A Foundational Population Genetics Investigation of the Sexual Systems of Solanum (Solanaceae) in the Australian Monsoon Tropics Suggests Dioecious Taxa May Benefit from Increased Genetic Admixture via Obligate Outcrossing
Abstract
1. Introduction
2. Results
2.1. Sequencing and Ipyrad Filtering
2.2. Summary Statistics
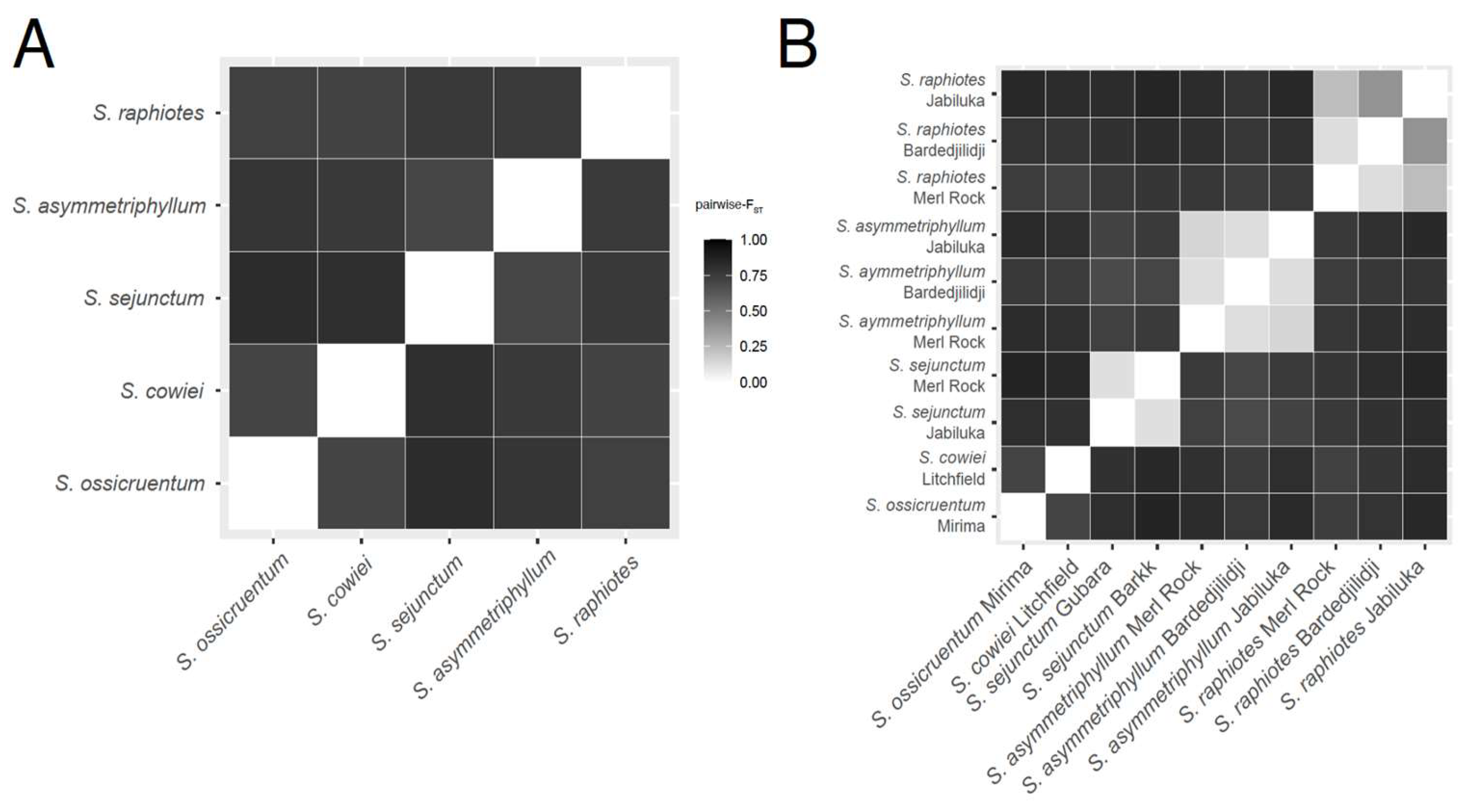
2.3. Pairwise-FST and AMOVA
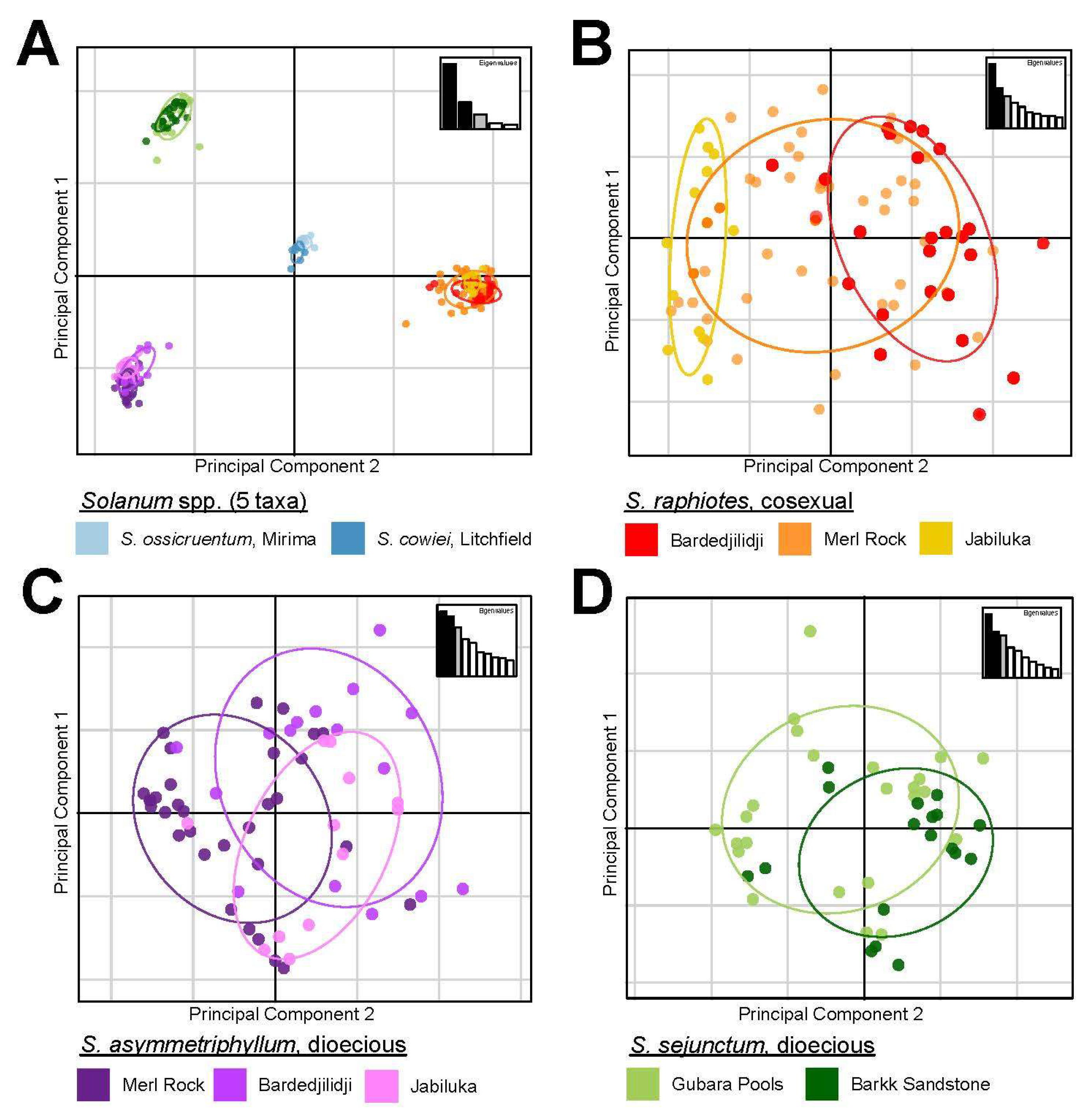
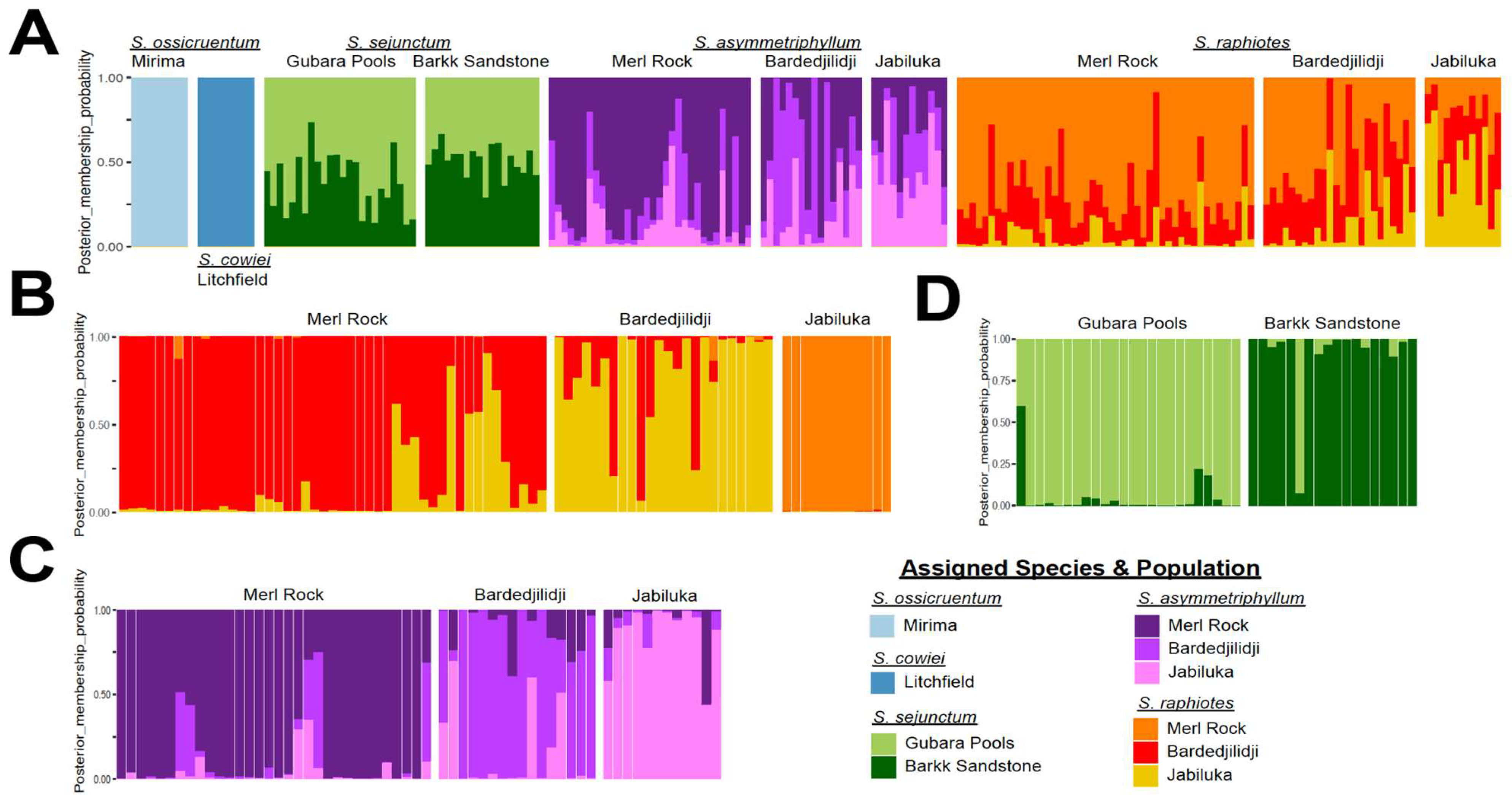
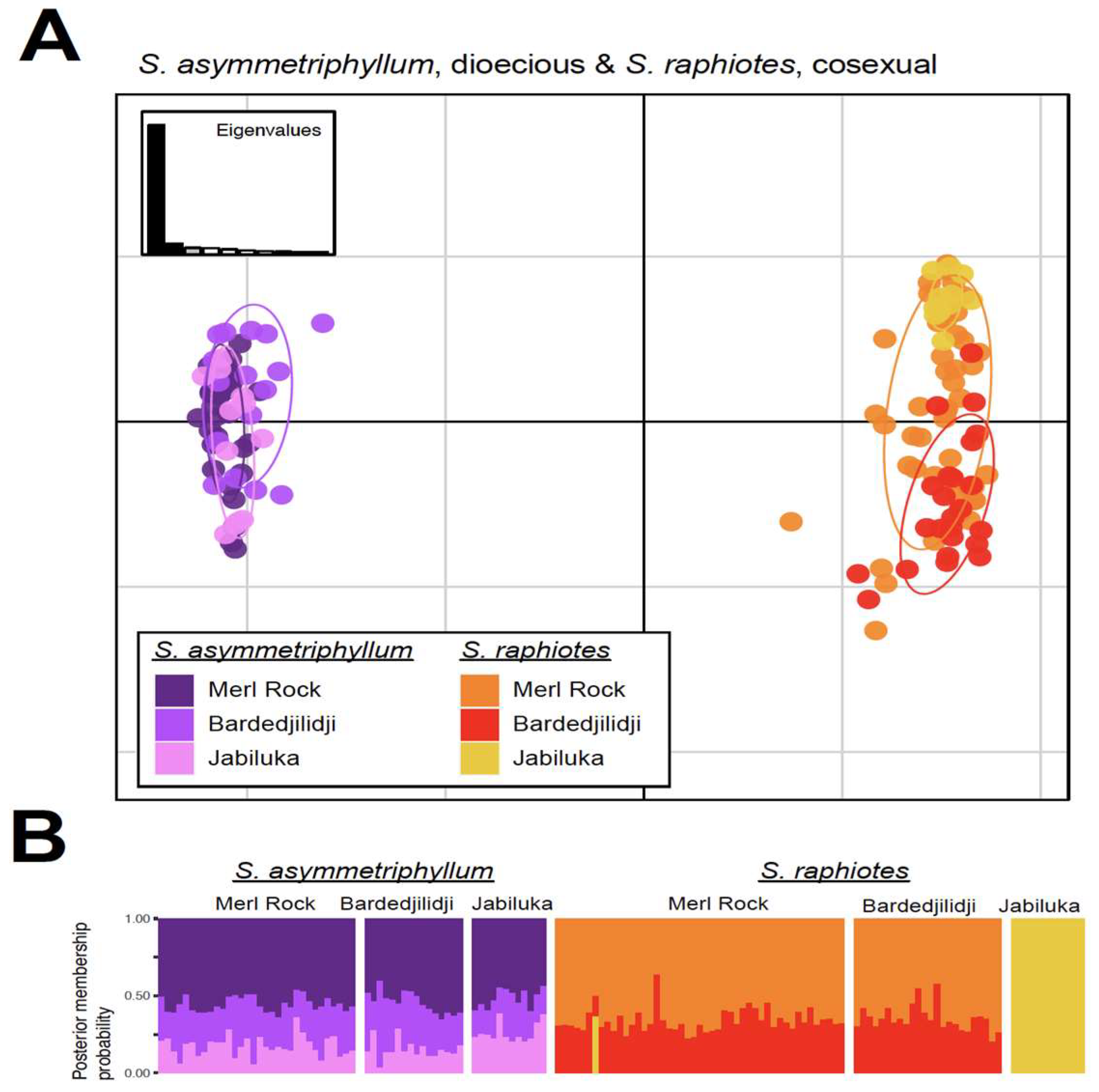
2.4. PCA and DAPC Analyses
2.5. Isolation by Distance
3. Discussion
3.1. Genetic Diversity and Structure in Dioecious Australian Solanum
3.2. Insights from a Shared Background of Elevated Inbreeding
4. Materials and Methods
4.1. Taxon Sampling and Field-Observed Life History
4.2. DNA Library Preparation and Sequencing
4.3. Sequenced Data Processing and ‘Hard-Filtering’
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Darwin, C. The Different Forms of Flowers on Plants of the Same Species; John Murray: London, UK, 1877. [Google Scholar]
- Darwin, C. The Effects of Cross and Self Fertilization in the Vegetable Kingdom; Ams Press Inc.: London, UK, 1876. [Google Scholar]
- Barrett, S.C. Understanding plant reproductive diversity. Philos. Trans. R. Soc. B Biol. Sci. 2010, 365, 99–109. [Google Scholar] [CrossRef] [PubMed]
- Bull, J.J.; Charnov, E.L. On irreversible evolution. Evolution 1985, 39, 1149–1155. [Google Scholar] [CrossRef] [PubMed]
- Heilbuth, J. Lower species richness in dioecious clades. Am. Nat. 2000, 156, 221–241. [Google Scholar] [CrossRef]
- Barrett, S.C. The evolution of plant reproductive systems: How often are transitions irreversible? Proc. R. Soc. B Biol. Sci. 2013, 280, 20130913. [Google Scholar] [CrossRef]
