Genome-Wide Identification of Cotton (Gossypium spp.) Trehalose-6-Phosphate Phosphatase (TPP) Gene Family Members and the Role of GhTPP22 in the Response to Drought Stress
Abstract
:1. Introduction
2. Results
2.1. Identification and Fundamental Analysis of TPP Genes
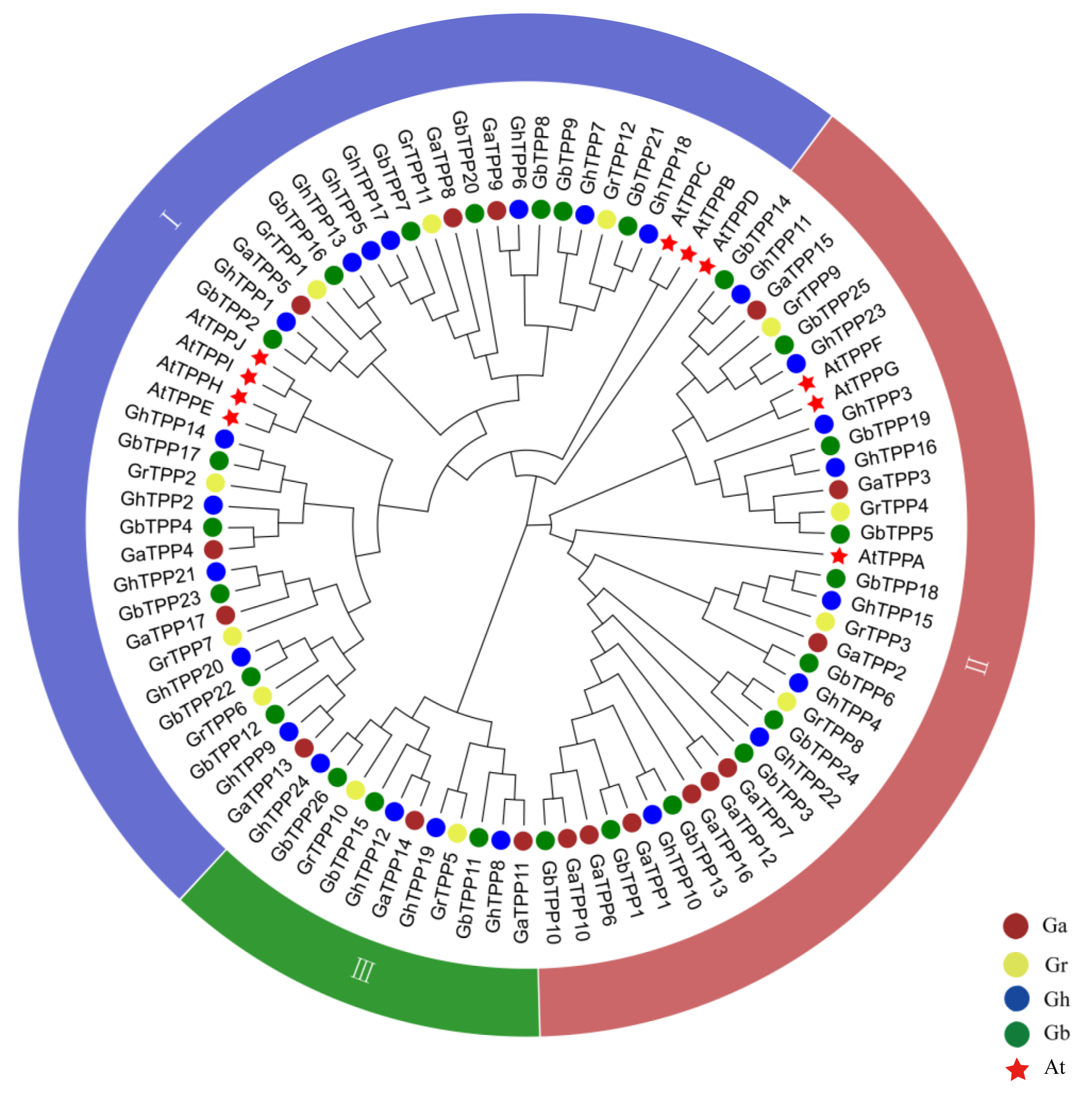
2.2. Phylogenetic Analysis of TPP Family Genes
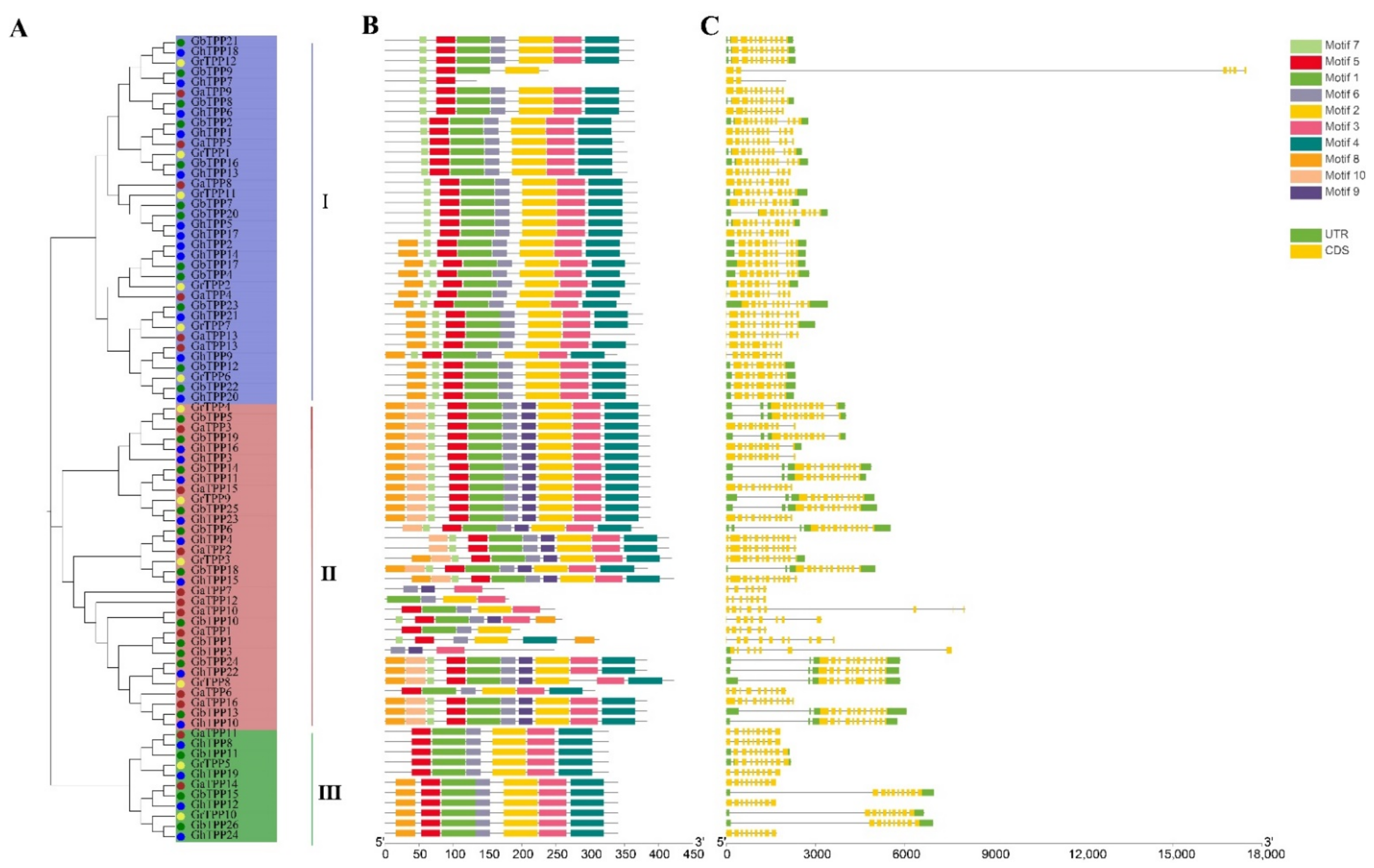
2.3. Conserved Motif and Gene Structure Analyses of TPP Proteins
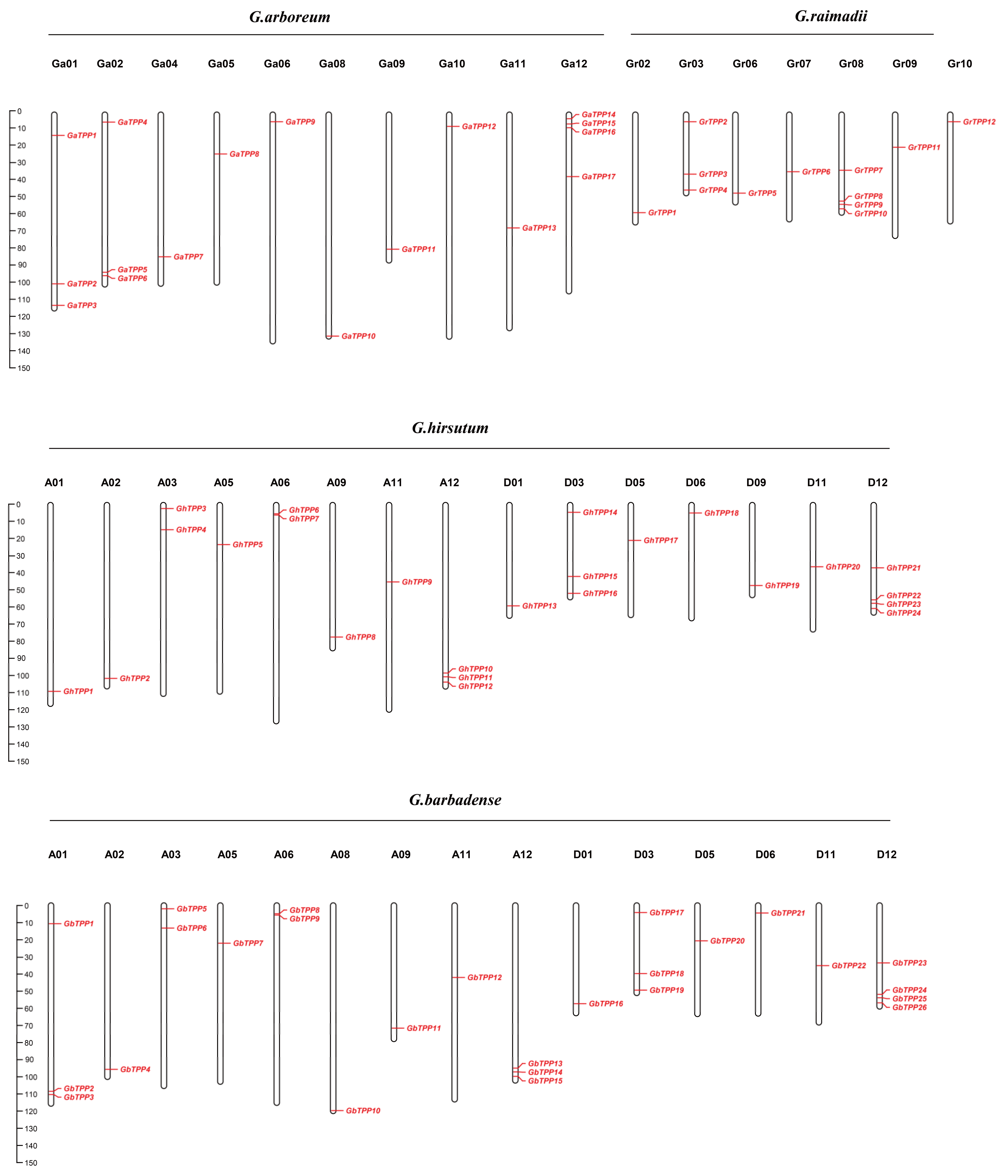
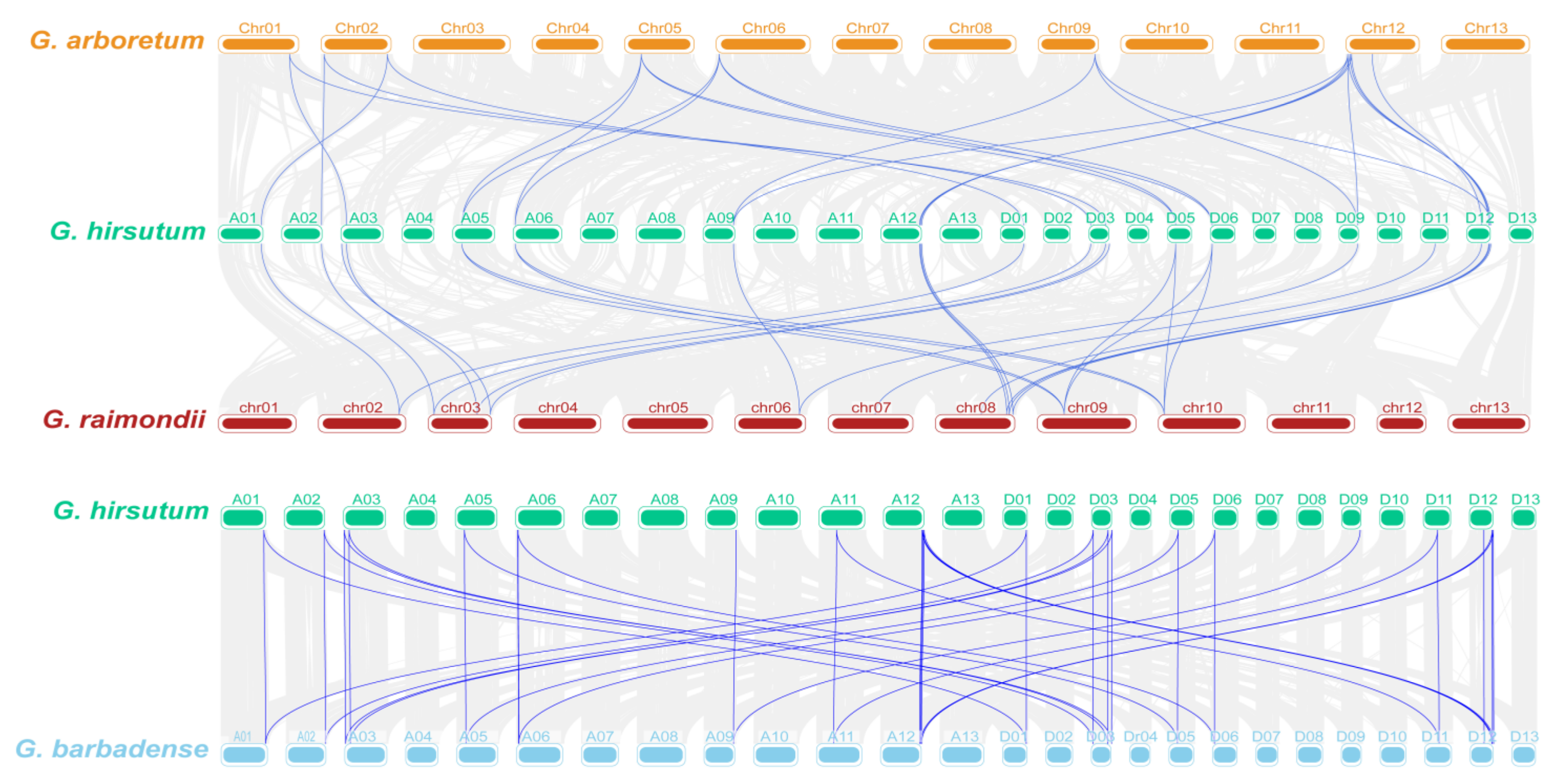
2.4. Chromosomal Distribution and Homology Analysis of TPP Genes
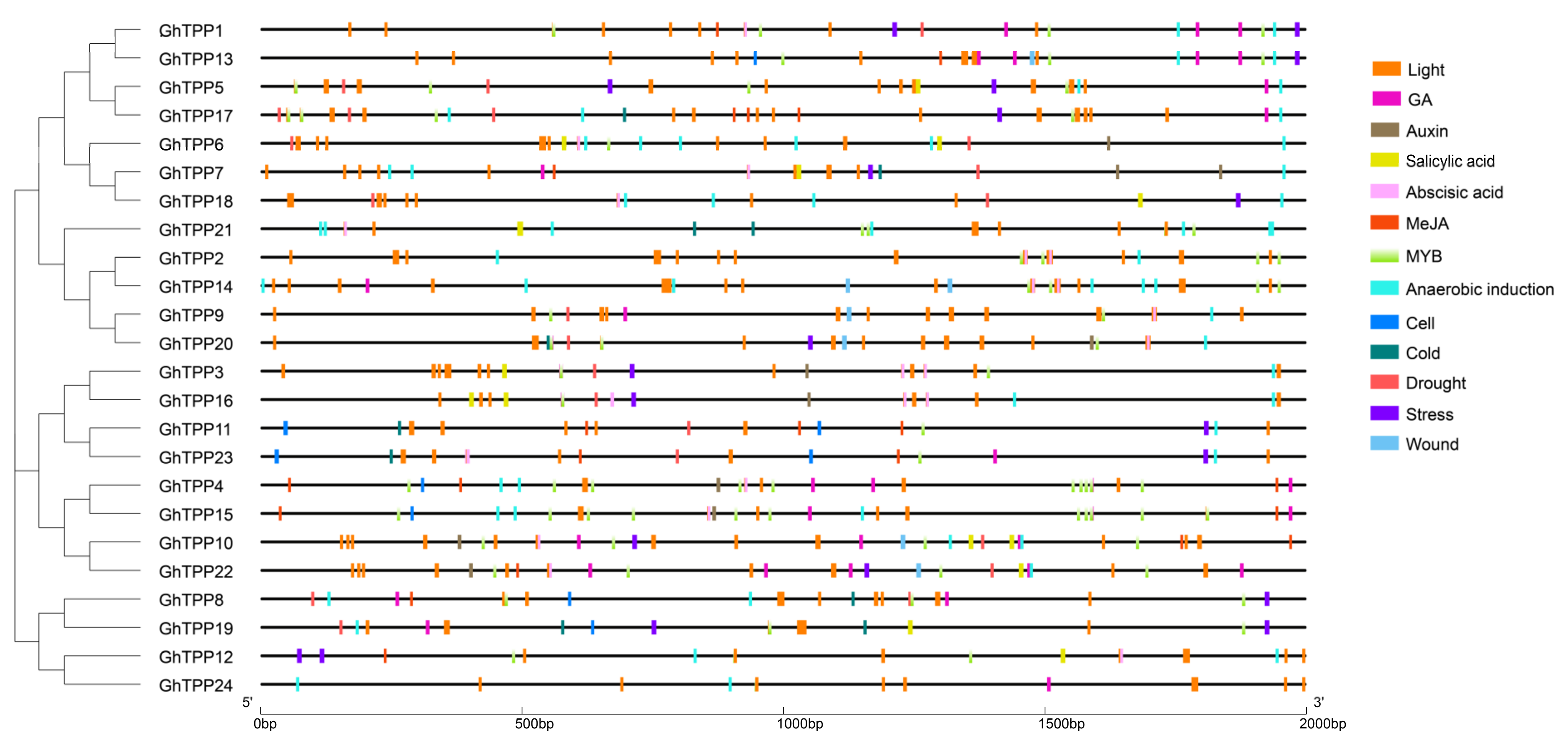
2.5. Prediction and Analysis of Cis-Acting Elements in Promoter Regions
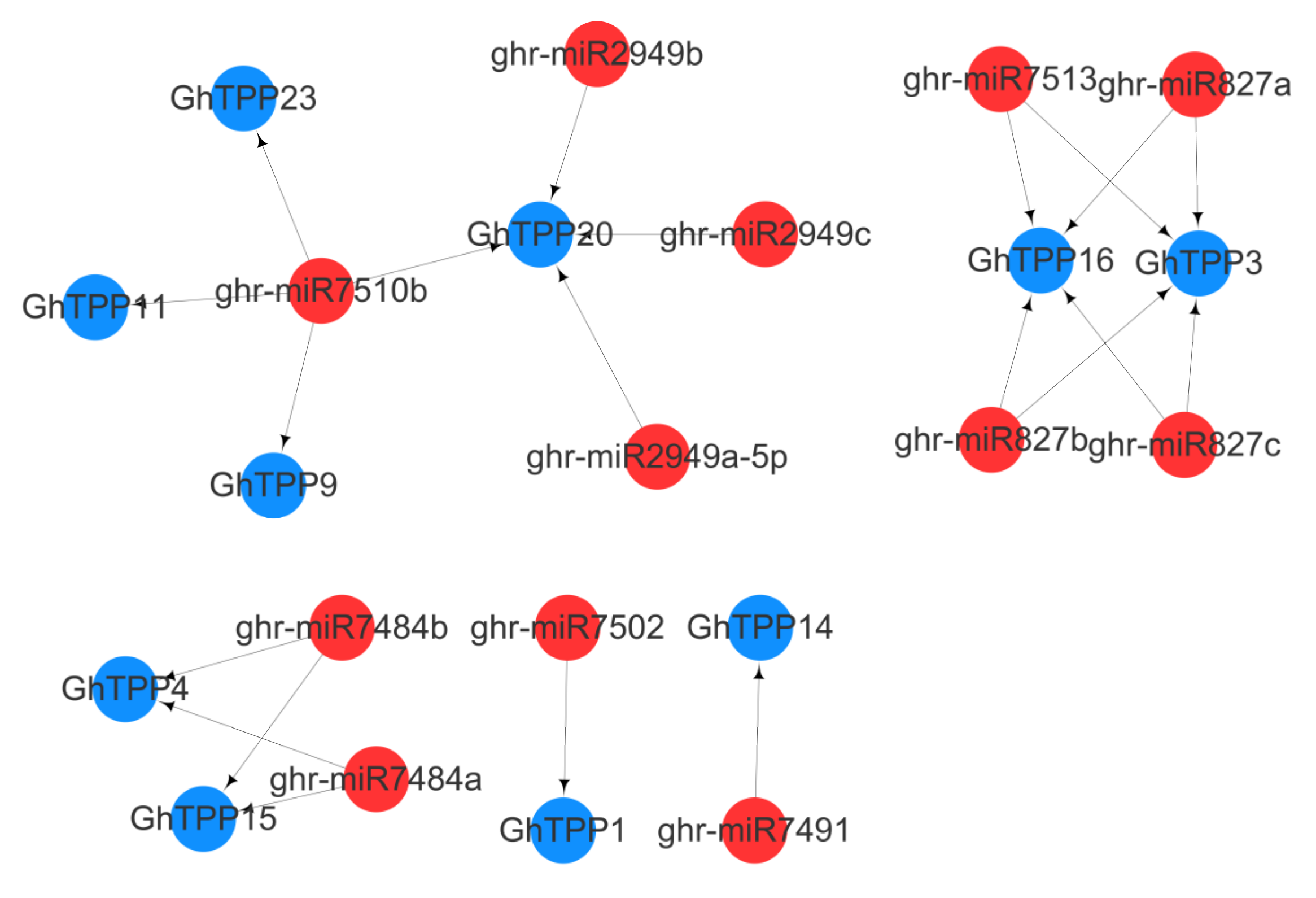
2.6. Prediction of miRNAs Targeting TPP Family Genes
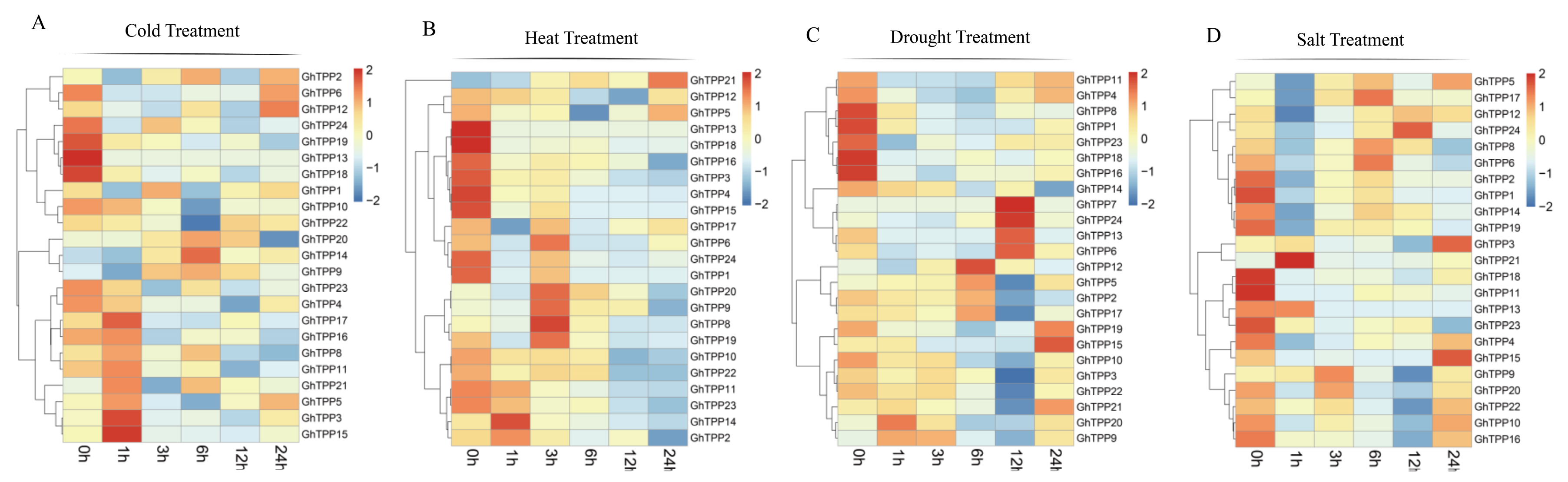
2.7. Analysis of the Expression Patterns of TPP Family Members in Response to Abiotic Stress
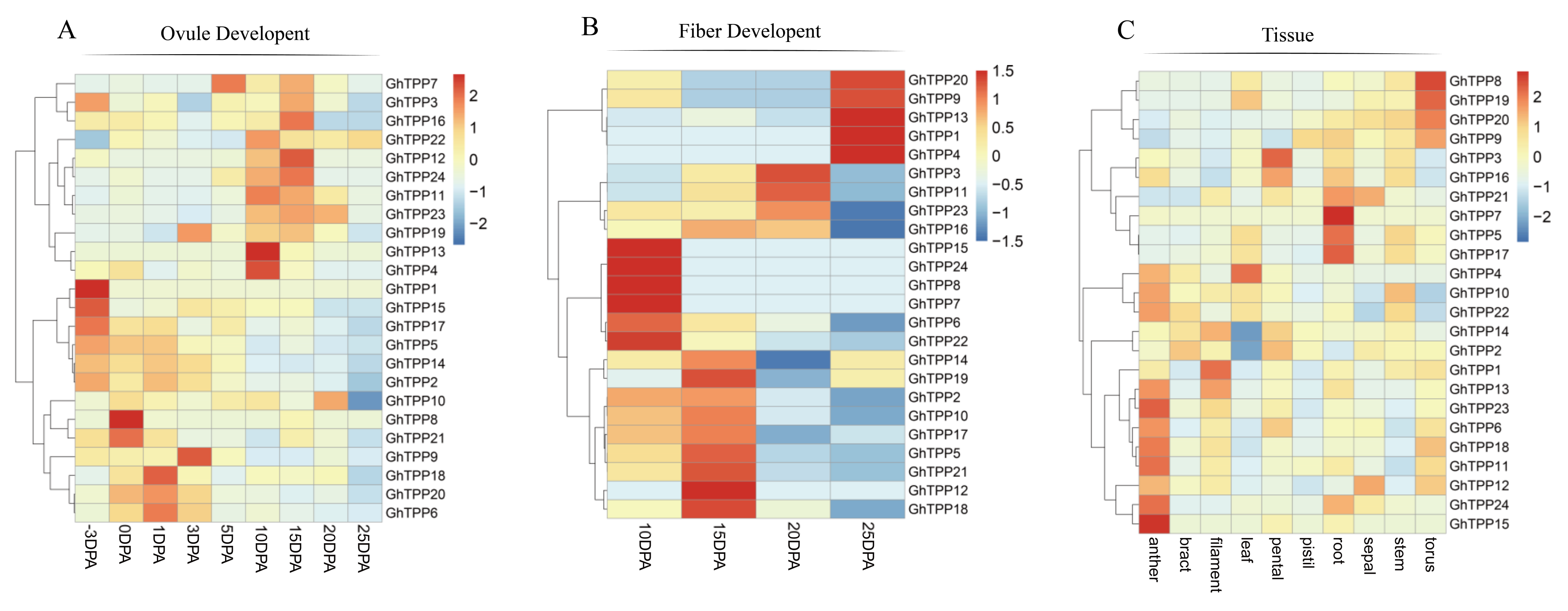
2.8. Expression Pattern Analysis in Different Tissues and at Different Growth Stages
