Desiccation Tolerance in Ramonda serbica Panc.: An Integrative Transcriptomic, Proteomic, Metabolite and Photosynthetic Study
Abstract
:1. Introduction
2. Results
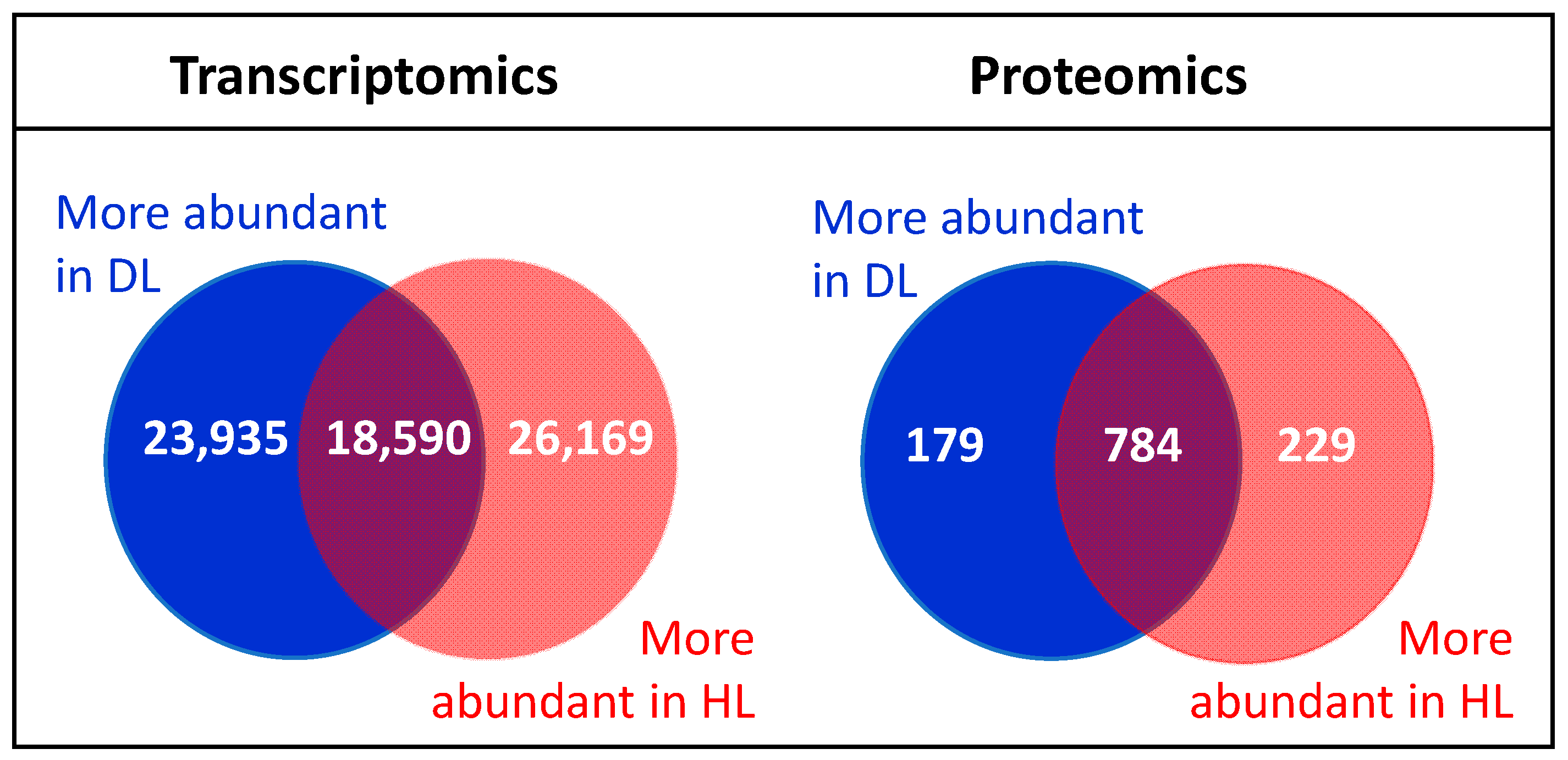
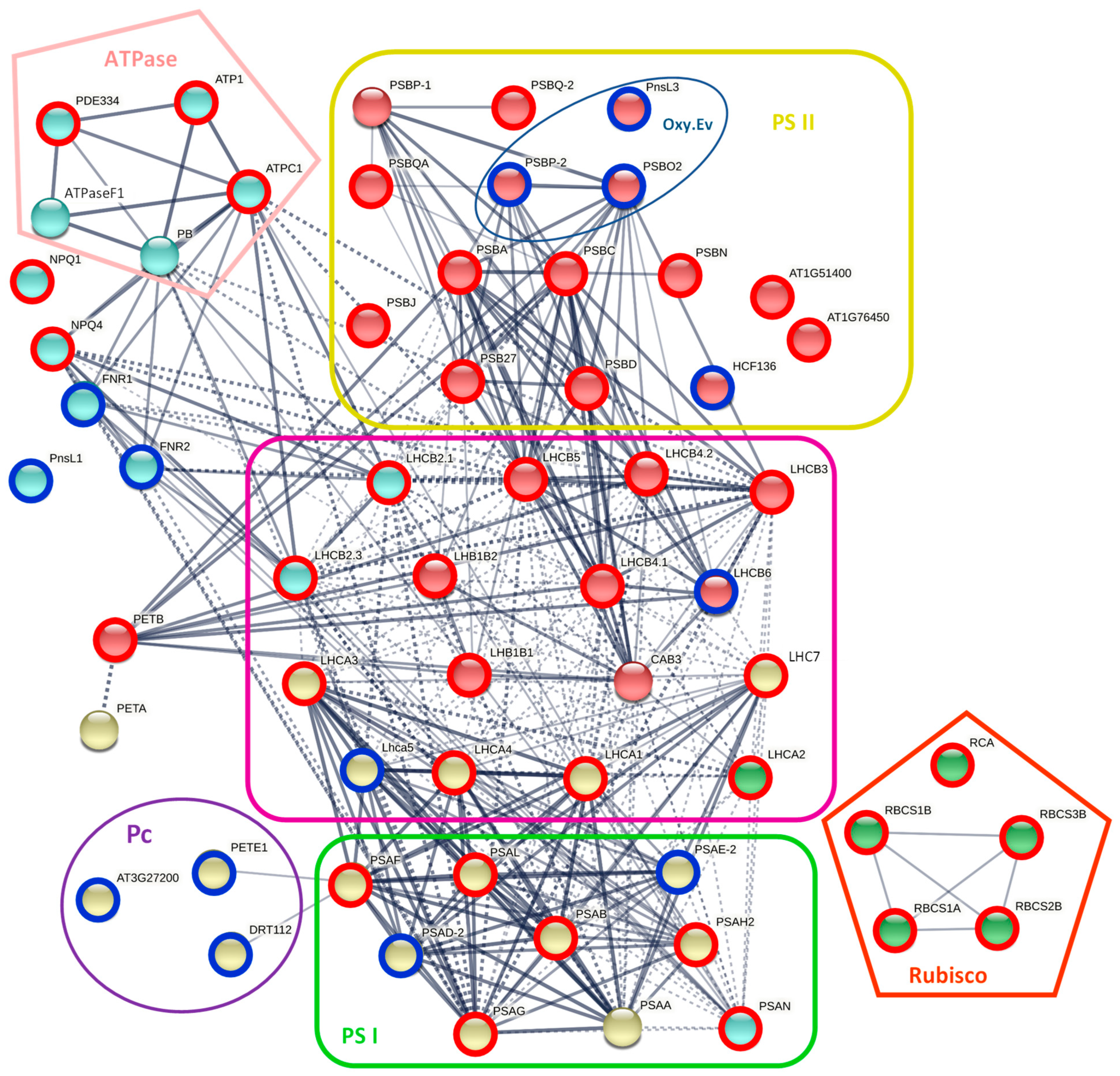
2.1. Changes in the Transcriptome and Proteome of R. serbica HL and DL
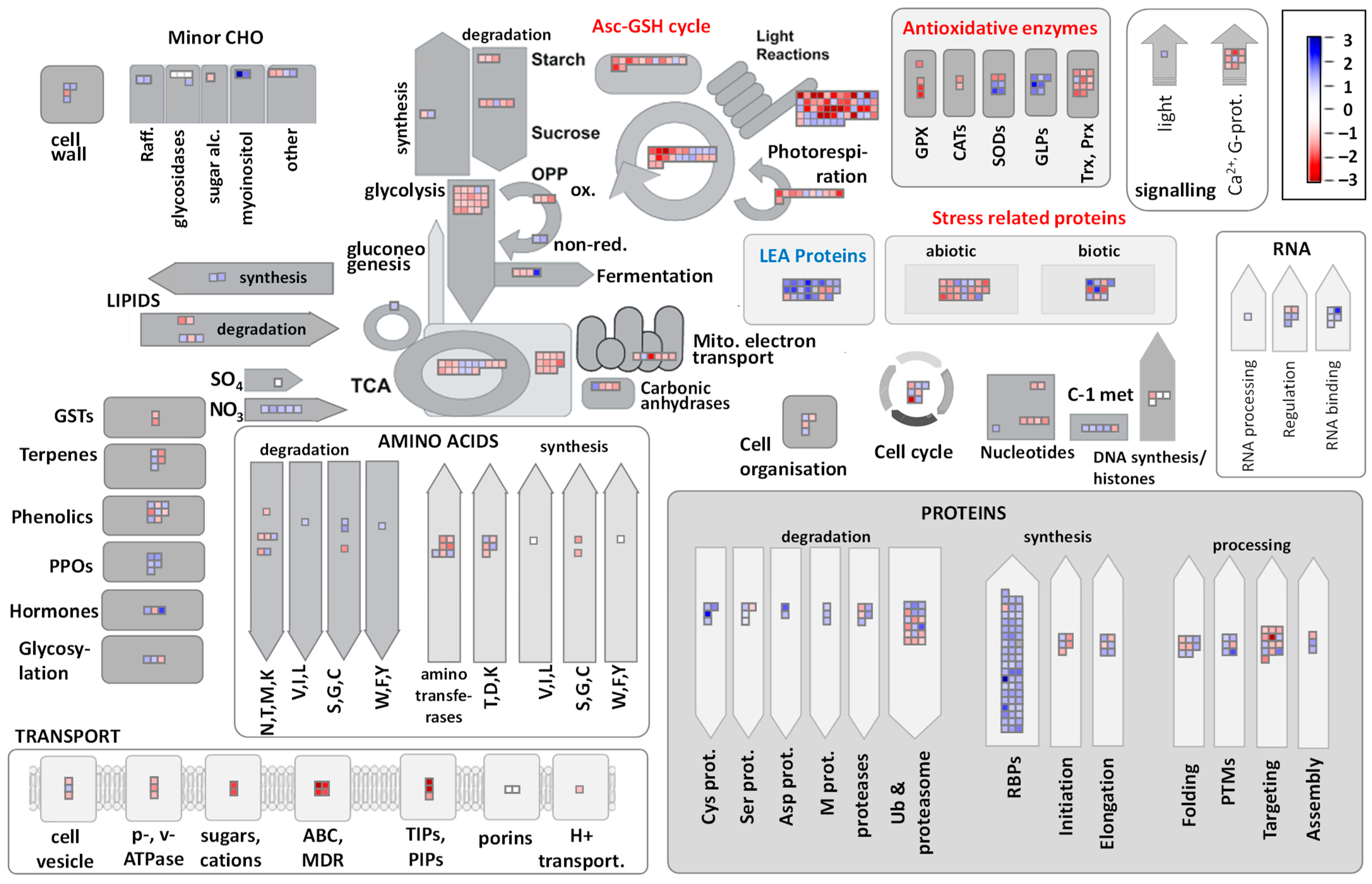
2.2. Functional Classification of DEGs and DAPs in R. serbica HL and DL
2.3. Major Metabolic Pathways Affected in DL of R. serbica
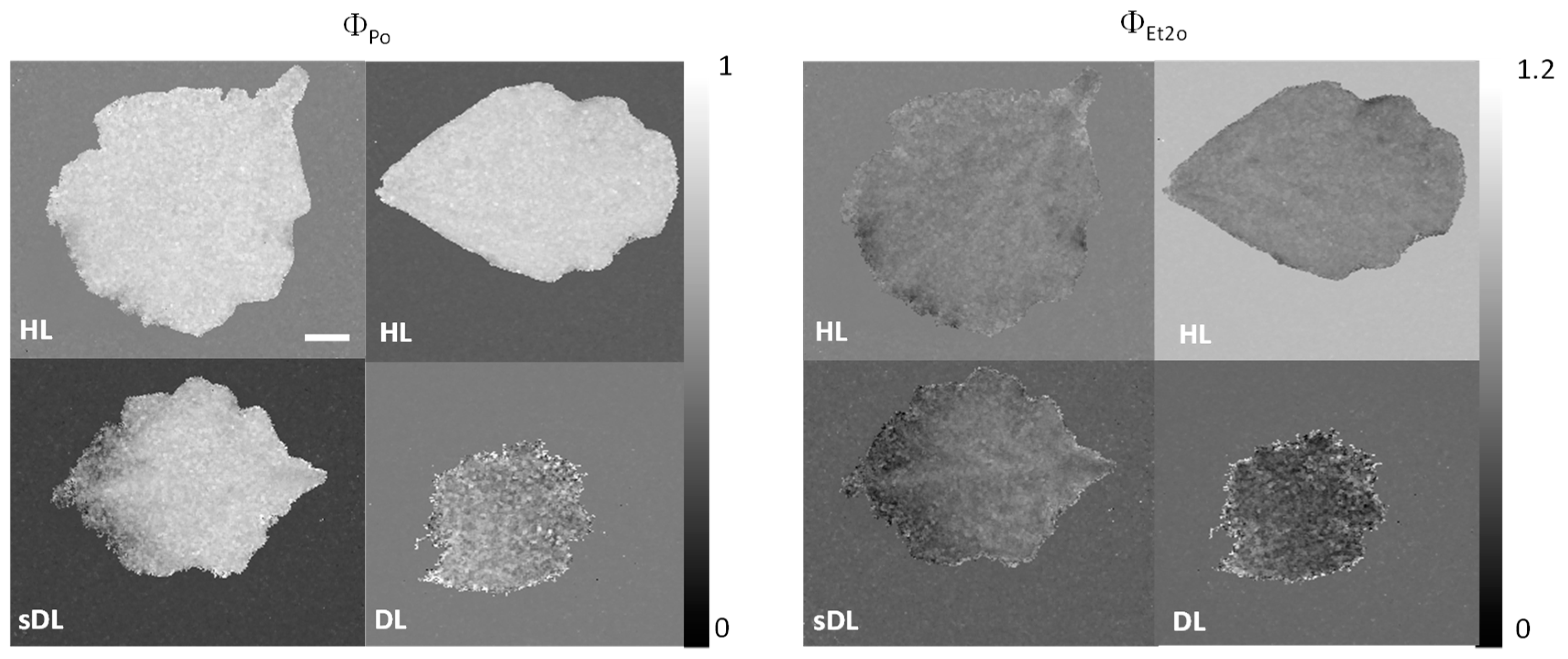
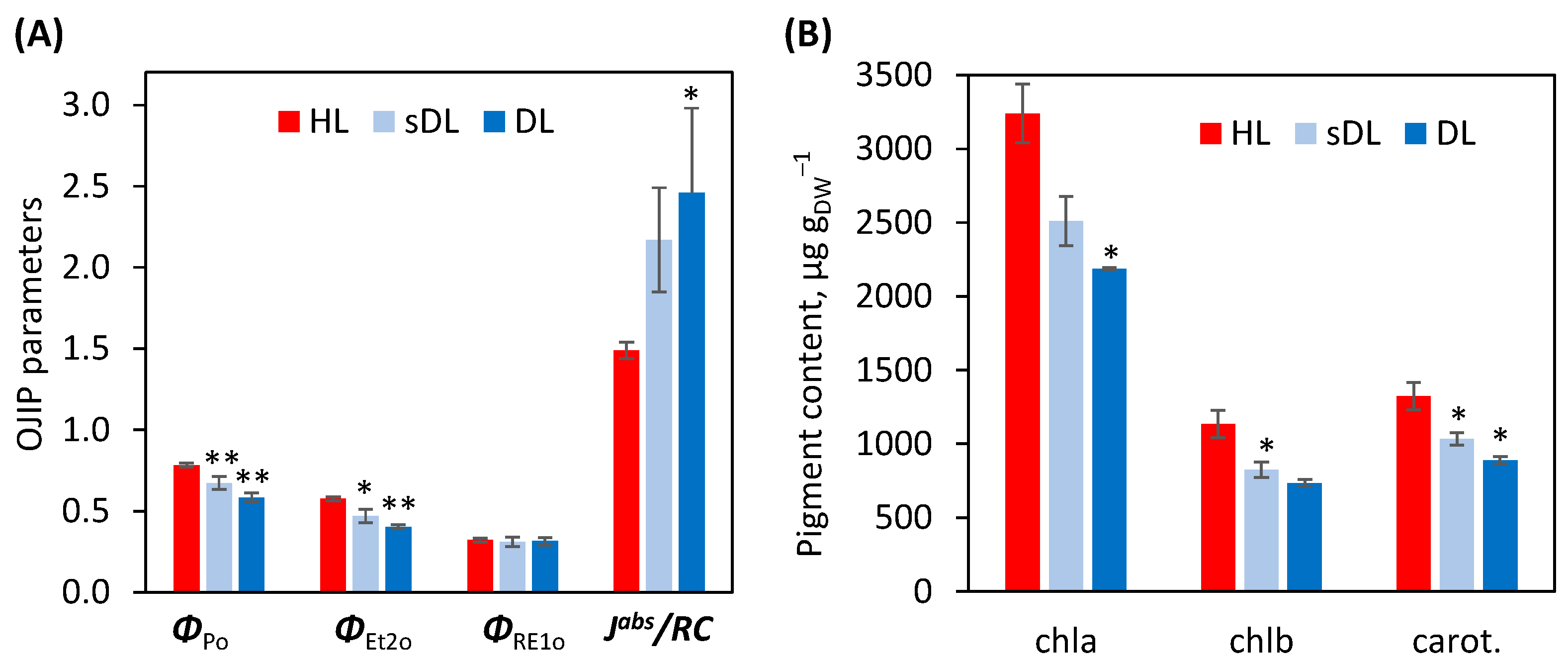
2.3.1. Photosynthesis
2.3.2. Carbohydrate Metabolism
2.3.3. Energy Production
2.3.4. Antioxidative Metabolism
2.3.5. Late Embryogenesis Abundant Proteins
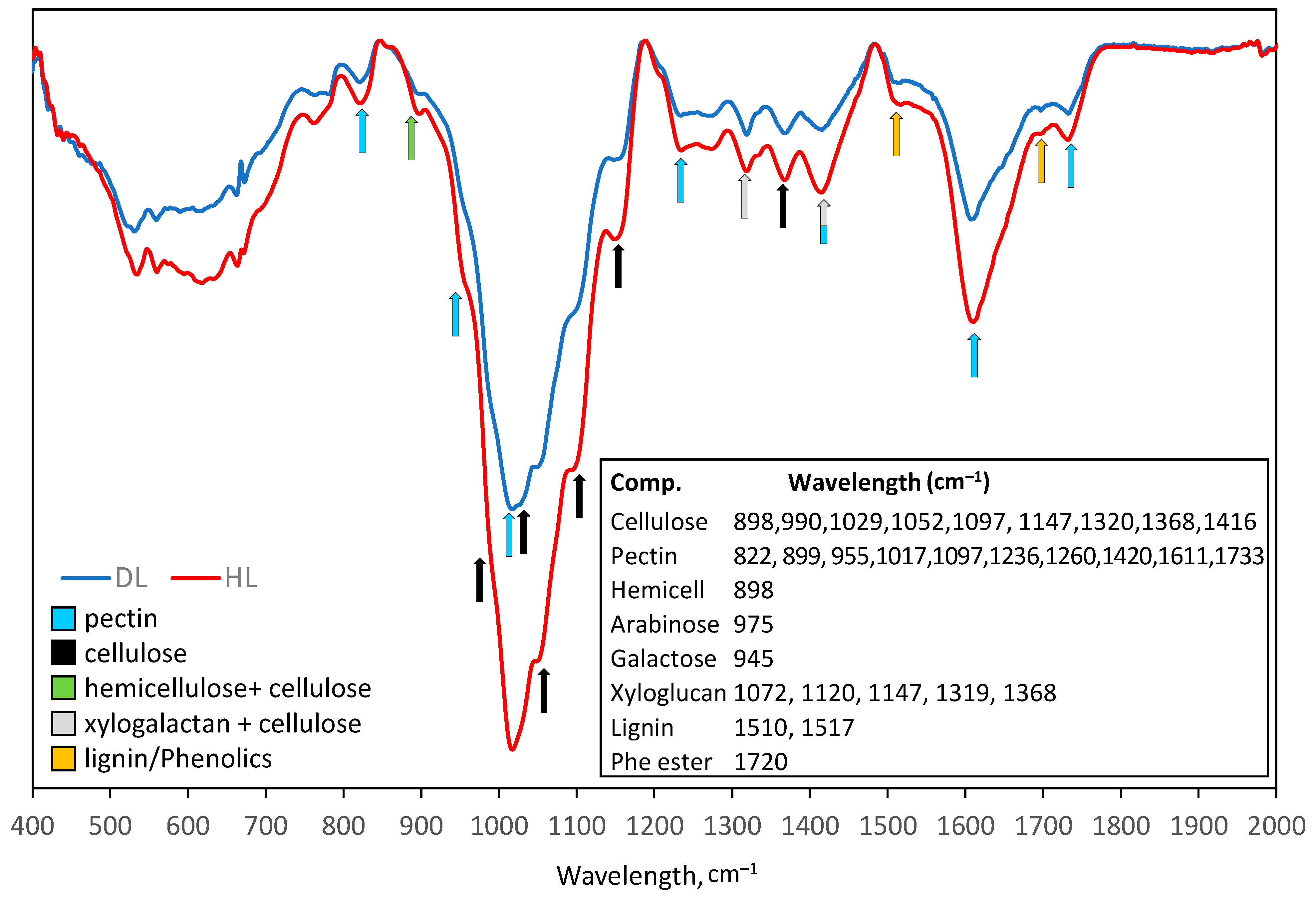
2.3.6. Cell Wall Remodeling
3. Discussion
3.1. Comparison of Proteomic and Transcriptomic Data
3.2. Cell Wall Remodeling
3.3. Germin-like Proteins
3.4. Photosynthesis
3.5. Energy Metabolism
3.6. Soluble Sugars and Proline
3.7. Chloroplastic Antioxidants and Phenolics
3.8. Late Embryogenesis Abundant Proteins
3.9. Proteostasis
4. Materials and Methods
4.1. Plant Material and Experimental Conditions
4.2. RNA Isolation, cDNA Library Construction, Sequencing, and De Novo Transcriptome Assembly
4.3. Functional Annotation and Gene Expression Analysis
4.4. Protein Extraction, TMT Labeling and SCX Fractionation
4.5. Liquid Chromatography Coupled to Tandem Mass Spectrometry
4.6. LC-MS/MS Data Analysis
4.7. Fast Chlorophyll Fluorescence Induction Kinetics (OJIP) and Analysis of Photosynthetic Pigments
4.8. Extraction and Measurement of Polyphenol Oxidases and Citrate Synthase Activities
4.9. Phenolics Analysis
4.10. Soluble Sugar Analysis
4.11. Cell Wall Isolation and Purification
4.12. Infrared (IR) Spectroscopy of Cell Wall Fractions
4.13. Phylogenetic Analysis and Conserved Motif Composition of R. serbica GLPs
4.14. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Rabara, R.C.; Tripathi, P.; Reese, R.N.; Rushton, D.L.; Alexander, D.; Timko, M.P.; Rushton, P.J. Tobacco drought stress responses reveal new targets for Solanaceae crop improvement. BMC Genom. 2015, 16, 484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farrant, J.M.; Hilhorst, H.W.M. What is dry? Exploring metabolism and molecular mobility at extremely low water contents. J. Exp. Bot. 2021, 72, 1507–1510. [Google Scholar] [CrossRef] [PubMed]
- Challabathula, D.; Zhang, Q.; Bartels, D. Protection of photosynthesis in desiccation-tolerant resurrection plants. J. Plant Physiol. 2018, 227, 84–92. [Google Scholar] [CrossRef] [PubMed]
- Scott, P. Resurrection plants and the secrets of eternal leaf. Ann. Bot. 2000, 85, 159–166. [Google Scholar] [CrossRef] [Green Version]
- Veljović-Jovanović, S.; Kukavica, B.; Stevanović, B.; Navari-Izzo, F. Senescence-and drought-related changes in peroxidase and superoxide dismutase isoforms in leaves of Ramonda serbica. J. Exp. Bot. 2006, 57, 1759–1768. [Google Scholar] [CrossRef] [PubMed]
- Challabathula, D.; Bartels, D. Desiccation tolerance in resurrection plants: New insights from transcriptome, proteome and metabolome analysis. Front. Plant Sci. 2013, 4, 482–497. [Google Scholar]