- Muenchow, G.E. Is dioecy associated with fleshy fruit? Am. J. Bot. 1987, 74, 287–293. [Google Scholar] [CrossRef]
- Vamosi, J.C.; Vamosi, S.M. The role of diversification in causing the correlates of dioecy. Evolution 2004, 58, 723–731. [Google Scholar]
- Muyle, A.; Martin, H.; Zemp, N.; Mollion, M.; Gallina, S.; Tavares, R.; Silva, A.; Bataillon, T.; Widmer, A.; Glémin, S.; et al. Dioecy is associated with high genetic diversity and adaptation rates in the plant genus Silene. Mol. Biol. Evol. 2021, 38, 805–818. [Google Scholar] [CrossRef]
- Renner, S.S.; Ricklefs, R.E. Dioecy and its correlates in the flowering plants. Am. J. Bot. 1995, 82, 595–606. [Google Scholar] [CrossRef]
- Sakai, A.K.; Wagner, W.L.; Ferguson, D.M.; Herbst, D.R. Origins of dioecy in the Hawaiian flora. Ecology 1995, 76, 2517–2529. [Google Scholar] [CrossRef]
- Martine, C.T.; Jordon-Thaden, I.E.; McDonnell, A.J.; Cantley, J.T.; Hayes, D.; Roche, M.; Frawley, E.S.; Gilman, I.S.; Tank, D. Phylogeny of the Australian Solanum dioicum group using seven nuclear genes: Testing Symon’s fruit and seed dispersal hypotheses. PLoS ONE 2019, 14, e0207564. [Google Scholar] [CrossRef]
- Whalen, M.D.; Costich, D.E. Andromonoecy in Solanum. In Solanaceae: Biology and Systematics; Columbia University Press: New York, NY, USA, 1986; pp. 284–302. [Google Scholar]
- Knapp, S.; Sagona, E.; Carbonell, A.; Chiarini, F. A revision of the Solanum elaeagnifolium clade (Elaeagnifolium clade; subgenus Leptostemonum, Solanaceae). PhytoKeys 2017, 84, 1–104. [Google Scholar] [CrossRef]
- Knapp, S. Solanaceae Source. PBI Solanum: A Worldwide Treatment. 2014. Available online: http://www.solanaceaesource.org/ (accessed on 31 May 2023).
- Symon, D. Dioecious solanums. Taxon 1970, 19, 909–910. [Google Scholar] [CrossRef]