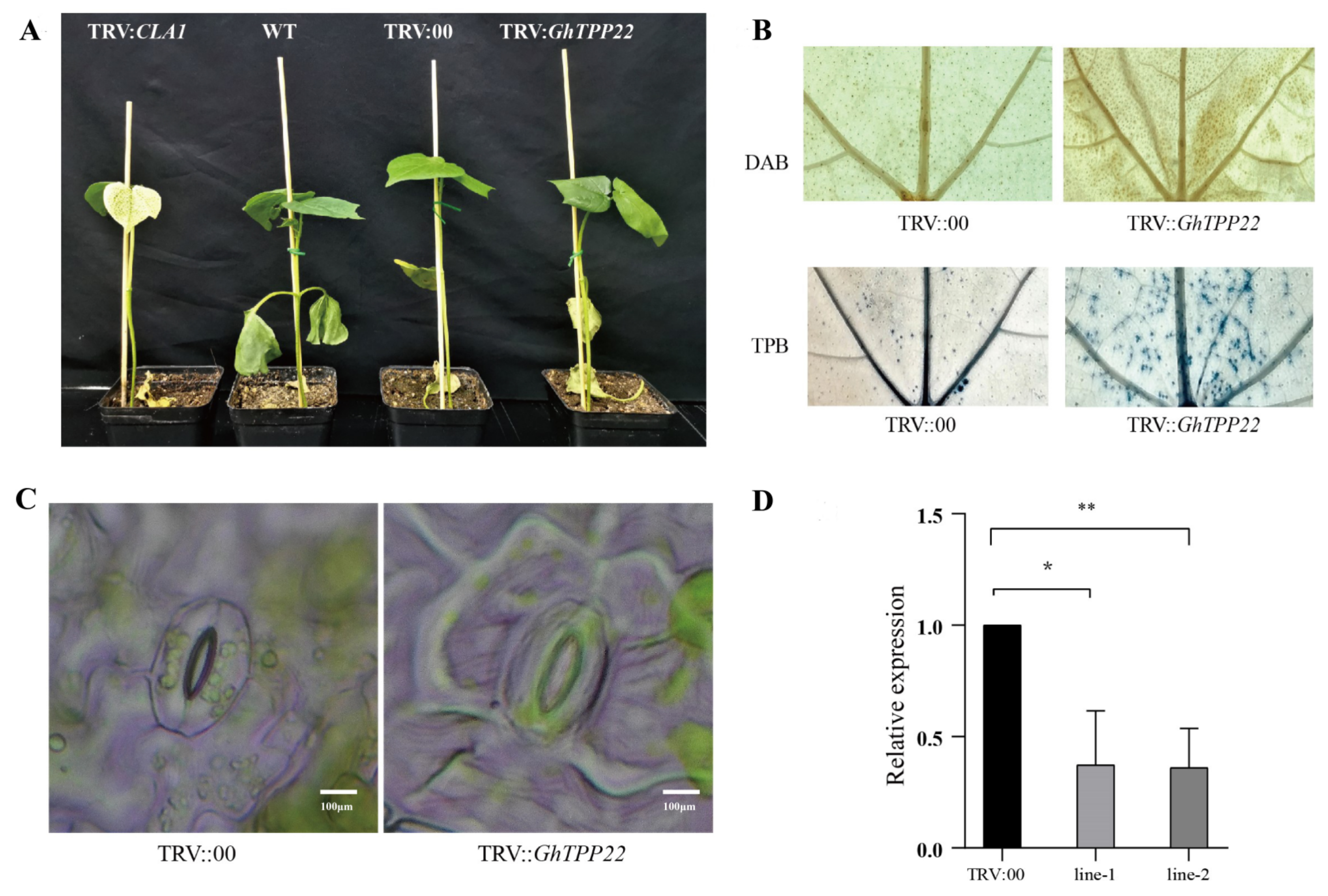
2.9. Analysis of the Function of GhTPP22 in Cotton in Response to Stress
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Identification and Physicochemical Properties of the TPP Gene Family in Cotton
5.2. Phylogenetic Analysis
5.3. Gene Structure and Conserved Motif Analysis
5.4. Analysis of the Promoters of the TPP Genes in G. Hirsutum
5.5. Homology Analysis of the TPP Gene Family Members
5.6. Expression Profile Analysis of the TPP Gene Family in Different Tissues of Cotton and under Stress Conditions
5.7. Prediction of microRNAs (miRNAs)
5.8. Functional Validation of the GhTPP22 Gene in G. Hirsutum
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Elbein, A.D.; Pan, Y.T.; Pastuszak, I.; Carroll, D. New insights on trehalose: A multifunctional molecule. Glycobiology 2003, 13, 17R–27R. [Google Scholar] [CrossRef] [PubMed]
- Paul, M.J.; Gonzalez-Uriarte, A.; Griffiths, C.A.; Hassani-Pak, K. The Role of Trehalose 6-Phosphate in Crop Yield and Resilience. Plant Physiol. 