Characterization of the PIN Auxin Efflux Carrier Gene Family and Its Expression during Zygotic Embryogenesis in Persea americana
Abstract
1. Introduction
2. Results
2.1. Identification of PIN Gene Family in Avocado
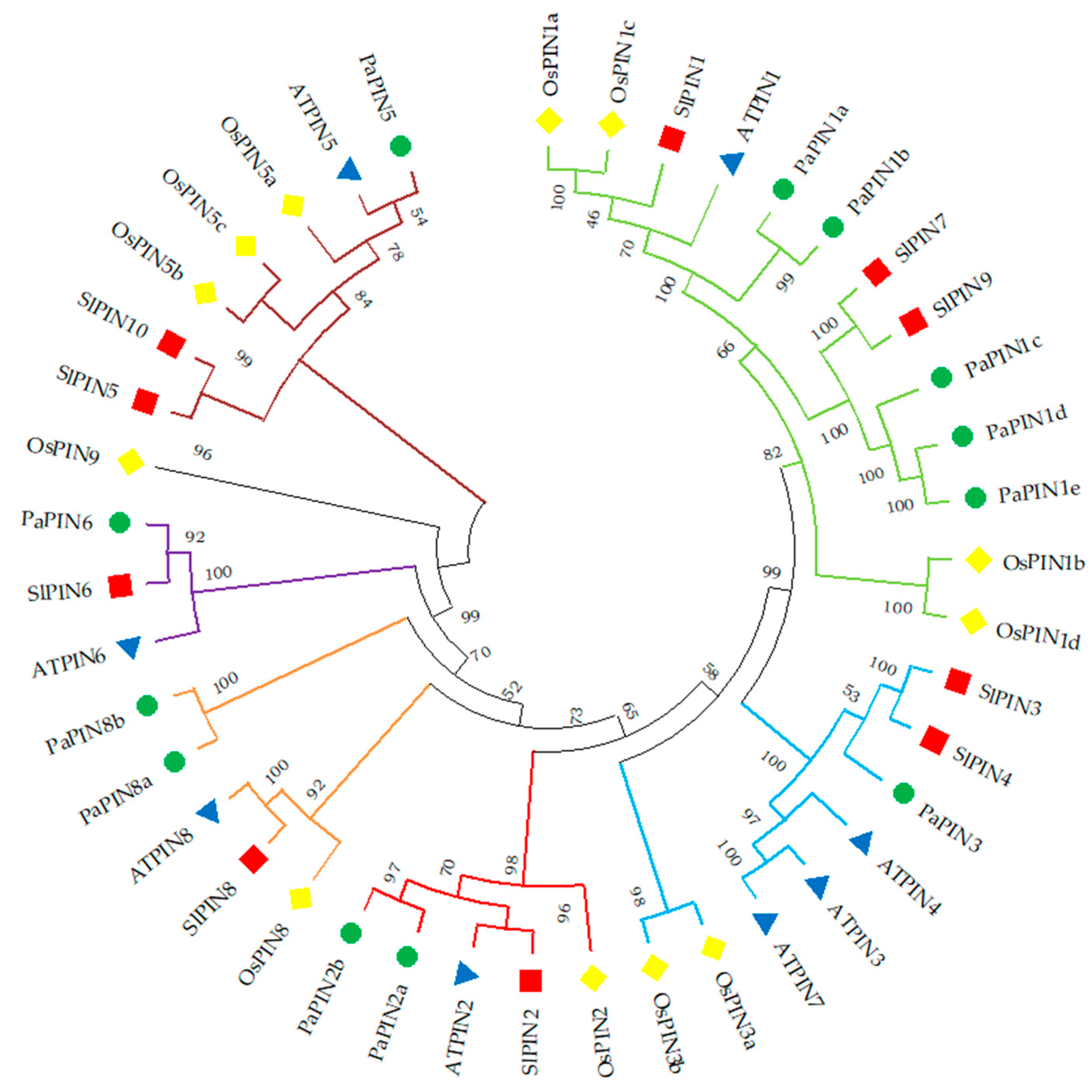
2.2. Phylogenetic Analysis of PaPIN Family
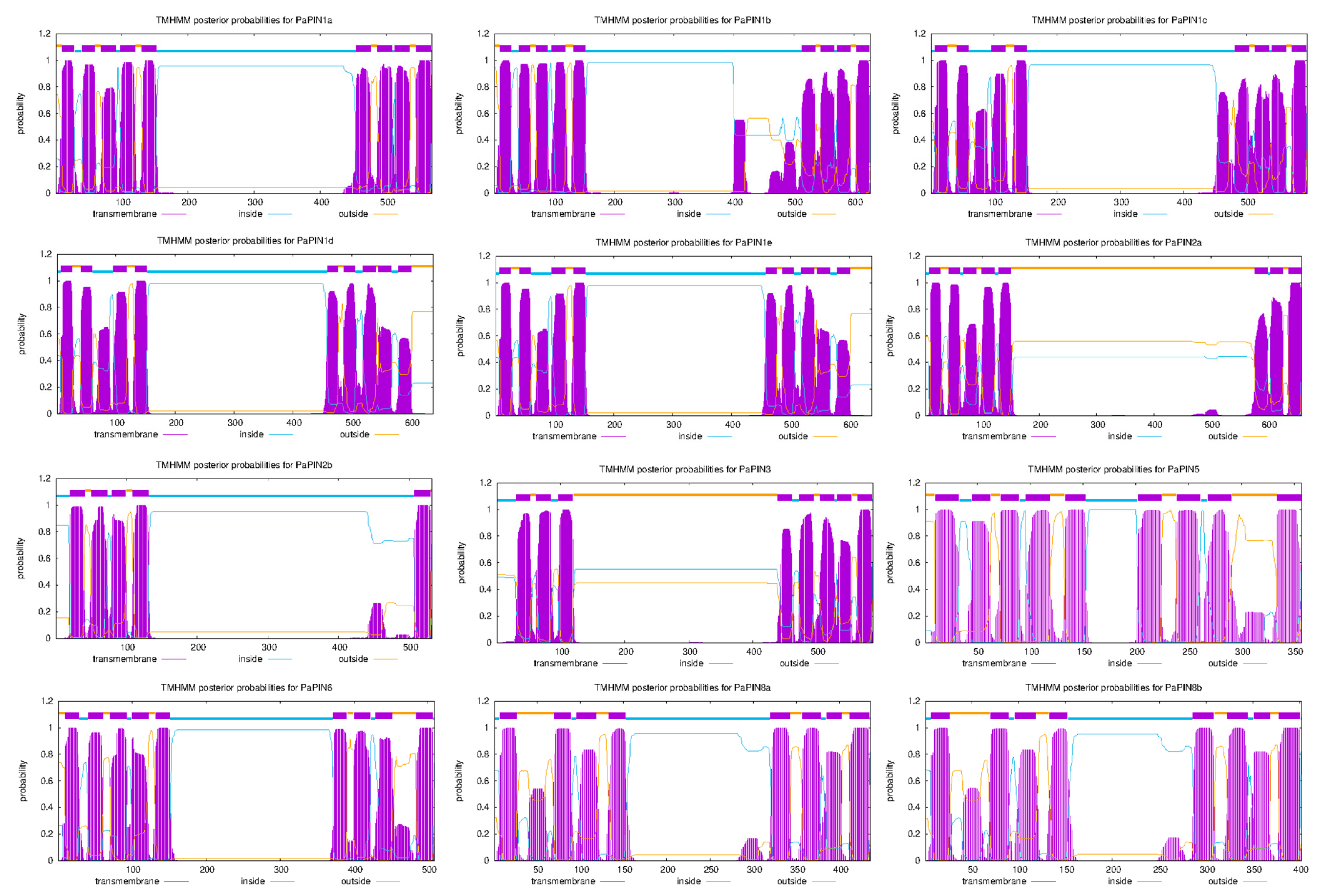
2.3. Gene Structure Analysis, Transmembrane Region Prediction and Conserved Motifs of PIN Genes in Avocado
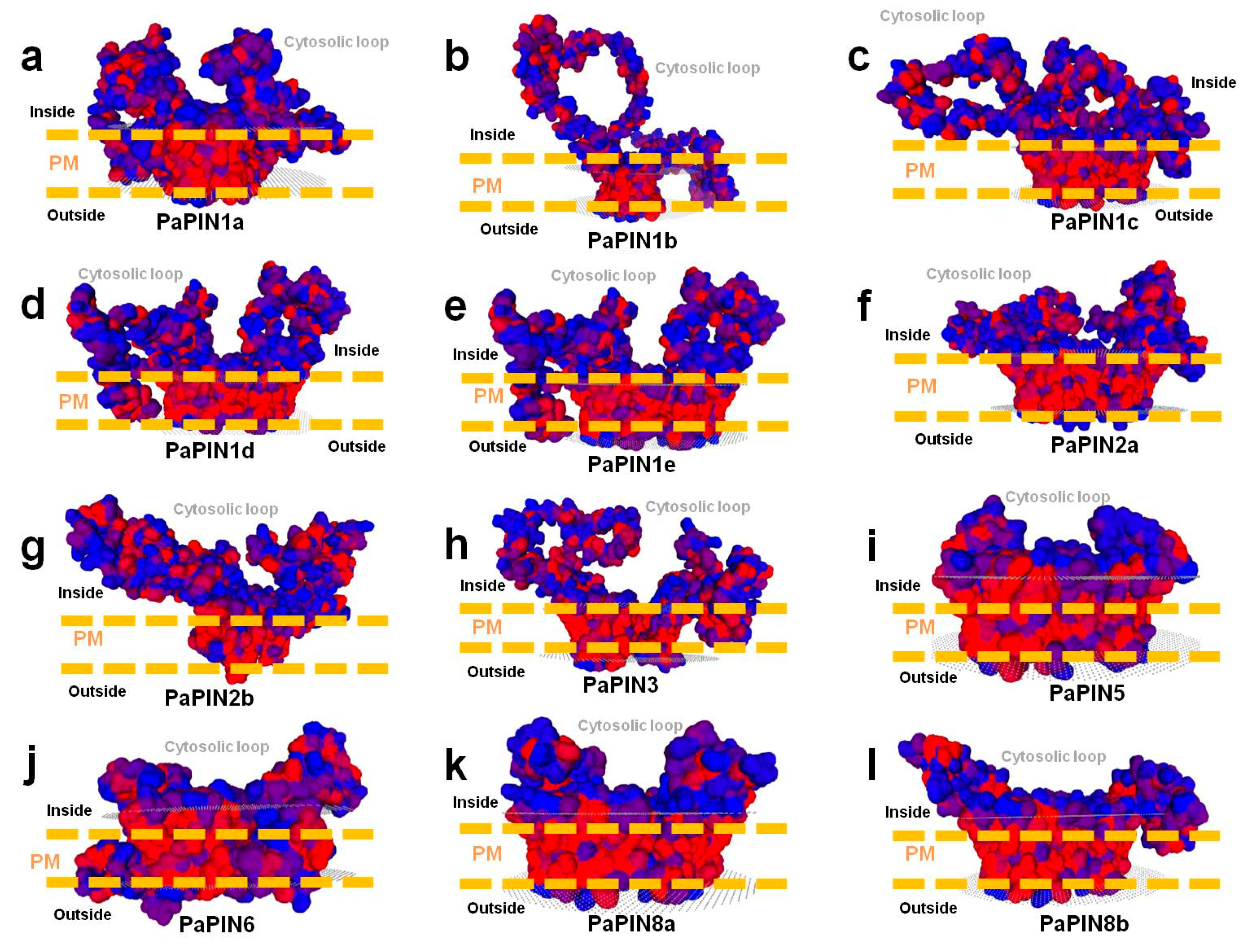
2.4. Building of 3D Structures, Molecular Modeling and Multiple-Sequence Alignment of PaPIN Proteins from P. americana
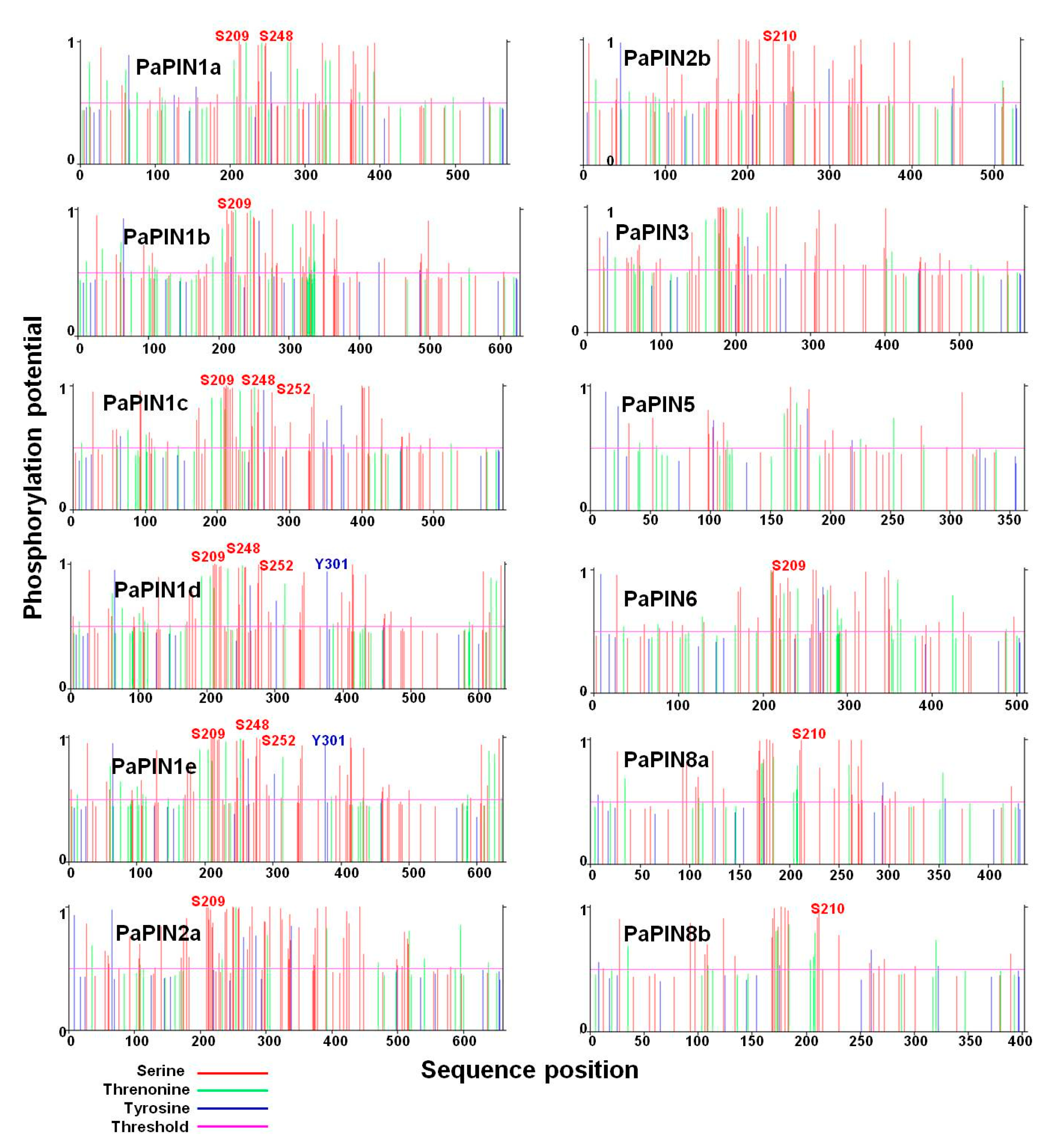
2.5. Prediction of the Phosphorylation Sites in the PaPIN Proteins
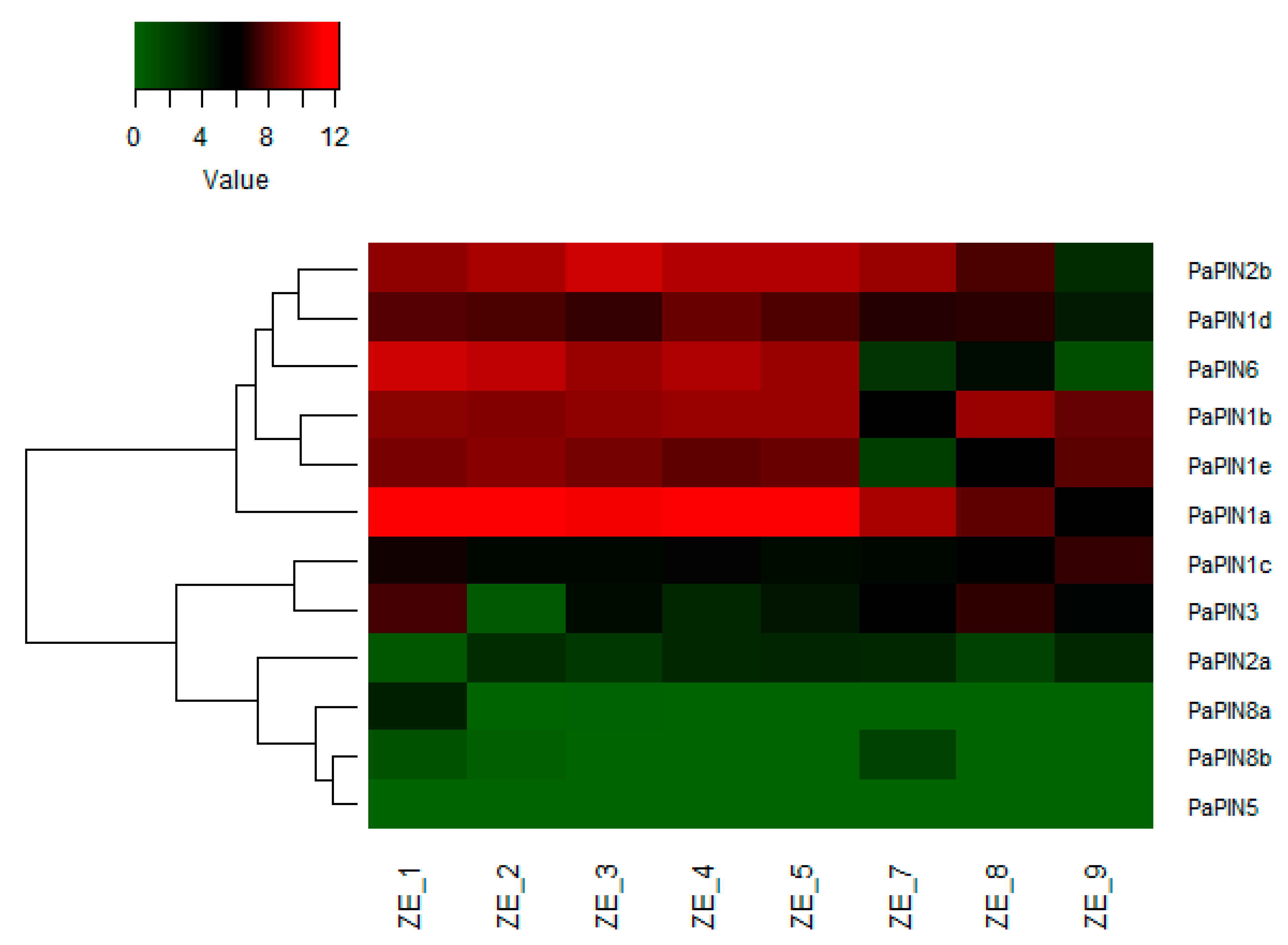
2.6. Differential Expression Analysis of PINs in Zygotic Embryo in Avocado
3. Discussion
4. Materials and Methods
4.1. Phylogenetic Tree Construction
4.2. Identification and Theoretical Characterization of the PaPIN Protein Family in Avocado
4.3. Gene Structure and Motif Analysis
4.4. Plant Material, RNA Extraction and Transcriptome Analysis
4.5. Building of 3D Structures, Molecular Modelling, Multiple-Sequence Alignment and Prediction of Transmembrane Segments of PaPIN1 Proteins from Persea americana
4.6. Prediction of the Phosphorylation Sites
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Geisler, M.M. A retro-perspective on auxin transport. Front. Plant Sci. 2021, 12, 756968. [Google Scholar] [CrossRef] [PubMed]
- Aloni, R. The hormonal signals that regulate plant vascular differentiation. In Vascular Differentiation and Plant Hormones; Aloni, R., Ed.; Springer: Cham, Switzerland, 2021; pp. 55–96. [Google Scholar]
- Weijers, D.; Nemhauser, J.; Yang, Z. Auxin: Small molecule, big impact. J. Exp. Bot. 2018, 69, 133–136. [Google Scholar] [CrossRef] [PubMed]
- Friml, J. Fourteen stations of auxin. Cold Spring Harb. Perspect. Biol. 2021, 14, a039859. [Google Scholar] [CrossRef] [PubMed]
- Robert, H.S.; Crhak Khaitova, L.; Mroue, S.; Benková, E. The importance of localized auxin production for morphogenesis of reproductive organs and embryos in Arabidopsis. J. Exp. Bot. 2015, 66, 5029–5042. [Google Scholar] [CrossRef]
- Yang, Z.; Xia, J.; Hong, J.; Zhang, C.; Wei, H.; Ying, W.; Sun, C.; Sun, L.; Mao, Y.; Gao, Y.; et al. Structural insights into auxin recognition and efflux by Arabidopsis PIN1. Nature 2022, 609, 611–615. [Google Scholar] [CrossRef]
- Carrillo-Carrasco, V.P.; Hernandez-Garcia, J.; Mutte, S.K.; Weijers, D. The birth of a giant: Evolutionary insights into the origin of auxin responses in plants. EMBO J. 2023, 42, e113018. [Google Scholar] [CrossRef]
- Weijers, D.; Wagner, D. Transcriptional responses to the auxin hormone. Annu. Rev. Plant Biol. 2016, 67, 539–574. [Google Scholar] [CrossRef]
- Hammes, U.Z.; Murphy, A.S.; Schwechheimer, C. Auxin transporters—A biochemical view. Cold Spring Harbor Perspect. Biol. 2021, 14, a039875. [Google Scholar] [CrossRef]
- Swarup, R.; Péret, B. AUX/LAX family of auxin influx carriers—An overview. Front. Plant Sci. 2012, 3, 225. [Google Scholar] [CrossRef]
- Péret, B.; Swarup, K.; Ferguson, A.; Seth, M.; Yang, Y.; Dhondt, S.; James, N.; Casimiro, I.; Perry, P.; Syed, A.; et al. AUX/LAX genes encode a family of auxin influx transporters that perform distinct functions during Arabidopsis development. Plant Cell 2012, 24, 2874–2885. [Google Scholar] [CrossRef]
- Geisler, M.; Aryal, B.; di Donato, M.; Hao, P. A critical view on ABC transporters and their interacting partners in auxin transport. Plant Cell Physiol. 2017, 58, 1601–1614. [Google Scholar] [CrossRef]
- Jenness, M.K.; Carraro, N.; Pritchard, C.A.; Murphy, A.S. The Arabidopsis ATP-BINDING CASSETTE transporter ABCB21 regulates auxin levels in cotyledons, the root pericycle, and leaves. Front. Plant Sci. 2019, 10, 806. [Google Scholar] [CrossRef]