- Symon, D.E. Sex forms in Solanum (Solanaceae) and the role of pollen collecting insects. In The Biology and Taxonomy of the Solanaceae; Hawkes, J., Lester, G., Skelding, N.A., Eds.; Academic Press for the Linnean Society: London, UK, 1979; pp. 385–397. [Google Scholar]
- Gagnon, E.; Hilgenhof, R.; Orejuela, A.; McDonnell, A.J.; Sablok, G.; Aubriot, X.; Giacomin, L.; Gouvêa, L.; Bohs, L.; Dodsworth, S.; et al. Phylogenomic data reveal hard polytomies across the backbone of the large genus Solanum (Solanaceae). Am. J. Bot. 2022, 109, 1–22. [Google Scholar]
- Barrett, R.L. Solanum zoeae (Solanaceae), a new species of bush tomato from the North Kimberley, Western Australia. Nuytsia 2013, 23, 5–21. [Google Scholar] [CrossRef]
- Martine, C.T.; Symon, D.E.; Evans, E.C. A new cryptically dioecious species of bush tomato (Solanum) from the Northern Territory, Australia. PhytoKeys 2013, 30, 23–32. [Google Scholar] [CrossRef]
- Anderson, G.J.; Anderson, M.K.J.; Patel, N. The ecology, evolution, and biogeography of dioecy in the genus Solanum: With paradigms from the strong dioecy in Solanum polygamum, to the unsuspected and cryptic dioecy in Solanum conocarpum. Am. J. Bot. 2015, 102, 471–486. [Google Scholar] [CrossRef] [PubMed]
- Martine, C.T.; Cantley, J.T.; Frawley, E.S.; Butler, A.R.; Jordon-Thaden, I.E. New functionally dioecious bush tomato from northwestern Australia, Solanum ossicruentum, may utilize “trample burr” dispersal. PhytoKeys 2016, 63, 19–29. [Google Scholar] [CrossRef]
- Williams, T.M.; Hayes, J.; Cantley, J.T.; McDonnell, A.J.; Jobson, P.; Martine, C.T. Solanum scalarium (Solanaceae), a newly-described dioecious bush tomato from Judbarra/Gregory National Park, Northern Territory, Australia. PhytoKeys 2022, 216, 103–116. [Google Scholar] [CrossRef]
- D’Arcy, W.G. Solanaceae studies II. Typification of the subdivisions of Solanum. Ann. Mo. Bot. Gard. 1972, 59, 262–278. [Google Scholar] [CrossRef]