- Liu, J.; Moyankova, D.; Lin, C.T.; Mladenov, P.; Sun, R.Z.; Djilianov, D.; Deng, X. Transcriptome reprogramming during severe dehydration contributes to physiological and metabolic changes in the resurrection plant Haberlea rhodopensis. BMC Plant Biol. 2018, 18, 351–367. [Google Scholar] [CrossRef]
- Farrant, J.M.; Moore, J.P. Programming desiccation-tolerance: From plants to seeds to resurrection plants. Curr. Opin. Plant Biol. 2011, 14, 340–345. [Google Scholar] [CrossRef]
- Chandra, J.; Keshavkant, S. Desiccation-induced ROS accumulation and lipid catabolism in recalcitrant. Physiol. Mol. Biol. Plants 2018, 24, 75–87. [Google Scholar] [CrossRef]
- Gechev, T.S.; Dinakar, C.; Benina, M.; Toneva, V.; Bartels, D. Molecular mechanisms of desiccation tolerance in resurrection plants. Cell. Mol. Life Sci. 2012, 69, 3175–3186. [Google Scholar] [CrossRef]
- Bartels, D. Desiccation tolerance studied in the resurrection plant Craterostigma plantagineum. Integr. Comp. Biol. 2005, 45, 696–701. [Google Scholar] [CrossRef] [PubMed]
- Olvera-Carrillo, Y.; Campos, F.; Reyes, J.L.; Garciarrubio, A.; Covarrubias, A.A. Functional analysis of the group 4 late embryogenesis abundant proteins reveals their relevance in the adaptive response during water deficit in Arabidopsis. Plant Physiol. 2010, 154, 373–390. [Google Scholar] [CrossRef] [Green Version]
- Farrant, J.M. Mechanisms of desiccation tolerance in angiosperm resurrection plants. Plant Stress 2007, 1, 72–84. [Google Scholar]
- Strasser, R.J.; Tsimilli-Michael, M.; Qiang, S.; Goltsev, V. Simultaneous in vivo recording of prompt and delayed fluorescence and 820-nm reflection changes during drying and after rehydration of the resurrection plant Haberlea rhodopensis. Biochim. Biophys. Acta–Bioenerg. 2010, 1797, 1313–1326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dirk, L.M.A.; Abdel, C.G.; Ahmad, I.; Neta, I.C.S.; Pereira, C.C.; Pereira, F.E.C.B.; Unêda-Trevisoli, S.H.; Pinheiro, D.G.; Downie, A.B. Late embryogenesis abundant protein-client protein interactions. Plants 2020, 9, 814. [Google Scholar] [CrossRef] [PubMed]
- Ginsawaeng, O.; Heise, C.; Sangwan, R.; Karcher, D.; Hernández-Sánchez, I.E.; Sampathkumar, A.; Zuther, E. Subcellular localization of seed-expressed LEA_4 proteins reveals liquid-liquid phase separation for LEA9 and for LEA48 homo- and LEA42-LEA48 heterodimers. Biomolecules 2021, 11, 1770. [Google Scholar] [CrossRef] [PubMed]
- Dražić, G.; Mihailović, N.; Stevanović, B. Chlorophyll metabolism in leaves of higher poikilohydric plants Ramonda serbica Panč. and Ramonda nathaliae Panč. et Petrov. during dehydration and rehydration. J. Plant Physiol. 1999, 154, 379–384. [Google Scholar] [CrossRef]
- Zhu, Y.; Wang, B.; Phillips, J.; Zhang, Z.N.; Du, H.; Xu, T.; Huang, L.C.; Zhang, X.F.; Xu, G.H.; Li, W.L.; et al. Global transcriptome analysis reveals acclimation–primed processes involved in the acquisition of desiccation tolerance in Boea hygrometrica. Plant Cell Physiol. 2015, 56, 1429–1441. [Google Scholar] [CrossRef] [Green Version]
- Artur, M.A.S.; Zhao, T.; Ligterink, W.; Schranz, E.; Hilhorst, H.W.M. Dissecting the genomic diversification of late embryogenesis abundant (LEA) protein gene families in plants. Genome Biol. Evol. 2019, 11, 459–471. [Google Scholar] [CrossRef]