- Mohan, A.; Dhaliwal, A.K.; Nagarajan, R.; Gill, K.S. Molecular characterization of auxin efflux carrier—ABCB1 in hexaploid wheat. Sci. Rep. 2019, 9, 17327. [Google Scholar] [CrossRef]
- Krecek, P.; Skupa, P.; Libus, J.; Naramoto, S.; Tejos, R.; Friml, J.; Zazimalová, E. The PIN-FORMED (PIN) protein family of auxin transporters. Genome Biol. 2009, 10, 249. [Google Scholar] [CrossRef]
- Zhou, J.J.; Luo, J. The PIN-FORMED auxin efflux carriers in plants. Int. J. Mol. Sci. 2018, 19, 2759. [Google Scholar] [CrossRef]
- Lam Ung, K.; Winkler, M.; Schulz, L.; Kolb, M.; Janacek, D.P.; Dedic, E.; Stokes, D.L.; Hammes, U.Z.; Pedersen, B.P. Structures and mechanism of the plant PIN-FORMED auxin transporter. Nature 2022, 609, 605–610. [Google Scholar] [CrossRef]
- Barbez, E.; Kubes, M.; Rolcik, J.; Beziat, C.; Pencik, A.; Wang, B.; Rosquete, M.R.; Zhu, J.; Dobrev, P.I.; Lee, Y.; et al. A novel putative auxin carrier family regulates intracellular auxin homeostasis in plants. Nature 2012, 485, 119–122. [Google Scholar] [CrossRef]
- Feraru, E.; Vosolsobe, S.; Feraru, M.I.; Petrášek, J.; Kleine-Vehn, J. Evolution and structural diversification of PILS putative auxin carriers in plants. Front. Plant Sci. 2012, 3, 227. [Google Scholar] [CrossRef]
- Krouk, G.; Lacombe, B.; Bielach, A.; Perrine-Walker, F.; Malinska, K.; Mounier, E.; Hoyerova, K.; Tillard, P.; Leon, S.; Ljung, K.; et al. Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev. Cell 2010, 18, 927–937. [Google Scholar] [CrossRef]
- Ranocha, P.; Dima, O.; Nagy, R.; Felten, J.; Corratgé-Faillie, C.; Novák, O.; Morreel, K.; Lacombe, B.; Martinez, Y.; Pfrunder, S.; et al. Arabidopsis WAT1 is a vacuolar auxin transport facilitator required for auxin homoeostasis. Nat. Commun. 2013, 4, 2625. [Google Scholar] [CrossRef]
- Marhava, P. Recent developments in the understanding of PIN polarity. New Phytol. 2022, 233, 624–630. [Google Scholar] [CrossRef] [PubMed]
- Vosolsobê, S.; Skokan, R.; Petrášek, J. The evolutionary origins of auxin transport: What we know and what we need to know. J. Exp. Bot. 2020, 71, 3287–3295. [Google Scholar] [CrossRef]
- Bennett, T.; Brockington, S.F.; Rothfels, C.; Graham, S.W.; Stevenson, D.; Kutchan, T.; Rolf, M.; Thomas, P.; Wong, G.K.-S.; Leyser, O.; et al. Paralogous radiations of PIN proteins with multiple origins of noncanonical PIN structure. Mol. Biol. Evol. 2014, 31, 2042–2060. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Hu, H.; Wang, G.; Li, J.; Chen, J.; Wu, P. Expression of PIN genes in rice (Oryza sativa L.): Tissue specificity and regulation by hormones. Mol. Plant 2009, 2, 823–831. [Google Scholar] [CrossRef] [PubMed]
- Pattison, R.J.; Catalá, C. Evaluating auxin distribution in tomato (Solanum lycopersicum) through an analysis of the PIN and AUX/LAX gene families. Plant J. 2012, 70, 585–598. [Google Scholar] [CrossRef]
- Wang, Y.; Chai, C.; Valliyodan, B.; Maupin, C.; Annen, B.; Nguyen, H.T. Genome-wide analysis and expression profiling of the PIN auxin transporter gene family in soybean (Glycine max). BMC Genom. 2015, 16, 951. [Google Scholar] [CrossRef]
- Liu, B.; Zhang, J.; Wang, L.; Li, J.; Zheng, H.; Chen, J.; Lu, M. A survey of Populus PIN-FORMED family genes reveals their diversified expression patterns. J. Exp. Bot. 2014, 65, 2437–2448. [Google Scholar] [CrossRef]
- Kumar, M.; Kherawat, B.S.; Dey, P.; Saha, D.; Singh, A.; Bhatia, S.K.; Ghodake, G.S.; Kadam, A.A.; Kim, H.U.; Manorama; et al. Genome-wide identification and characterization of PIN-FORMED (PIN) gene family reveals role in developmental and various stress conditions in Triticum aestivum L. Int. J. Mol. Sci. 2021, 22, 7396. [Google Scholar] [CrossRef]
- Huang, X.; Bai, X.; Guo, T.; Xie, Z.; Laimer, M.; Du, D.; Gbokie, T.; Zhang, Z.; He, C.; Lu, Y.; et al. Genome-wide analysis of the PIN auxin efflux carrier gene family in Coffee. Plants 2020, 9, 1061. [Google Scholar] [CrossRef]
