; Naseem, M.; Kintl, A.; Ejaz, M.; Naveed, M.; et al. Rhizosphere Bacteria in Plant Growth Promotion, Biocontrol, and Bioremediation of Contaminated Sites: A Comprehensive Review of Effects and Mechanisms. Int. J. Mol. Sci. 2021, 22, 10529. [Google Scholar] [CrossRef] [PubMed]
- Ajijah, N.; Fiodor, A.; Pandey, A.K.; Rana, A.; Pranaw, K. Plant Growth-Promoting Bacteria (PGPB) with Biofilm-Forming Ability: A Multifaceted Agent for Sustainable Agriculture. Diversity 2023, 15, 112. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nadarajah, K.; Abdul Rahman, N.S.N. The Microbial Connection to Sustainable Agriculture. Plants 2023, 12, 2307. https://doi.org/10.3390/plants12122307
Nadarajah K, Abdul Rahman NSN. The Microbial Connection to Sustainable Agriculture. Plants. 2023; 12(12):2307. https://doi.org/10.3390/plants12122307
Chicago/Turabian StyleNadarajah, Kalaivani, and Nur Sabrina Natasha Abdul Rahman. 2023. "The Microbial Connection to Sustainable Agriculture" Plants 12, no. 12: 2307. https://doi.org/10.3390/plants12122307
APA StyleNadarajah, K., & Abdul Rahman, N. S. N. (2023). The Microbial Connection to Sustainable Agriculture. Plants, 12(12), 2307. https://doi.org/10.3390/plants12122307