- Thimm, O.; Bläsing, O.; Gibon, Y.; Nagel, A.; Meyer, S.; Krüger, P.; Stitt, M. MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004, 37, 914–939. [Google Scholar] [CrossRef]
- Pantelić, A.; Stevanović, S.; Komić, S.M.; Kilibarda, N.; Vidović, M. In silico characterisation of the late embryogenesis abundant (LEA) protein families and their role in desiccation tolerance in Ramonda serbica Panc. Int. J. Mol. Sci. 2022, 23, 3547. [Google Scholar] [CrossRef] [PubMed]
- Živanović, B.; Milić-Komić, S.; Nikolić, N.; Mutavdžić, D.; Srećković, T.; Veljović-Jovanović, S.; Prokić, L. Differential response of two tomato genotypes, wild type cv. Ailsa Craig and its ABA-deficient mutant flacca to short-termed drought cycles. Plants 2021, 10, 2308. [Google Scholar] [CrossRef]
- Alonso-Simón, A.; García-Angulo, P.; Mélida, H.; Encina, A.; Álvarez, J.M.; Acebes, J.L. The use of FTIR spectroscopy to monitor modifications in plant cell wall architecture caused by cellulose biosynthesis inhibitors. Plant Signal. Behav. 2011, 6, 1104–1110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ross, A.B.; Langer, J.D.; Jovanovic, M. Proteome turnover in the spotlight: Approaches, applications, and perspectives. Mol. Cell. Proteom. 2021, 20, 100016. [Google Scholar] [CrossRef] [PubMed]
- Vogel, C.; Marcotte, E.M. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 2012, 13, 227–232. [Google Scholar] [CrossRef] [PubMed]
- Fernie, A.R.; Stitt, M. On the discordance of metabolomics with proteomics and transcriptomics: Coping with increasing complexity in logic, chemistry, and network interactions scientific correspondence. Plant Physiol. 2012, 158, 1139–1145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, C.; Cheng, S.; Zhang, Y.; Sun, Y.; Fernie, A.R.; Kang, K.; Panagiotou, G.; Lo, C.; Lim, B.L. Transcriptomic, proteomic and metabolic changes in Arabidopsis thaliana leaves after the onset of illumination. BMC Plant Biol. 2016, 16, 43. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.; Legay, S.; Sergeant, K.; Zorzan, S.; Leclercq, C.C.; Charton, S.; Giarola, V.; Liu, X.; Challabathula, D.; Renaut, J.; et al. Molecular insights into plant desiccation tolerance: Transcriptomics, proteomics and targeted metabolite profiling in Craterostigma plantagineum. Plant J. 2021, 107, 377–398. [Google Scholar] [CrossRef]
- Rakić, T.; Lazarević, M.; Jovanović, Z.S.; Radović, S.; Siljak–Yakovlev, S.; Stevanović, B.; Stevanović, V. Resurrection plants of the genus Ramonda: Prospective survival strategies—Unlock further capacity of adaptation, or embark on the path of evolution? Front. Plant Sci. 2014, 4, 550–560. [Google Scholar] [CrossRef] [Green Version]
- Moore, J.P.; Nguema-Ona, E.E.; Vicré-Gibouin, M.; Sørensen, I.; Willats, W.G.; Driouich, A.; Farrant, J.M. Arabinose-rich polymers as an evolutionary strategy to plasticize resurrection plant cell walls against desiccation. Planta 2013, 237, 739–754. [Google Scholar] [CrossRef]
- Jung, N.U. Molecular and Biochemical Studies of the Craterostigma plantagineum Cell Wall during Dehydration and Rehydration. Ph.D. Thesis, Mathematisch-Naturwissenschaftlichen Fakultät, Rheinischen Friedrich-Wilhelms-Universität Bonn, Bonn, Germany, 11 February 2020. [Google Scholar]