- Knapp, S. A revision of the Solanum sessile species group (section Geminata pro parte: Solanaceae). Bot. J. Linn. Soc. 1991, 105, 179–210. [Google Scholar] [CrossRef]
- Knapp, S.; Persson, V.; Blackmore, S. Pollen morphology and functional dioecy in Solanum (Solanaceae). Plant Syst. Evol. 1998, 210, 113–139. [Google Scholar] [CrossRef]
- Whalen, M.D. Conspectus of species groups in Solanum subgenus Leptostemonum. Gentes Herb. 1984, 12, 179–282. [Google Scholar]
- Martine, C.T.; Anderson, G.J. Dioecy, pollination, and seed dispersal in Australian spiny Solanum. In Proceedings of the VIth International Solanaceae Conference: Acta Horticulturae, Madison, WI, USA, 23–27 July 2006; ISHS: Leuven, Belgium, 2007; Volume 745, pp. 269–283. [Google Scholar]
- McDonnell, A.J.; Wetreich, H.B.; Cantley, J.T.; Jobson, P.C.; Martine, C.T. Solanum plastisexum, an enigmatic new bush tomato from the Australian Monsoon Tropics exhibiting breeding system fluidity. PhytoKeys 2019, 124, 39–55. [Google Scholar] [CrossRef] [PubMed]
- Martine, C.T.; Anderson, G.J.; Les, D.H. Gender-bending aubergines: Molecular phylogenetics of cryptically dioecious Solanum in Australia. Aust. Syst. Bot. 2009, 22, 107–120. [Google Scholar] [CrossRef]
- Bean, A.R. A taxonomic revision of the Solanum echinatum group (Solanaceae). Phytotaxa 2012, 57, 33–50. [Google Scholar] [CrossRef]
- Anderson, G.J. Dioecious Solanum of hermaphroditic origin is an example of a broad convergence. Nature 1979, 282, 836–838. [Google Scholar] [CrossRef]
- Anderson, G.J.; Symon, D.E. Functional dioecy and andromonoecy in Solanum. Evolution 1989, 43, 204–219. [Google Scholar] [CrossRef]
- Symon, D.E. A revision of the genus Solanum in Australia. J. Adel. Bot. Gard. 1981, 4, 1–367. [Google Scholar]