2018, 177, 12–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bohnert, H.J. What makes desiccation tolerable? Genome Biol. 2000, 1, 1–4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Avonce, N.; Mendoza-Vargas, A.; Morett, E.; Iturriaga, G. Insights on the evolution of trehalose biosynthesis. BMC Evol. Biol. 2006, 6, 109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iordachescu, M.; Imai, R. Trehalose Biosynthesis in Response to Abiotic Stresses. J. Integr. Plant Biol. 2008, 50, 1223–1229. [Google Scholar] [CrossRef]
- Van Houtte, H.; Vandesteene, L.; López-Galvis, L.; Lemmens, L.; Kissel, E.; Carpentier, S.; Feil, R.; Avonce, N.; Beeckman, T.; Lunn, J.E.; et al. Overexpression of the Trehalase Gene AtTRE1 Leads to Increased Drought Stress Tolerance in Arabidopsis and Is Involved in Abscisic Acid-Induced Stomatal Closure. Plant Physiol. 2013, 161, 1158–1171. [Google Scholar] [CrossRef] [Green Version]
- Glinski, M.; Weckwerth, W. Differential Multisite Phosphorylation of the Trehalose-6-phosphate Synthase Gene Family in Arabidopsis thaliana: A mass spectrometry-based process for multiparallel peptide library phosphorylation analysis. Mol. Cell. Proteom. 2005, 4, 1614–1625. [Google Scholar] [CrossRef] [Green Version]