- Vicré, M.; Lerouxel, O.; Farrant, J.; Lerouge, P.; Driouich, A. Composition and desiccation-induced alterations of the cell wall in the resurrection plant Craterostigma wilmsii. Physiol. Plant. 2004, 120, 229–239. [Google Scholar] [CrossRef] [PubMed]
- Wormit, A.; Usadel, B. The multifaceted role of pectin methylesterase inhibitors (PMEIs). Int. J. Mol. Sci. 2018, 19, 2878. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, N.U.; Giarola, V.; Chen, P.; Knox, J.P.; Bartels, D. Craterostigma plantagineum cell wall composition is remodelled during desiccation and the glycine-rich protein CpGRP1 interacts with pectins through clustered arginines. Plant J. 2019, 100, 661–676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levesque-Tremblay, G.; Pelloux, J.; Braybrook, S.A.; Müller, K. Tuning of pectin methylesterification: Consequences for cell wall biomechanics and development. Planta 2015, 242, 791–811. [Google Scholar] [CrossRef]
- Wang, L.; Shang, H.; Liu, Y.; Zheng, M.; Wu, R.; Phillips, J.; Bartels, D.; Deng, X. A role for a cell wall localized glycine-rich protein in dehydration and rehydration of the resurrection plant Boea hygrometrica. Plant Biol. 2009, 11, 837–848. [Google Scholar] [CrossRef]
- Giarola, V.; Krey, S.; von den Driesch, B.; Bartels, D. The Craterostigma plantagineum glycine-rich protein CpGRP1 interacts with a cell wall-associated protein kinase 1 (CpWAK1) and accumulates in leaf cell walls during dehydration. New Phytol. 2016, 210, 535–550. [Google Scholar] [CrossRef] [Green Version]
- Pei, Y.; Li, X.; Zhu, Y.; Ge, X.; Sun, Y.; Liu, N.; Jia, Y.; Li, F.; Hou, Y. GhABP19, a novel germin-like protein from Gossypium hirsutum, plays an important role in the regulation of resistance to Verticillium and Fusarium wilt pathogens. Front. Plant Sci. 2019, 10, 583–601. [Google Scholar] [CrossRef] [Green Version]
- Veljović-Jovanović, S.; Kukavica, B.; Vidović, M.; Morina, F.; Menckhoff, L. Class III peroxidases: Functions, localization and redox regulation of isoenzymes. In Antioxidants and Antioxidant Enzymes in Higher Plants; Gupta, D.K., Palma, J.M., Corpas, F.J., Eds.; Springer: Cham, Switzerland; New York, NY, USA, 2018; pp. 269–300. [Google Scholar]
- Pristov, J.B.; Mitrović, A.; Spasojević, I. A comparative study of antioxidative activities of cell-wall polysaccharides. Carbohydr. Res. 2011, 346, 2255–2259. [Google Scholar] [CrossRef]
- Kukavica, B.; Mojović, M.; Vučinić, Ž.; Maksimović, V.; Takahama, U.; Veljović-Jovanović, S. Generation of hydroxyl radical in isolated pea root cell wall, and the role of cell wall-bound peroxidase, Mn-SOD and phenolics in their production. Plant Cell Physiol. 2009, 50, 304–317. [Google Scholar] [CrossRef] [Green Version]
- Collett, H.M.; Butowt, R.; Smith, J.; Farrant, J.; Illing, N. Photosynthetic genes are differentially transcribed during the dehydration-rehydration cycle in the resurrection plant, Xerophyta humilis. J. Exp. Bot. 2003, 54, 2543–2595. [Google Scholar] [CrossRef]
- Yang, E.J.; Oh, Y.A.; Lee, E.S.; Park, A.R.; Cho, S.K.; Yoo, Y.J.; Park, O.K. Oxygen-evolving enhancer protein 2 is phosphorylated by glycine-rich protein 3/wall-associated kinase 1 in Arabidopsis. Biochem. Biophys. Res. Commun. 2003, 305, 862–868. [Google Scholar] [CrossRef]