- Ndem-Galbert, J.R.; Hall, J.; McDonnell, A.J.; Martine, C.T. Differential reward in “male” versus “female” pollen of functionally dioecious Solanum (Solanaceae). Am. J. Bot. 2021, 108, 2282–2293. [Google Scholar] [CrossRef]
- Miller, J.S.; Diggle, P.K. Correlated evolution of fruit size and sexual expression in andromonoecious Solanum sections Acanthophora and Lasiocarpa (Solanaceae). Am. J. 2007, 94, 1706–1715. [Google Scholar]
- Eaton, D.A.; Overcast, I. ipyrad: Interactive assembly and analysis of RADseq datasets. Bioinformatics 2020, 36, 2592–2594. [Google Scholar] [CrossRef]
- Weir, B.S.; Cockerham, C.C. Estimating F-statistics for the analysis of population structure. Evolution 1984, 38, 1358–1370. [Google Scholar]
- Hilgenhof, R.; Gagnon, E.; Knapp, S.; Aubriot, X.; Tepe, E.J.; Bohs, L.; Giacomin, L.L.; Gouvêa, Y.F.; Stehmann, J.R.; Martine, C.T.; et al. Morphological trait evolution in Solanum (Solanaceae): Evolutionary lability of key taxonomic characters. bioRxiv 2023. bioRxiv:2023.02. 24.529849. [Google Scholar]
- Marino, C. Cleaning the Variable Mess: A Population Genomics Approach to Understanding the Evolutionary History of a Complicated Plant Group (2023). Honors Thesis 645, Bucknell University, Lewisburg, PA, USA, 2023. Available online: https://digitalcommons.bucknell.edu/honors_theses/645 (accessed on 31 May 2023).
- Brennan, K.; Martine, C.T.; Symon, D. Solanum sejunctum (Solanaceae), a new functionally dioecious species from Kakadu National Park, Northern Territory, Australia. Beagle Rec. Mus. Art Gall. North. Territ. 2006, 22, 1–7. [Google Scholar] [CrossRef]
- Bean, A.R. 2012 Onwards. Solanum species of eastern and northern Australia. Version: 25th January 2023. Available online: http://www.delta-intkey.com (accessed on 31 May 2023).