- Prasad, Y.U.; Alexander, I.; Regina, F.; You, D.G.; Dirk, W.; Patrick, G.; Maria, P.; Petronia, C.; Hans-Michael, H.; Mark, S.; et al. The sucrose-trehalose 6-phosphate (Tre6P) nexus: Specificity and mechanisms of sucrose signalling by Tre6P. J. Exp. Bot. 2014, 65, 1051–1068. [Google Scholar]
- Wang, S.; Ouyang, K.; Wang, K. Genome-Wide Identification, Evolution, and Expression Analysis of TPS and TPP Gene Families in Brachypodium distachyon. Plants 2019, 8, 362. [Google Scholar] [CrossRef] [Green Version]
- Acosta-Pérez, P.; Camacho-Zamora, B.D.; Espinoza-Sánchez, E.A.; Gutiérrez-Soto, G.; Zavala-García, F.; Abraham-Juárez, M.J.; Sinagawa-García, S.R. Characterization of Trehalose-6-phosphate Synthase and Trehalose-6-phosphate Phosphatase Genes and Analysis of its Differential Expression in Maize (Zea mays) Seedlings under Drought Stress. Plants 2020, 9, 315. [Google Scholar] [CrossRef] [Green Version]
- Krasensky-Wrzaczek, J.; Broyart, C.; Rabanal, F.; Jonak, C. The Redox-Sensitive Chloroplast Trehalose-6-Phosphate Phosphatase AtTPPD Regulates Salt Stress Tolerance. Antioxid. Redox Signal. 2014, 21, 1289–1304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, Q.; Wang, S.; Dao, Y.; Wang, J.; Wang, K.; Wang, S. Arabidopsis thaliana trehalose-6-phosphate phosphatase gene TPPI enhances drought tolerance by regulating stomatal apertures. J. Exp. Bot. 2020, 71, 4285–4297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, G.; Li, X.; Ye, N.; Huang, M.; Feng, L.; Li, H.; Zhang, J. OsTPP1 regulates seed germination through the crosstalk with abscisic acid in rice. New Phytol. 2021, 230, 1925–1939. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.-L.; Ni, J.; Chen, M.-S.; Xu, Z.-F. Ectopic Expression of Jatropha curcas TREHALOSE-6-PHOSPHATE PHOSPHATASE J Causes Late-Flowering and Heterostylous Phenotypes in Arabidopsis but not in Jatropha. Int. J. Mol. Sci. 2019, 20, 2165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smeekens, S. From Leaf to Kernel: Trehalose-6-Phosphate Signaling Moves Carbon in the Field. Plant Physiol. 2015, 169, 912–913. [Google Scholar] [CrossRef] [Green Version]