- Jiang, G.; Wang, Z.; Shang, H.; Yang, W.; Hu, Z.; Phillips, J.; Deng, X. Proteome analysis of leaves from the resurrection plant Boea hygrometrica in response to dehydration and rehydration. Planta 2007, 225, 1405–1420. [Google Scholar] [CrossRef] [PubMed]
- Farrant, J.M. A comparison of mechanisms of desiccation tolerance among three angiosperm resurrection plant species. Plant Ecol. 2000, 151, 29–39. [Google Scholar] [CrossRef]
- Heber, U.; Bilger, W.; Bligny, R.; Lange, O.L. Photo-tolerance of lichens, mosses and higher plants in an alpine environment: Analysis of photoreactions. Planta 2000, 211, 770–780. [Google Scholar] [CrossRef]
- Huang, W.; Yang, S.-J.; Zhang, S.-B.; Zhang, J.-L.; Cao, K.-F. Cyclic electron flow plays an important role in photoprotection for the resurrection plant Paraboea rufescens under drought stress. Planta 2012, 235, 819–828. [Google Scholar] [CrossRef]
- Tan, T.; Sun, Y.; Luo, S.; Zhang, C.; Zhou, H.; Lin, H. Efficient modulation of photosynthetic apparatus confers desiccation tolerance in the resurrection plant Boea hygrometrica. Plant Cell Physiol. 2017, 58, 1976–1990. [Google Scholar] [CrossRef]
- Mladenov, P.; Finazzi, G.; Bligny, R.; Moyankova, D.; Zasheva, D.; Boisson, A.-M.; Brugière, S.; Krasteva, V.; Alipieva, K.; Simova, S.; et al. In vivo spectroscopy and NMR metabolite fingerprinting approaches to connect the dynamics of photosynthetic and metabolic phenotypes in resurrection plant Haberlea rhodopensis during desiccation and recovery. Front. Plant Sci. 2015, 6, 564–578. [Google Scholar] [CrossRef] [Green Version]
- Markovska, Y.; Tsonev, T.; Kimenov, G. Regulation of cam and respiratory recycling by water supply in higher poikilohydric plants—Haberlea rhodopensis Friv. and Ramonda serbica Panc, at transition from biosis to anabiosis and vice versa. Bot. Acta 1997, 110, 18–24. [Google Scholar] [CrossRef]
- Kirch, H.H.; Nair, A.; Bartels, D. Novel ABA-and dehydration-inducible aldehyde dehydrogenase genes isolated from the resurrection plant Craterostigma plantagineum and Arabidopsis thaliana. Plant J. 2001, 28, 555–567. [Google Scholar] [CrossRef]
- Živković, T.; Quartacci, M.F.; Stevanović, B.; Marinone, F.; Navari-Izzo, F. Low molecular weight substances in the poikilohydric plant Ramonda serbica during dehydration and rehydration. Plant Sci. 2005, 168, 105–111. [Google Scholar] [CrossRef]
- Ingram, J.; Bartels, D. The molecular basis of dehydration tolerance in plants. Annu. Rev. Plant Physiol. 1996, 47, 377–403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vidović, M.; Morina, F.; Milić, S.; Albert, A.; Zechmann, B.; Tosti, T.; Winkler, J.B.; Jovanović, S.V. Carbon allocation from source to sink leaf tissue in relation to flavonoid biosynthesis in variegated Pelargonium zonale under UV-B radiation and high PAR intensity. Plant Physiol. Biochem. 2015, 93, 44–55. [Google Scholar] [CrossRef] [PubMed]
- Nishizawa, A.; Yabuta, Y.; Shigeoka, S. Galactinol and raffinose constitute a novel function to protect plants from oxidative damage. Plant Physiol. 2008, 147, 1251–1263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dietz, K.J.; Jacob, S.; Oelze, M.L.; Laxa, M.; Tognetti, V.; de Miranda, S.M.; Baier, M.; Finkemeier, I. The function of peroxiredoxins in plant organelle redox metabolism. J. Exp. Bot. 2016, 57, 1697–1709. [Google Scholar] [CrossRef] [PubMed]