- Martine, C.T.; Lavoie, E.; Tippery, N.L.; Vogt, F.D.; Les, D.H. Solanum from Litchfield National Park is a relative of S. dioicum. North. Territ. Nat. 2011, 23, 29–38. [Google Scholar]
- Ennos, R.A. Detection and measurement of selection: Genetic and ecological approaches. In Plant Population Genetics, Breeding and Genetic Resources; Brown, A.H.D., Clegg, M.T., Kahler, A.L., Wier, B.S., Eds.; Sinauer Associates: Sunderland, MA, USA, 1989; pp. 200–214. [Google Scholar]
- Farris, M.A.; Mitton, J.B. Population density, outcrossing rate, and heterozygote superiority in ponderosa pine. Evolution 1984, 38, 1151–1154. [Google Scholar] [CrossRef]
- Bowman, D.M.; Brown, G.K.; Braby, M.F.; Brown, J.R.; Cook, L.G.; Crisp, M.D.; Ford, F.; Haberle, S.; Hughes, J.; Isagi, Y.; et al. Biogeography of the Australian monsoon tropics. J. Biogeogr. 2010, 37, 201–216. [Google Scholar] [CrossRef]
- Haug, G.H.; Tiedemann, R. Effect of the formation of the Isthmus of Panama on Atlantic Ocean thermohaline circulation. Nature 1998, 393, 673–676. [Google Scholar] [CrossRef]
- O’Dea, A.; Lessios, H.A.; Coates, A.G.; Eytan, R.I.; Restrepo-Moreno, S.A.; Cione, A.L.; Collins, L.S.; De Queiroz, A.; Farris, D.W.; Norris, R.D.; et al. Formation of the Isthmus of Panama. Sci. Adv. 2016, 2, e1600883. [Google Scholar] [CrossRef]
- Bowman, D.M.J.S. The Australian summer monsoon: A biogeographic perspective. Aust. Geogr. Stud. 2002, 40, 261–277. [Google Scholar] [CrossRef]
- Felderhof, L.; Gillieson, D. Comparison of fire patterns and fire frequency in two tropical savanna bioregions. Austral Ecol. 2006, 31, 736–746. [Google Scholar] [CrossRef]
- Byrne, M.; Yeates, D.K.; Joseph, L.; Kearne, M.; Bowler, J.; Williams, M.A.J.; Cooper, S.; Donnellan, S.C.; Keogh, J.S.; Leys, R.; et al. Birth of a biome: Insights into the assembly and maintenance of the Australian arid zone biota. Mol. Ecol. 2008, 17, 4398–4417. [Google Scholar] [CrossRef] [PubMed]
- Vigilante, T. Analysis of explorer’s records of Aboriginal landscape burning in the Kimberley region of Western Australia. Aust. Geogr. Stud. 2001, 39, 135–155. [Google Scholar] [CrossRef]
- Russell-Smith, J.; Yates, C.; Edwards, A.; Allan, G.E.; Cook, G.D.; Cooke, P.; Craig, R.; Heath, B.; Smith, R. Contemporary fire regimes of northern Australia, 1997–2001: Change since Aboriginal occupancy, challenges for sustainable management. Int. J. Wildland Fire 2003, 12, 283–297. [Google Scholar] [CrossRef]
- Russell-Smith, J.; Ryan, P.G.; Klessa, D.; Waight, G.; Harwood, R. Fire regimes, fire-sensitive vegetation and fire management of the sandstone Arnhem Plateau, monsoonal northern Australia. J. Appl. Ecol. 1998, 35, 829–846. [Google Scholar] [CrossRef]
- Bowman, D.M.J.S. The impact of Aboriginal landscape burning on the Australian biota. New Phytol. 1998, 140, 385–410. [Google Scholar] [CrossRef]
- Bowman, D.M.J.S.; Price, O.; Whitehead, P.J.; Walsh, A. The ‘wilderness effect’ and the decline of Callitris intratropica on the Arnhem Land Plateau, northern Australia. Aust. J. Bot. 2001, 49, 665–672. [Google Scholar] [CrossRef]
- Bowman, D.M.J.S.; Walsh, A.; Prior, L.D. Landscape analysis of Aboriginal fire management in central Arnhem Land, north Australia. J. Biogeogr. 2004, 31, 207–223. [Google Scholar] [CrossRef]
- Kelly, L.T.; Giljohann, K.M.; Duane, A.; Aquilué, N.; Archibald, S.; Batllori, E.; Bennett, A.F.; Buckland, S.T.; Canelles, Q.; Clarke, M.F.; et al. Fire and biodiversity in the Anthropocene. Science 2020, 370, eabb0355. [Google Scholar] [CrossRef] [PubMed]
- Bowman, D.M.J.S.; Franklin, D.C.; Price, O.F.; Brook, B.W. Land management affects grass biomass in the Eucalyptus tetrodonta savannas of monsoonal Australia. Austral Ecol. 2007, 32, 446–452. [Google Scholar] [CrossRef]
- Russell-Smith, J.; Bowman, D.M.J.S. Conservation of monsoon rainforest isolates in the Northern Territory, Australia. Biol. Conserv. 1992, 59, 51–63. [Google Scholar] [CrossRef]
- Russell-Smith, J.; Lucas, D.E.; Brock, J.; Bowman, D.M.J.S. Allosyncarpia-dominated rain forest in monsoon northern Australia. J. Veg. Sci. 1993, 4, 67–82. [Google Scholar] [CrossRef]
- Russell-Smith, J.; Craig, R.; Gill, A.M.; Smith, R.; Williams, J.E. Australian Fire Regimes: Contemporary Patterns (April 1998–March 2000) and Changes since European Settlement; Department of the Environment and Heritage: Canberra, Australia, 2002.