- Chen, L.; Cai, M.; Chen, M.; Ke, W.; Pan, Y.; Huang, J.; Zhang, J.; Peng, C. Genome-wide characterization of PIN auxin efflux carrier gene family in Mikania micrantha. Int. J. Mol. Sci. 2022, 23, 10183. [Google Scholar] [CrossRef]
- Xiao, S.M.; Chu, Y.; Chen, Y.J.; Zhao, Q.H.; Liao, B.S.; Zhang, J.J.; Gao, Y.; Xu, J.; Chen, S.L. Genome-wide identification and transcriptional profiling analysis of PIN/PILS auxin transporter gene families in Panax ginseng. Chin. Herb. Med. 2022, 14, 48–57. [Google Scholar] [CrossRef]
- Sanko-Sawczenko, I.; Lotocka, B.; Czarnocka, W. Expression analysis of PIN genes in root tips and nodules of Medicago truncatula. Int. J. Mol. Sci. 2016, 17, 1197. [Google Scholar] [CrossRef]
- Yue, R.; Tie, S.; Sun, T.; Zhang, L.; Yang, Y.; Qi, J.; Yan, S.; Han, X.; Wang, H.; Shen, C. Genome-wide identification and expression profiling analysis of ZmPIN, ZmPILS, ZmLAX and ZmABCB auxin transporter gene families in maize (Zea mays L.) under various abiotic stresses. PLoS ONE 2015, 10, e0118751. [Google Scholar] [CrossRef]
- Long, Y.; Chen, Q.; Qu, Y.; Liu, P.; Jiao, Y.; Cai, Y.; Deng, X.; Zheng, K. Identification and functional analysis of PIN family genes in Gossypium barbadense. PeerJ 2022, 10, e14236. [Google Scholar] [CrossRef]
- Cardoso, H.; Campos, C.; Grzebelus, D.; Egas, C.; Peixe, A. Understanding the role of PIN auxin carrier genes under biotic and abiotic stresses in Olea europaea L. Biology 2022, 11, 1040. [Google Scholar] [CrossRef]
- Nodzynski, T.; Vanneste, S.; Zwiewka, M.; Pernisová, M.; Hejátko, J.; Friml, J. Enquiry into the topology of plasma membrane-localized PIN auxin transport components. Mol. Plant 2016, 9, 1504–1519. [Google Scholar] [CrossRef]
- Simon, S.; Skupa, P.; Viaene, T.; Zwiewka, M.; Tejos, R.; Klíma, P.; Carná, M.; Rolcík, J.; De Rycke, R.; Moreno, I.; et al. PIN6 auxin transporter at endoplasmic reticulum and plasma membrane mediates auxin homeostasis and organogenesis in Arabidopsis. New Phytol. 2016, 211, 65–74. [Google Scholar] [CrossRef]
- Estrella-Maldonado, H.; Fuentes Ortíz, G.; Chan León, A.C.; Rodríguez Zapata, L.C.; Talavera May, C.; Gil, F.; Barredo Pool, F.; Idrovo Espín, F.M.; Santamaría, J.M. The papaya CpAUX1/LAX and CpPIN genes: Structure, phylogeny and expression analysis related to root formation on in vitro plantlets. Plant Cell Tissue Organ Cult. 2016, 126, 187–204. [Google Scholar] [CrossRef]
- Viaene, T.; Delwiche, C.F.; Rensing, S.A.; Friml, J. Origin and evolution of PIN auxin transporters in the green lineage. Trends Plant Sci. 2013, 18, 5–10. [Google Scholar] [CrossRef]
- Zhang, C.; Dong, W.; Huang, Z.A.; Cho, M.; Yu, Q.; Wu, C.; Yu, C. Genome-wide identification and expression analysis of the CaLAX and CaPIN gene families in pepper (Capsicum annuum L.) under various abiotic stresses and hormone treatments. Genome 2018, 61, 121–130. [Google Scholar] [CrossRef]
- Yu, C.; Dong, W.; Zhan, Y.; Huang, Z.A.; Li, Z.; Kim, I.S.; Zhang, C. Genome-wide identification and expression analysis of ClLAX, ClPIN and ClABCB genes families in Citrullus lanatus under various abiotic stresses and grafting. BMC Genet. 2017, 18, 33. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Bayaer, E.; Qi, Y. Advances in the biological functions of auxin transporters in rice. Agriculture 2022, 12, 989. [Google Scholar] [CrossRef]
- Forestan, C.; Farinati, S.; Varotto, S. The maize PIN gene family of auxin transporters. Front. Plant Sci. 2012, 3, 16. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; He, P.; Yang, Z.; Huang, G.; Wang, L.; Pang, C.; Xiao, H.; Zhao, P.; Yu, J.; Xiao, G. A genome-scale analysis of the PIN gene family reveals its functions in cotton fiber development. Front. Plant Sci. 2017, 8, 461. [Google Scholar] [CrossRef]