- Augusti, A.; Scartazza, A.; Navari-Izzo, F.; Sgherri, C.L.M.; Stevanović, B.; Brugnoli, E. Photosystem II photochemical efficiency, zeaxanthin and antioxidant contents in the poikilohydric Ramonda serbica during dehydration and rehydration. Photosynth. Res. 2001, 67, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Vidović, M.; Franchin, C.; Morina, F.; Veljović-Jovanović, S.; Masi, A.; Arrigoni, G. Efficient protein extraction for shotgun proteomics from hydrated and desiccated leaves of resurrection Ramonda serbica plants. Anal. Bioanal. Chem. 2020, 412, 8299–8312. [Google Scholar] [CrossRef]
- Vidović, M.; Morina, F.; Veljović-Jovanović, S. Stimulation of various phenolics in plants under ambient UV-B radiation. In UV-B Radiation: From Environmental Stressor to Regulator of Plant Growth; Singh, V.P., Singh, S., Prasad, S.M., Parihar, P., Eds.; Wiley-Blackwell: Hoboken, NJ, USA, 2017; pp. 9–56. [Google Scholar]
- Veljović-Jovanović, S.; Kukavica, B.; Navari-Izzo, F. Characterization of polyphenol oxidase changes induced by desiccation of Ramonda serbica leaves. Physiol. Plant. 2008, 132, 407–416. [Google Scholar] [CrossRef]
- Sgherri, C.; Stevanovic, B.; Navari-Izzo, F. Role of phenolics in the antioxidative status of the resurrection plant Ramonda serbica during dehydration and rehydration. Physiol. Plant. 2004, 122, 478–485. [Google Scholar] [CrossRef]
- Takahama, U. Oxidation of vacuolar and apoplastic phenolic substrates by peroxidase: Physiological significance of the oxidation reactions. Phytochem. Rev. 2004, 3, 207–219. [Google Scholar] [CrossRef]
- Boeckx, T.; Winters, A.L.; Webb, K.J.; Kingston-Smith, A.H. Polyphenol oxidase in leaves: Is there any significance to the chloroplastic localization? J. Exp. Bot. 2015, 66, 3571–3579. [Google Scholar] [CrossRef] [Green Version]
- Bremer, A.; Wolff, M.; Thalhammer, A.; Hincha, D.K. Folding of intrinsically disordered plant LEA proteins is driven by glycerol-induced crowding and the presence of membranes. FEBS J. 2017, 284, 919–936. [Google Scholar] [CrossRef] [PubMed]
- Cuevas-Velazquez, C.L.; Reyes, J.L.; Covarrubias, A.A. Group 4 late embryogenesis abundant proteins as a model to study intrinsically disordered proteins in plants. Plant Signal. Behav. 2017, 12, 10893–10903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Candat, A.; Paszkiewicz, G.; Neveu, M.; Gautier, R.; Logan, D.C.; Avelange-Macherel, M.H.; Macherel, D. The ubiquitous distribution of late embryogenesis abundant proteins across cell compartments in Arabidopsis offers tailored protection against abiotic stress. Plant Cell 2014, 26, 3148–3166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koag, M.C.; Wilkens, S.; Fenton, R.D.; Resnik, J.; Vo, E.; Close, T.J. The K-segment of maize DHN1 mediates binding to anionic phospholipid vesicles and concomitant structural changes. Plant Physiol. 2009, 150, 1503–1514. [Google Scholar] [CrossRef] [Green Version]
- Hara, M.; Shinoda, Y.; Tanaka, Y.; Kuboi, T. DNA binding of citrus dehydrin promoted by zinc ion. Plant Cell Environ. 2009, 32, 532–541. [Google Scholar] [CrossRef]
- Liu, X.; Wang, Z.; Wang, L.; Wu, R.; Phillips, J.; Deng, X. LEA 4 group genes from the resurrection plant Boea hygrometrica confer dehydration tolerance in transgenic tobacco. Plant Sci. 2009, 176, 90–98. [Google Scholar] [CrossRef]