- Bowman, D.M.J.S.; Panton, W.J. Decline of Callitris intratropica in the Northern Territory: Implications for pre- and post-colonisation fire regimes. J. Biogeogr. 1993, 20, 373–381. [Google Scholar] [CrossRef]
- Bowman, D.M.J.S. Preliminary observations on the mortality of Allosyncarpia ternata stems on the Arnhemland plateau, northern Australia. Aust. For. 1994, 57, 62–64. [Google Scholar] [CrossRef]
- Price, O.; Bowman, D.M.J.S. Fire-stick forestry: A matrix model in support of skillful fire management of Callitris intratropica R.T. Baker by north Australian Aborigines. J. Biogeogr. 1994, 21, 573–580. [Google Scholar] [CrossRef]
- Martine, C.T.; Vanderpool, D.; Anderson, G.J.; Les, D.H. Phylogenetic relationships of andromonoecious and dioecious Australian species of Solanum subgenus Leptostemonum section Melongena: Inferences from ITS sequence data. Syst. Bot. 2006, 31, 410–420. [Google Scholar] [CrossRef]
- Särkinen, T.; Bohs, L.; Olmstead, R.G.; Knapp, S. A phylogenetic framework for evolutionary study of the nightshades (Solanaceae): A dated 1000-tip tree. BMC Evol. Biol. 2013, 13, 214. [Google Scholar] [CrossRef]
- Hayes, D.S. Ex Situ Interspecies Crossing Rates Infer Importance of Geographic Barriers in Speciation among Closely Related Solanum Species of the Australian Monsoon Tropics. Honors Thesis, Bucknell University, Lewisburg, PA, USA, 2018. p. 447. Available online: https://digitalcommons.bucknell.edu/honors_theses/447 (accessed on 31 May 2023).
- Zizis, D.; Williams, T.M.; Martine, C.T. Heading for a Breakdown: Assessing Evolution through the Hybridization of Two Sexual Systems. Honors Thesis 650, Bucknell University, Lewisburg, PA, USA, 2023. Available online: https://digitalcommons.bucknell.edu/honors_theses/650 (accessed on 31 May 2023).
- Doyle, J.J.; Doyle, J.L. Isolation of Plant DNA from Fresh Tissue. Am. J. Plant Sci. 1990, 12, 13–15. [Google Scholar]
- Peterson, B.K.; Weber, J.N.; Kay, E.H.; Fisher, H.S.; Hoekstra, H.E. Double digest RADseq: An inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS ONE 2012, 7, e37135. [Google Scholar] [CrossRef] [PubMed]
- Jordon-Thaden, I.E.; Beck, J.; Rushworth, C.; Windham, M.; Diaz, N.; Cantley, J.T.; Martine, C.T.; Rothfels, C. A basic ddRADseq two-enzyme protocol performs well with herbarium and silica-dried tissues across four genera. Appl. Plant Sci. 2020, 8, e11344. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; De Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021; Available online: https://www.R-project.org/ (accessed on 31 May 2023).
- Goudet, J. Hierfstat, a package for R to compute and test hierarchical F-statistics. Mol. Ecol. Notes 2005, 5, 184–186. [Google Scholar] [CrossRef]
- Paradis, E. Pegas: An R package for population genetics with an integrated–modular approach. Bioinformatics 2010, 26, 419–420. [Google Scholar] [CrossRef] [PubMed]
- Jombart, T.; Ahmed, I. adegenet 1.3-1: New tools for the analysis of genome-wide SNP data. Bioinformatics 2011, 27, 3070–3071. [Google Scholar] [CrossRef]