- Ge, L.; Chao, D.-Y.; Shi, M.; Zhu, M.-Z.; Gao, J.-P.; Lin, H.-X. Overexpression of the trehalose-6-phosphate phosphatase gene OsTPP1 confers stress tolerance in rice and results in the activation of stress responsive genes. Planta 2008, 228, 191–201. [Google Scholar] [CrossRef]
- Satoh-Nagasawa, N.; Nagasawa, N.; Malcomber, S.; Sakai, H.; Jackson, D. A trehalose metabolic enzyme controls inflorescence architecture in maize. Nat. Int. Wkly. J. Sci. 2006, 441, 227–230. [Google Scholar] [CrossRef]
- Nuccio, M.L.; Wu, J.; Mowers, R.; Zhou, H.-P.; Meghji, M.; Primavesi, L.; Paul, M.; Chen, X.; Gao, Y.; Haque, E.; et al. Expression of trehalose-6-phosphate phosphatase in maize ears improves yield in well-watered and drought conditions. Nat. Biotechnol. 2015, 33, 862–869. [Google Scholar] [CrossRef]
- Li, P.; Ma, S.; Bohnert, H.J. Coexpression characteristics of trehalose-6-phosphate phosphatase subfamily genes reveal different functions in a network context. Physiol. Plant. 2008, 133, 544–556. [Google Scholar] [CrossRef]