- He, P.; Zhao, P.; Wang, L.; Zhang, Y.; Wang, X.; Xiao, H.; Yu, J.; Xiao, G. The PIN gene family in cotton (Gossypium hirsutum): Genome-wide identification and gene expression analyses during root development and abiotic stress responses. BMC Genom. 2017, 18, 507. [Google Scholar] [CrossRef]
- Xie, X.; Qin, G.; Si, P.; Luo, Z.; Gao, J.; Chen, X.; Zhang, J.; Wei, P.; Xia, Q.; Lin, F.; et al. Analysis of Nicotiana tabacum PIN genes identifies NtPIN4 as a key regulator of axillary bud growth. Physiol. Plant. 2017, 160, 222–239. [Google Scholar] [CrossRef]
- Liu, Y.; Wei, H. Genome-wide identification and evolution of the PIN-FORMED (PIN) gene family in Glycine max. Genome 2017, 60, 564–571. [Google Scholar] [CrossRef]
- Bai, Y.; Dou, Y.; Xie, Y.; Zheng, H.; Gao, J. Phylogeny, transcriptional profile, and auxin-induced phosphorylation modification characteristics of conserved PIN proteins in Moso bamboo (Phyllostachys edulis). Int. J. Biol. Macromol. 2023, 234, 123671. [Google Scholar] [CrossRef]
- Forestan, C.; Meda, S.; Varotto, S. ZmPIN1-mediated auxin transport is related to cellular differentiation during maize embryogenesis and endosperm development. Plant Physiol. 2010, 152, 1373–1390. [Google Scholar] [CrossRef]
- Xu, M.; Zhu, L.; Shou, H.; Wu, P. A PIN1 family gene, OsPIN1, involved in auxin-dependent adventitious root emergence and tillering in rice. Plant Cell Physiol. 2005, 46, 1674–1681. [Google Scholar] [CrossRef]
- Wang, H.; Ouyang, Q.; Yang, C.; Zhang, Z.; Hou, D.; Liu, H.; Xu, H. Mutation of OsPIN1b by CRISPR/Cas9 reveals a role for auxin transport in modulating rice architecture and root gravitropism. Int. J. Mol. Sci. 2022, 23, 8965. [Google Scholar] [CrossRef]
- Zhang, Y.; Han, S.; Lin, Y.; Qiao, J.; Han, N.; Li, Y.; Feng, Y.; Li, D.; Qi, Y. Auxin transporter OsPIN1b, a novel regulator of leaf inclination in rice (Oryza sativa L.). Plants 2023, 12, 409. [Google Scholar] [CrossRef]
- Lee, H.; Ganguly, A.; Lee, R.D.; Park, M.; Cho, H.T. Intracellularly localized PIN-FORMED8 promotes lateral root emergence in Arabidopsis. Front. Plant Sci. 2020, 10, 1808. [Google Scholar] [CrossRef]
- Larsen, P.B.; Cancel, J.; Rounds, M.; Ochoa, V. Arabidopsis ALS1 encodes a root tip and stele localized half type ABC transporter required for root growth in an aluminum toxic environment. Planta 2007, 225, 1447–1458. [Google Scholar] [CrossRef]
- Friml, J.; Benková, E.; Blilou, I.; Wisniewska, J.; Hamann, T.; Ljung, K.; Woody, S.; Sandberg, G.; Scheres, B.; Jürgens, G.; et al. AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell 2002, 108, 661–673. [Google Scholar] [CrossRef]
- Friml, J.; Vieten, A.; Sauer, M.; Weijers, D.; Schwarz, H.; Hamann, T.; Offringa, R.; Jurgens, G. Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 2003, 426, 147–153. [Google Scholar] [CrossRef]
- Baba, A.I.; Rigó, G.; Ayaydin, F.; Rehman, A.U.; Andrási, N.; Zsigmond, L.; Valkai, I.; Urbancsok, J.; Vass, I.; Pasternak, T.; et al. Functional analysis of the Arabidopsis thaliana CDPK-related kinase family: AtCRK1 regulates responses to continuous light. Int. J. Mol. Sci. 2018, 19, 1282. [Google Scholar] [CrossRef]
- Baba, A.I.; Valkai, I.; Labhane, N.M.; Koczka, L.; Andrási, N.; Klement, É.; Darula, Z.; Medzihradszky, K.F.; Szabados, L.; Fehér, A.; et al. CRK5 protein kinase contributes to the progression of embryogenesis of Arabidopsis thaliana. Int. J. Mol. Sci. 2019, 20, 6120. [Google Scholar] [CrossRef]
- Rigó, G.; Ayaydin, F.; Tietz, O.; Zsigmond, L.; Kovács, H.; Páy, A.; Salchert, K.; Darula, Z.; Medzihradszky, K.F.; Szabados, L.; et al. Inactivation of plasma membrane-localized CDPK-RELATED KINASE5 decelerates PIN2 exocytosis and root gravitropic response in Arabidopsis. Plant Cell 2013, 25, 1592–1608. [Google Scholar] [CrossRef]
- Baba, A.I.; Andrási, N.; Valkai, I.; Gorcsa, T.; Koczka, L.; Darula, Z.; Medzihradszky, K.F.; Szabados, L.; Fehér, A.; Rigó, G.; et al. AtCRK5 protein kinase exhibits a regulatory role in hypocotyl hook development during skotomorphogenesis. Int. J. Mol. Sci. 2019, 20, 3432. [Google Scholar] [CrossRef]