- Olvera-Carrillo, Y.; Luis Reyes, J.; Covarrubias, A.A. Late embryogenesis abundant proteins: Versatile players in the plant adaptation to water limiting environments. Plant Signal. Behav. 2011, 6, 586–589. [Google Scholar] [CrossRef] [Green Version]
- Chakrabortee, S.; Tripathi, R.; Watson, M.; Schierle, G.S.; Kurniawan, D.P.; Kaminski, C.F.; Wise, M.J.; Tunnacliffe, A. Intrinsically disordered proteins as molecular shields. Mol. Biosyst. 2012, 8, 210–219. [Google Scholar] [CrossRef] [Green Version]
- Belott, C.; Janis, B.; Menze, M.A. Liquid-liquid phase separation promotes animal desiccation tolerance. Proc. Natl. Acad. Sci. USA 2020, 117, 27676–27684. [Google Scholar] [CrossRef]
- Rohrig, H.; Schmidt, J.; Colby, T.; Brautigam, A.; Hufnagel, P.; Bartels, D. Desiccation of the resurrection plant Craterostigma plantagineum induces dynamic changes in protein phosphorylation. Plant Cell Environ. 2006, 29, 1606–1617. [Google Scholar] [CrossRef] [Green Version]
- Biundo, A.; Braunschmid, V.; Pretzler, M.; Kampatsikas, I.; Darnhofer, B.; Birner-Gruenberger, R.; Rompel, A.; Ribitsch, D.; Guebitz, G.M. Polyphenol oxidases exhibit promiscuous proteolytic activity. Commun. Chem. 2020, 3, 62. [Google Scholar] [CrossRef]
- Harten, J.B.; Eickmeier, W.G. Enzyme dynamics of the resurrection plant Selaginella lepidophylla (Hook. & Grev.) spring during rehydration. Plant Phys. 1986, 82, 61–64. [Google Scholar]
- Bradford, M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Ebinezer, L.B.; Franchin, C.; Trentin, A.R.; Carletti, P.; Trevisan, S.; Agrawal, G.K.; Quaggiotti, S.; Arrigoni, G.; Masi, A. Quantitative proteomics of maize roots treated with a protein hydrolysate: A comparative study with transcriptomics highlights the molecular mechanisms responsive to biostimulants. J. Agric. Food Chem. 2020, 68, 7541–7553. [Google Scholar] [CrossRef]
- Emanuelsson, O.; Brunak, S.; von Heijne, G.; Nielsen, H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2007, 2, 953–971. [Google Scholar] [CrossRef]
- Küpper, H.; Benedikty, Z.; Morina, F.; Andresen, E.; Mishra, A.; Trtilek, M. Analysis of OJIP chlorophyll fluorescence kinetics and QA reoxidation kinetics by direct fast imaging. Plant Physiol. 2019, 179, 369–381. [Google Scholar] [CrossRef] [Green Version]
- Lichtenthaler, H.K.; Wellburn, A.R. Determinations of total carotenoids and chlorophylls a and b of leaf extracts in different solvents. Biochem. Soc. Trans. 1983, 11, 591–592. [Google Scholar] [CrossRef] [Green Version]


| HL | DL | HL | DL | ||
|---|---|---|---|---|---|
| Hexose (mmol g−1 DW) | Oligosaccharides (µmol g−1 DW) | ||||
| Glucose | 0.94 ± 0.15 | 2.26 ± 0.34 ** | Isomaltotriose | 2.7 ± 0.6 | 7.8 ± 2.7 |
| Fructose | 0.71 ± 0.10 | 2.06 ± 0.40 ** | Maltotriose | 6.3 ± 1.5 | 6.7 ± 1.7 |
| Galactose | 0.19 ± 0.02 | 0.79 ± 0.10 * | Raffinose | 6.4 ± 2.9 | 16.4 ± 2.8 * |
| Pentose (µmol g−1 DW) | Melezitose | 2.9 ± 1.2 | 10.0 ± 2.0 * | ||
| Arabinose | 64.4 ± 19.9 | 168.6 ± 57.2 | Panose | 0.23 ± 0.12 | 0.82 ± 0.32 |
| Ribose | 26.5 ± 6.5 | 105.3 ± 21.1 * | Stachyose | 0.70 ± 0.21 | 4.54 ± 0.81 ** |
| Rhamnose | 2.6 ± 0.4 | 25.1 ± 5.6 * | Sugar alcohols (µmol g−1 DW) | ||
| Xylose | 12.3 ± 3.3 | 34.7 ± 6.9 * | Erythritol | 75.23 ± 27.5 | 213.7 ± 50.5 * |