- Kamvar, Z.N.; Tabima, J.F.; Grünwald, N.J. Poppr: An R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2014, 2, e281. [Google Scholar] [CrossRef]
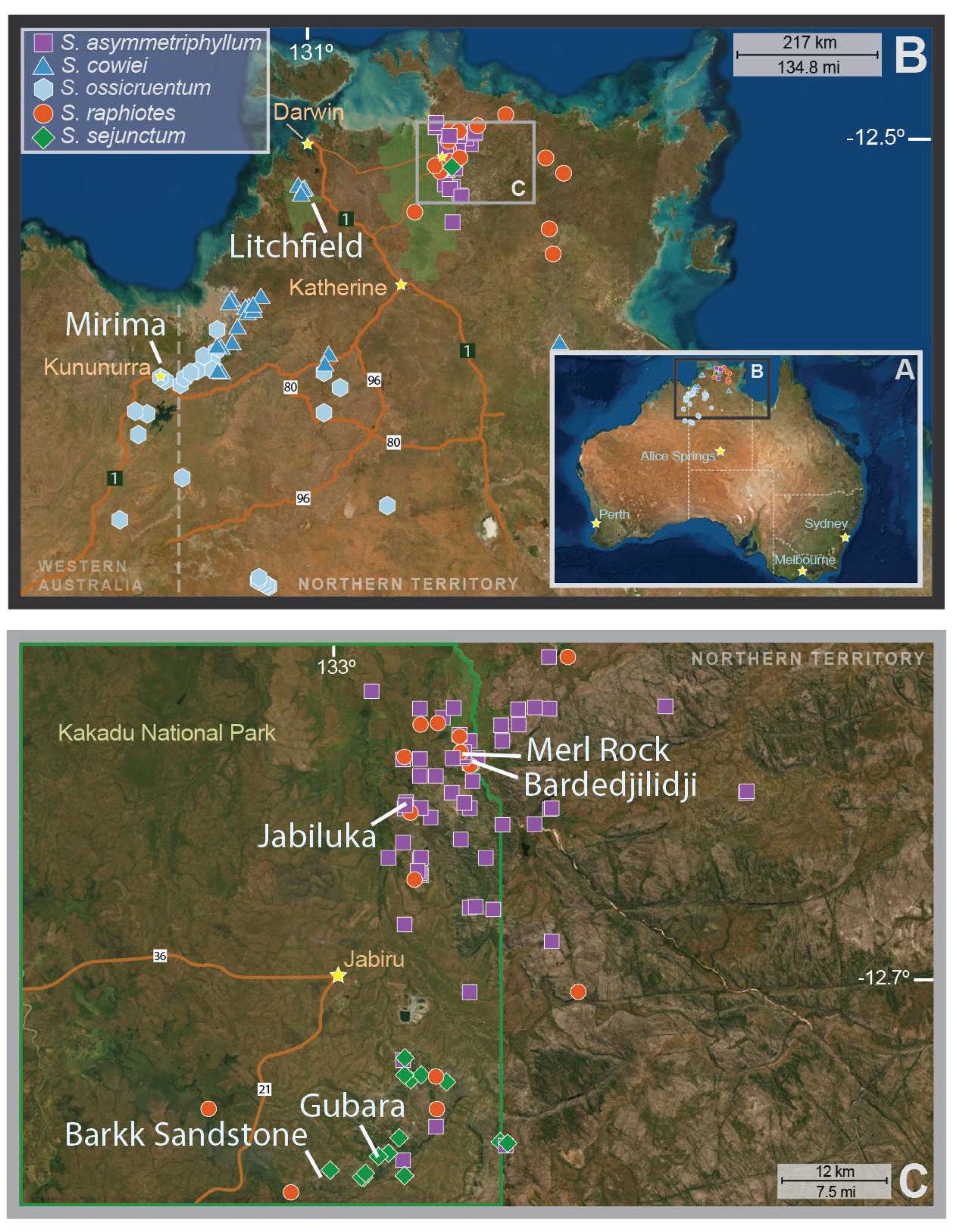
| Species | Sexual System | Population | n | Latitude | Longitude | Reference Voucher (Herbarium) | Ho | He | FIS |
|---|---|---|---|---|---|---|---|---|---|
| S. ossicruentum | Dioecious | Mirima | 9 | 15.76378 | 128.75175 | CTM 4011 (BUPL) | 0.0014 | 0.0924 | 0.9849 |
| S. cowiei | Dioecious | Florence Falls | 9 | 13.21958 | 130.73645 | CTM 1751 (BUPL) | 0.0004 | 0.1101 | 0.9968 |
| S. sejunctum | Dioecious | 43 | 0.0000 | 0.1215 | 1.0000 | ||||
| Gubara Pools | 24 | 12.82928 | 132.8756 | CTM 1739 (BUPL) | 0.0000 | 0.1288 | 1.0000 | ||
| Barkk Sandstone | 19 | 12.85907 | 132.81788 | CTM 1729 (BUPL) | 0.0000 | 0.0953 | 1.0000 | ||
| S. asymmetriphyllum | Dioecious | 49 | 0.0004 | 0.1582 | 0.9973 | ||||
| Merl Rock | 32 | 12.42622 | 132.96022 | CTM 1702 (BUPL) | 0.0001 | 0.1333 | 0.9993 | ||
| Bardedjilidji | 16 | 12.43727 | 132.96803 | CTM 1721 (BUPL) | 0.0006 | 0.1825 | 0.9967 | ||
| Jabaluka | 12 | 12.47702 | 132.90275 | CTM 1700 (BUPL) | 0.0011 | 0.1286 | 0.9920 | ||
| S. raphiotes | Cosexual | 83 | 0.0009 | 0.1778 | 0.9951 | ||||
| Merl Rock | 47 | 12.42622 | 132.96022 | CTM 1709 (BUPL) | 0.0004 | 0.1735 | 0.9977 | ||
| Bardedjilidji | 24 | 12.43727 | 132.96803 | CTM 1714 (BUPL) | 0.0004 | 0.1439 | 0.9974 | ||
| Jabaluka | 12 | 12.47702 | 132.90275 | CTM 737 (CONN) | 0.0038 | 0.1043 | 0.9635 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cantley, J.T.; Jordon-Thaden, I.E.; Roche, M.D.; Hayes, D.; Kate, S.; Martine, C.T. A Foundational Population Genetics Investigation of the Sexual Systems of Solanum (Solanaceae) in the Australian Monsoon Tropics Suggests Dioecious Taxa May Benefit from Increased Genetic Admixture via Obligate Outcrossing. Plants 2023, 12, 2200. https://doi.org/10.3390/plants12112200
Cantley JT, Jordon-Thaden IE, Roche MD, Hayes D, Kate S, Martine CT. A Foundational Population Genetics Investigation of the Sexual Systems of Solanum (Solanaceae) in the Australian Monsoon Tropics Suggests Dioecious Taxa May Benefit from Increased Genetic Admixture via Obligate Outcrossing. Plants. 2023; 12(11):2200. https://doi.org/10.3390/plants12112200
Chicago/Turabian StyleCantley, Jason T., Ingrid E. Jordon-Thaden, Morgan D. Roche, Daniel Hayes, Stephanie Kate, and Christopher T. Martine. 2023. "A Foundational Population Genetics Investigation of the Sexual Systems of Solanum (Solanaceae) in the Australian Monsoon Tropics Suggests Dioecious Taxa May Benefit from Increased Genetic Admixture via Obligate Outcrossing" Plants 12, no. 11: 2200. https://doi.org/10.3390/plants12112200
APA StyleCantley, J. T., Jordon-Thaden, I. E., Roche, M. D., Hayes, D., Kate, S., & Martine, C. T. (2023). A Foundational Population Genetics Investigation of the Sexual Systems of Solanum (Solanaceae) in the Australian Monsoon Tropics Suggests Dioecious Taxa May Benefit from Increased Genetic Admixture via Obligate Outcrossing. Plants, 12(11), 2200. https://doi.org/10.3390/plants12112200