- Thomas, C.; Tampé, R. Structural and mechanistic principles of ABC transporters. Annu. Rev. Biochem. 2020, 89, 605–636. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Yu, N.; Ju, M.; Fan, B.; Zhang, Y.; Zhu, E.; Zhang, M.; Zhang, K. ABC transporter OsABCG18 controls the shootward transport of cytokinins and grain yield in rice. J. Exp. Bot. 2019, 70, 6277–6291. [Google Scholar] [CrossRef] [PubMed]
- Geisler, M.; Blakeslee, J.J.; Bouchard, R.; Lee, O.R.; Vincenzetti, V.; Bandyopadhyay, A.; Titapiwatanakun, B.; Peer, W.A.; Bailly, A.; Richards, E.L.; et al. Cellular efflux of auxin catalyzed by the Arabidopsis MDR/PGP transporter AtPGP1. Plant J. 2005, 44, 179–194. [Google Scholar] [CrossRef] [PubMed]
- Kubes, M.; Yang, H.; Richter, G.L.; Cheng, Y.; Mtodzinska, E.; Wang, X.; Blakeslee, J.J.; Carraro, N.; Petrášek, J.; Zazimalová, E.; et al. The Arabidopsis concentration-dependent influx/efflux transporter ABCB4 regulates cellular auxin levels in the root epidermis. Plant J. 2012, 69, 640–654. [Google Scholar] [CrossRef]
- Goodman, C.D.; Casati, P.; Walbot, V. A multidrug resistance-associated protein involved in anthocyanin transport in Zea mays. Plant Cell 2004, 16, 1812–1826. [Google Scholar] [CrossRef]
- Song, W.Y.; Yamaki, T.; Yamaji, N.; Ko, D.; Jung, K.H.; Fujii-Kashino, M.; An, G.; Martinoia, E.; Lee, Y.; Ma, J.F. A rice ABC transporter, OsABCC1, reduces arsenic accumulation in the grain. Proc. Natl. Acad. Sci. USA 2014, 111, 15699–15704. [Google Scholar] [CrossRef]
- Loyola-Vargas, V.M.; Broeckling, C.D.; Badri, D.V.; Vivanco, J.M. Effect of transporters on the secretion of phytochemicals by the roots of Arabidopsis thaliana. Planta 2007, 225, 301–310. [Google Scholar] [CrossRef]
- Sauer, M.; Kleine-Vehn, J. PIN-FORMED and PIN-LIKES auxin transport facilitators. Development 2019, 146, dev168088. [Google Scholar] [CrossRef]
- Ganguly, A.; Sasayama, D.; Cho, H.T. Regulation of the polarity of protein trafficking by phosphorylation. Mol. Cells 2012, 33, 423–430. [Google Scholar] [CrossRef]
- Michniewicz, M.; Zago, M.K.; Abas, L.; Weijers, D.; Schweighofer, A.; Meskiene, I.; Heisler, M.G.; Ohno, C.; Zhang, J.; Huang, F.; et al. Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell 2007, 130, 1044–1056. [Google Scholar] [CrossRef]
- Wang, B.; Henrichs, S.; Geisler, M. The AGC kinase, PINOID, blocks interactive ABCB/PIN auxin transport. Plant Signal. Behav. 2012, 7, 1515–1517. [Google Scholar] [CrossRef]
- Enders, T.A.; Strader, L.C. Auxin activity: Past, present, and future. Am. J. Bot. 2015, 102, 180–196. [Google Scholar] [CrossRef]
- Smit, M.E.; Weijers, D. The role of auxin signaling in early embryo pattern formation. Curr. Opin. Plant Biol. 2015, 28, 99–105. [Google Scholar] [CrossRef]
- Winnicki, K. The winner takes it all: Auxin—The main player during plant embryogenesis. Cells 2020, 9, 606. [Google Scholar] [CrossRef]
- Liu, H.; Luo, Q.; Tan, C.; Song, J.; Zhang, T.; Men, S. Biosynthesis- and transport-mediated dynamic auxin distribution during seed development controls seed size in Arabidopsis. Plant J. 2023, 113, 1259–1277. [Google Scholar] [CrossRef]
- Petrášek, J.; Friml, J. Auxin transport routes in plant development. Development 2009, 136, 2675–2688. [Google Scholar] [CrossRef]
- Uc-Chuc, M.Á.; Pérez-Hernández, C.A.; Galaz-Ávalos, R.M.; Brito-Argáez, L.; Aguilar-Hernández, V.; Loyola-Vargas, V.M. YUCCA-mediated biosynthesis of the auxin IAA is required during the somatic embryogenic induction process in Coffea canephora. Int. J. Mol. Sci. 2020, 21, 4751. [Google Scholar] [CrossRef]
- Yu, Z.; Zhang, F.; Friml, J.; Ding, Z. Auxin signaling: Research advances over the past 30 years. J. Int. Plant Biol. 2022, 64, 371–392. [Google Scholar] [CrossRef]