| Disaccharides (µmol g−1 DW) | Sorbitol | 48.9 ± 16.2 | 155.0 ± 60.7 | ||
| Sucrose | 317.4 ± 60.1 | 1367.5 ± 461.3 | Galactitol | 122.0 ± 41.7 | 296.9 ± 171.5 |
| Trehalose | 29.2 ± 4.7 | 79.1 ± 31.8 | Arabinitol | 12.3 ± 4.6 | 66.5 ± 18.4 * |
| Melibiose | 2.4 ± 0.7 | 5.7 ± 1.9 | Mannitol | 6.2 ± 1.2 | 23.5 ± 8.8 |
| Maltose | 13.6 ± 4.2 | 62.4 ± 18.9 * | |||
| Isomaltose | 2.7 ± 0.6 | 7.8 ± 2.7 * | Total soluble sugars (mmol g−1 DW) | ||
| Turanose | 90.4 ± 28.0 | 312.2 ± 76.6 * | 2.7 ± 0.3 | 8.2 ± 1.0 ** | |
| Treatment | CS, U g–1DW | PPO, ΔA410 mg–1prot |
|---|---|---|
| HL (90% RWC) | 0.72 ± 0.08 | 52.3 ± 4.7 |
| sDL (50% RWC) | 0.95 ± 0.03 | 84.9 ± 6.8 ** |
| DL (15–20% RWC) | 1.07 ± 0.04 * | 142.6 ± 3.0 ** |
| LEAP Family Subgroup | Rs_code | Fold Change (DL/HL) | Subcellular Localisation * |
|---|---|---|---|
| LEA1.3 | Rs_170082 | 1.6 | nucleus |
| LEA2.2 | Rs_151841 | −1.3 | nucleus |
| LEA2.2 | Rs_70056 | −1.4 | chloroplasts; cytosol |
| LEA2.2 | Rs_70057 | −1.6 | cytosol |
| LEA4.3 | Rs_139539 | 2.3 | chloroplasts; mitochondrion |
| LEA4 ** | Rs_191445 | 2.3 | mitochondrion |
| LEA4.3 | Rs_67538 | 2.1 | chloroplast |
| LEA4 ** | Rs_121834 | 2.1 | mitochondrion |
| LEA4.3 | Rs_136891 | 2.0 | nucleus |
| LEA4.3 | Rs_3451 | 2.0 | chloroplast |
| LEA4.3 | Rs_67537 | 1.9 | nucleus |
| LEA4.3 | Rs_67210 | 1.9 | nucleus, mitochondrion |
| LEA4.3 | Rs_67211 | 1.7 | mitochondrion |
| LEA4.3 | Rs_67212 | 1.7 | mitochondrion |
| LEA4.3 | Rs_67539 | 1.6 | chloroplast |
| LEA4.3 | Rs_205247 | 1.6 | chloroplast |
| LEA4.3 | Rs_205105 | 1.6 | chloroplast |
| LEA4.3 | Rs_52085 | 1.5 | mitochondrion |
| LEA4.3 | Rs_140058 | 1.4 | nucleus |
| LEA4 ** | Rs_187459 | 1.4 | mitochondrion, nucleus |
| Other (LEA4) | Rs_67929 | 1.4 | cytosol |
| LEA4.2 | Rs_108587 | 1.4 | nucleus, mitochondrion |
| LEA4 ** | Rs_165950 | 1.3 | nucleus |
| DEH ** | Rs_194906 | 1.3 | nucleus |
| DEH1 | Rs_107018 | 1.3 | nucleus, mitochondrion |
| SMP | Rs_104365 | −1.6 | nucleus; cytosol |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vidović, M.; Battisti, I.; Pantelić, A.; Morina, F.; Arrigoni, G.; Masi, A.; Jovanović, S.V. Desiccation Tolerance in Ramonda serbica Panc.: An Integrative Transcriptomic, Proteomic, Metabolite and Photosynthetic Study. Plants 2022, 11, 1199. https://doi.org/10.3390/plants11091199
Vidović M, Battisti I, Pantelić A, Morina F, Arrigoni G, Masi A, Jovanović SV. Desiccation Tolerance in Ramonda serbica Panc.: An Integrative Transcriptomic, Proteomic, Metabolite and Photosynthetic Study. Plants. 2022; 11(9):1199. https://doi.org/10.3390/plants11091199
Chicago/Turabian StyleVidović, Marija, Ilaria Battisti, Ana Pantelić, Filis Morina, Giorgio Arrigoni, Antonio Masi, and Sonja Veljović Jovanović. 2022. "Desiccation Tolerance in Ramonda serbica Panc.: An Integrative Transcriptomic, Proteomic, Metabolite and Photosynthetic Study" Plants 11, no. 9: 1199. https://doi.org/10.3390/plants11091199
APA StyleVidović, M., Battisti, I., Pantelić, A., Morina, F., Arrigoni, G., Masi, A., & Jovanović, S. V. (2022). Desiccation Tolerance in Ramonda serbica Panc.: An Integrative Transcriptomic, Proteomic, Metabolite and Photosynthetic Study. Plants, 11(9), 1199. https://doi.org/10.3390/plants11091199