- Luschnig, C. Auxin transport: ABC proteins join the club. Trends Plant Sci. 2002, 7, 329–332. [Google Scholar] [CrossRef]
- Hilleary, R. The sum is greater than the parts: Co-dependent auxin efflux is mediated by ABCBs and PINs. Plant Cell 2022, 34, 2114–2115. [Google Scholar] [CrossRef]
- López Encina, C.; Parisi, A.; O’Brien, C.; Mitter, N. Enhancing somatic embryogenesis in avocado (Persea americana Mill.) using a two-step culture system and including glutamine in the culture medium. Sci. Hortic. 2014, 165, 44–50. [Google Scholar] [CrossRef]
- O’Brien, C.; Hiti-Bandaralage, J.C.A.; Hayward, A.; Mitter, N. Avocado (Persea americana Mill.). In Step Wise Protocols for Somatic Embryogenesis of Important Woody Plants: Volume II; Jain, S.M., Gupta, P., Eds.; Springer: Cham, Switzerland, 2018; pp. 305–328. [Google Scholar]
- Márquez-López, R.E.; Pérez-Hernández, C.A.; Kú-González, Á.; Galaz-Ávalos, R.M.; Loyola-Vargas, V.M. Localization and transport of indole-3-acetic acid during somatic embryogenesis in Coffea canephora. Protoplasma 2018, 255, 695–708. [Google Scholar] [CrossRef] [PubMed]
- Rendón-Anaya, M.; Ibarra-Laclette, E.; Méndez-Bravo, A.; Lan, T.; Zheng, C.; Carretero-Paulet, L.; Perez-Torres, C.A.; Chacón-López, A.; Hernandez-Guzmán, G.; Chang, T.H.; et al. The avocado genome informs deep angiosperm phylogeny, highlights introgressive hybridization, and reveals pathogen-influenced gene space adaptation. Proc. Natl. Acad. Sci. USA 2019, 116, 17081–17089. [Google Scholar] [CrossRef] [PubMed]
- Hori, K.; Maruyama, F.; Fujisawa, T.; Togashi, T.; Yamamoto, N.; Seo, M.; Sato, S.; Yamada, T.; Mori, H.; Tajima, N.; et al. Klebsormidium flaccidum genome reveals primary factors for plant terrestrial adaptation. Nat. Commun. 2014, 5, 3978. [Google Scholar] [CrossRef]
- Blilou, I.; Xu, J.; Wildwater, M.; Willemsen, V.; Paponov, I.; Friml, J.; Heidstra, R.; Aida, M.; Palme, K.; Scheres, B. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 2005, 433, 39–44. [Google Scholar] [CrossRef]
- Paponov, I.A.; Teale, W.D.; Trebar, M.; Blilou, I.; Palme, K. The PIN auxin efflux facilitators: Evolutionary and functional perspectives. Trends Plant Sci. 2005, 10, 170–177. [Google Scholar] [CrossRef]
- Li, Z.; Li, P.; Zhang, J. Expression analysis of PIN-formed auxin efflux transporter genes in maize. Plant Signal. Behav. 2019, 14, 1632689. [Google Scholar] [CrossRef]
- Vieten, A.; Vanneste, S.; Wisniewska, J.; Benková, E.; Benjamins, R.; Beeckman, T.; Luschnig, C.; Friml, J. Functional redundancy of PIN proteins is accompanied by auxin-dependent cross-regulation of PIN expression. Development 2005, 132, 4521–4531. [Google Scholar] [CrossRef]
- Skokan, R.; Medvecká, E.; Viaene, T.; Vosolsobe, S.; Zwiewka, M.; Müller, K.; Skupa, P.; Karady, M.; Zhang, Y.; Janacek, D.P.; et al. PIN-driven auxin transport emerged early in streptophyte evolution. Nat. Plants 2019, 5, 1114–1119. [Google Scholar] [CrossRef]
- Qi, L.; Chen, L.; Wang, C.; Zhang, S.; Yang, Y.; Liu, J.; Li, D.; Song, J.; Wang, R. Characterization of the auxin efflux transporter PIN proteins in pear. Plants 2020, 9, 349. [Google Scholar] [CrossRef]
- Ganguly, A.; Park, M.; Kesawat, M.S.; Cho, H.T. Functional analysis of the hydrophilic loop in intracellular trafficking of Arabidopsis PIN-FORMED proteins. Plant Cell 2014, 26, 1570–1585. [Google Scholar] [CrossRef]
- Bernales, M.; Monsalve, L.; Ayala-Raso, A.; Valdenegro, M.; Martínez, J.P.; Travisany, D.; Defilippi, B.; González-Agüero, M.; Cherian, S.; Fuentes, L. Expression of two indole-3-acetic acid (IAA)-amido synthetase (GH3) genes during fruit development of raspberry (Rubus idaeus Heritage). Sci. Hortic. 2019, 246, 168–175. [Google Scholar] [CrossRef]
- Ayil-Gutiérrez, B.A.; Galaz-Ávalos, R.M.; Peña-Cabrera, E.; Loyola-Vargas, V.M. Dynamics of the concentration of IAA and some of its conjugates during the induction of somatic embryogenesis in Coffea canephora. Plant Signal. Behav. 2013, 8, e26998. [Google Scholar] [CrossRef]
- Ding, Z.; Wang, B.; Moreno, I.; Dupláková, N.; Simon, S.; Carraro, N.; Reemmer, J.; Pencìk, A.; Chen, X.; Tejos, R.; et al. ER-localized auxin transporter PIN8 regulates auxin homeostasis and male gametophyte development in Arabidopsis. Nat. Commun. 2012, 3, 941. [Google Scholar] [CrossRef]
- Huang, F.; Kemel Zago, M.; Abas, L.; van Marion, A.; Galvan-Ampudia, C.S.; Offringa, R. Phosphorylation of conserved PIN motifs directs Arabidopsis PIN1 polarity and auxin transport. Plant Cell 2010, 22, 1129–1142. [Google Scholar] [CrossRef]
- Barbosa, I.C.R.; Hammes, U.Z.; Schwechheimer, C. Activation and polarity control of PIN-FORMED auxin transporters by phosphorylation. Trends Plant Sci. 2018, 23, 523–538. [Google Scholar] [CrossRef]
- Nic-Can, G.I.; Loyola-Vargas, V.M. The role of the auxins during somatic embryogenesis. In Somatic Embryogenesis: Fundamental Aspects and Applications; Loyola-Vargas, V.M., Ochoa-Alejo, N., Eds.; Springer: Cham, Switzerland, 2016; pp. 171–181. [Google Scholar]
- Quintana-Escobar, A.O.; Nic-Can, G.I.; Galaz-Ávalos, R.M.; Loyola-Vargas, V.M.; Góngora-Castillo, E. Transcriptome analysis of the induction of somatic embryogenesis in Coffea canephora and the participation of arf and AUX/IAA genes. PeerJ 2019, 7, e7752. [Google Scholar] [CrossRef]
- Song, S.; Wang, Z.; Ren, Y.; Sun, H. Full-length transcriptome analysis of the ABCB, PIN/PIN-LIKES, and AUX/LAX families involved in somatic embryogenesis of Lilium pumilum DC. Fisch. Int. J. Mol. Sci. 2020, 21, 453. [Google Scholar] [CrossRef]
- Estrella-Maldonado, H.; Posada-Pérez, L.; Talavera, M.C.; Barredo, P.F.; Gómez-Kosky, R.; Santamaría, J.M. The expression of CpAUX1/LAXs and most of the long-distance CpPINs genes increases as the somatic embryogenesis process develops in C. papaya cv. “Red MaradoL”. J. Plant Growth Regul. 2018, 37, 502–516. [Google Scholar] [CrossRef]
- Weijers, D.; Schlereth, A.; Ehrismann, J.S.; Schwank, G.; Kientz, M.; Jürgens, G. Auxin triggers transient local signaling for cell specification in Arabidopsis embryogenesis. Dev. Cell 2006, 10, 265–270. [Google Scholar] [CrossRef]
- Robert, H.S.; Grones, P.; Stepanova, A.N.; Robles, L.M.; Lokerse, A.S.; Alonso, J.M.; Weijers, D.; Friml, J. Local auxin sources orient the apical-basal axis in Arabidopsis embryos. Curr. Biol. 2013, 23, 2506–2512. [Google Scholar] [CrossRef] [PubMed]
- Wabnik, K.; Robert, H.S.; Smith, R.S.; Friml, J. Modeling framework for the establishment of the apical-basal embryonic axis in plants. Curr. Biol. 2013, 23, 2513–2518. [Google Scholar] [CrossRef] [PubMed]
- Robert, H.S.; Grunewald, W.; Sauer, M.; Cannoot, B.; Soriano, M.; Swarup, R.; Weijers, D.; Bennett, M.; Boutilier, K.; Friml, J. Plant embryogenesis requires AUX/LAX-mediated auxin influx. Development 2015, 142, 702–711. [Google Scholar] [CrossRef] [PubMed]
- Dhonukshe, P.; Tanaka, H.; Goh, T.; Ebine, K.; Mahonen, A.P.; Prasad, K.; Blilou, I.; Geldner, N.; Xu, J.; Uemura, T.; et al. Generation of cell polarity in plants links endocytosis, auxin distribution and cell fate decisions. Nature 2008, 456, 962–966. [Google Scholar] [CrossRef]
- Mravec, J.; Skupa, P.; Bailly, A.; Hoyerová, K.; Krecek, P.; Bielach, A.; Petrášek, J.; Zhang, J.; Gaykova, V.; Stierhof, Y.D.; et al. Subcellular homeostasis of phytohormone auxin is mediated by the ER-localized PIN5 transporter. Nature 2009, 459, 1136–1140. [Google Scholar] [CrossRef]
- Su, N.; Zhu, A.; Tao, X.; Ding, Z.J.; Chang, S.; Ye, F.; Zhang, Y.; Zhao, C.; Chen, Q.; Wang, J.; et al. Structures and mechanisms of the Arabidopsis auxin transporter PIN3. Nature 2022, 609, 616–621. [Google Scholar] [CrossRef]
- Dory, M.; Hatzimasoura, E.; Kállai, B.M.; Nagy, S.K.; Jäger, K.; Darula, Z.; Nádai, T.V.; Mészáros, T.; López-Juez, E.; Barnabás, B.; et al. Coevolving MAPK and PID phosphosites indicate an ancient environmental control of PIN auxin transporters in land plants. FEBS Lett. 2018, 592, 89–102. [Google Scholar] [CrossRef]
- Tan, S.; Luschnig, C.; Friml, J. Pho-view of auxin: Reversible protein phosphorylation in auxin biosynthesis, transport and signaling. Mol. Plant 2021, 14, 151–165. [Google Scholar] [CrossRef]
- Hajny, J.; Prát, T.; Rydza, N.; Rodriguez, L.; Tan, S.; Verstraeten, I.; Domjan, D.; Mazur, E.; Smakowska-Luzan, E.; Smet, W.; et al. Receptor kinase module targets PIN-dependent auxin transport during canalization. Science 2020, 370, 550–557. [Google Scholar] [CrossRef]
- Zourelidou, M.; Absmanner, B.; Weller, B.; Barbosa, I.C.; Willige, B.C.; Fastner, A.; Streit, V.; Port, S.A.; Colcombet, J.; de la Fuente van Bentem, S.; et al. Auxin efflux by PIN-FORMED proteins is activated by two different protein kinases, D6 PROTEIN KINASE and PINOID. eLife 2014, 3, e02860. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic tress. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L.L. Predicting transmembrane protein topology with a hidden markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef]
- Djami-Tchatchou, A.T.; Straker, C.J. The isolation of high quality RNA from the fruit of avocado (Persea americana Mill.). S. Afr. J. Bot. 2012, 78, 44–46. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Meth. 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]




| Deducted Polypeptide | ||||||||
|---|---|---|---|---|---|---|---|---|
| Gene | ORF Length (bp) | No. of Exons | Length (aa) | Molecular Weight (Da) | pI | GRAVY | No. of Trans-Membrane | Subcellular Localization |
| PaPIN1a | 1710 | 6 | 569 | 61,971.01 | 8.66 | 0.226 | 9 | Chloroplast |
| PaPIN1b | 1887 | 6 | 628 | 68,306.87 | 8.10 | 0.198 | 9 | Plasma membrane |
| PaPIN1c | 1791 | 6 | 596 | 64,915.32 | 9.09 | 0.226 | 8 | Plasma membrane |
| PaPIN1d | 1917 | 6 | 638 | 69,598.27 | 8.85 | 0.102 | 9 | Plasma membrane |
| PaPIN1e | 1917 | 6 | 638 | 69,598.27 | 8.85 | 0.102 | 9 | Plasma membrane |
| PaPIN2a | 1983 | 6 | 660 | 71,911.03 | 9.45 | 0.086 | 8 | Plasma membrane |
| PaPIN2b | 1599 | 5 | 532 | 58,012.56 | 6.57 | 0.111 | 5 | Plasma membrane |
| PaPIN3 | 1767 | 6 | 588 | 63,460.40 | 9.24 | 0.137 | 8 | Plasma membrane |
| PaPIN5 | 1074 | 5 | 357 | 39,164.18 | 6.95 | 0.722 | 9 | Vacuolar |
| PaPIN6 | 1530 | 7 | 509 | 55,370.96 | 8.91 | 0.410 | 9 | Plasma membrane |
| PaPIN8a | 1311 | 6 | 436 | 48,444.16 | 9.12 | 0.404 | 8 | Plasma membrane |
| PaPIN8b | 1209 | 6 | 402 | 44,630.07 | 9.43 | 0.529 | 8 | Plasma membrane |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Monroy-González, Z.; Uc-Chuc, M.A.; Quintana-Escobar, A.O.; Duarte-Aké, F.; Loyola-Vargas, V.M. Characterization of the PIN Auxin Efflux Carrier Gene Family and Its Expression during Zygotic Embryogenesis in Persea americana. Plants 2023, 12, 2280. https://doi.org/10.3390/plants12122280
Monroy-González Z, Uc-Chuc MA, Quintana-Escobar AO, Duarte-Aké F, Loyola-Vargas VM. Characterization of the PIN Auxin Efflux Carrier Gene Family and Its Expression during Zygotic Embryogenesis in Persea americana. Plants. 2023; 12(12):2280. https://doi.org/10.3390/plants12122280
Chicago/Turabian StyleMonroy-González, Zurisadai, Miguel A. Uc-Chuc, Ana O. Quintana-Escobar, Fátima Duarte-Aké, and Víctor M. Loyola-Vargas. 2023. "Characterization of the PIN Auxin Efflux Carrier Gene Family and Its Expression during Zygotic Embryogenesis in Persea americana" Plants 12, no. 12: 2280. https://doi.org/10.3390/plants12122280
APA StyleMonroy-González, Z., Uc-Chuc, M. A., Quintana-Escobar, A. O., Duarte-Aké, F., & Loyola-Vargas, V. M. (2023). Characterization of the PIN Auxin Efflux Carrier Gene Family and Its Expression during Zygotic Embryogenesis in Persea americana. Plants, 12(12), 2280. https://doi.org/10.3390/plants12122280