- Hu, Y.; Chen, J.; Fang, L.; Zhang, Z.; Ma, W.; Niu, Y.; Ju, L.; Deng, J.; Zhao, T.; Lian, J.; et al. Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nat. Genet. 2019, 51, 739–748. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Tu, L.; Yuan, D.; Zhu, D.; Shen, C.; Li, J.; Liu, F.; Pei, L.; Wang, P.; Zhao, G.; et al. Reference genome sequences of two cultivated allotetraploid cottons, Gossypium hirsutum and Gossypium barbadense. Nat. Genet. 2018, 51, 224–229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, L.; Wang, Q.; Hu, Y.; Jia, Y.; Chen, J.; Liu, B.; Zhang, Z.; Guan, X.; Chen, S.; Zhou, B.; et al. Genomic analyses in cotton identify signatures of selection and loci associated with fiber quality and yield traits. Nat. Genet. 2017, 49, 1089–1098. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Zhang, Y.; Wu, L.; Zhang, G.; Sun, Z.; Li, Z.; Jiang, Y.; Ke, H.; Chen, B.; Liu, Z.; et al. High-quality genome assembly and resequencing of modern cotton cultivars provide resources for crop improvement. Nat. Genet. 2021, 53, 1385–1391. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Wang, Z.; Li, F.; Ye, W.; Wang, J.; Song, G.; Yue, Z.; Cong, L.; Shang, H.; Zhu, S.; et al. The draft genome of a diploid cotton Gossypium raimondii. Nat. Genet. 2012, 44, 1098–1103. [Google Scholar] [CrossRef]
- Du, X.; Huang, G.; He, S.; Yang, Z.; Sun, G.; Ma, X.; Li, N.; Zhang, X.; Sun, J.; Liu, M.; et al. Resequencing of 243 diploid cotton accessions based on an updated A genome identifies the genetic basis of key agronomic traits. Nat. Genet. 2018, 50, 796–802. [Google Scholar] [CrossRef]
- Babushok, D.V.; Ostertag, E.M.; Kazazian, H.H., Jr. Current topics in genome evolution: Molecular mechanisms of new gene formation. Cell Mol. Life Sci. 2006, 64, 542–554. [Google Scholar] [CrossRef]
- Flagel, L.E.; Wendel, J.F. Evolutionary rate variation, genomic dominance and duplicate gene expression evolution during allotetraploid cotton speciation. New Phytol. 2009, 186, 184–193. [Google Scholar] [CrossRef]
- Heidari, P.; Faraji, S.; Ahmadizadeh, M.; Ahmar, S.; Mora-Poblete, F. New Insights Into Structure and Function of TIFY Genes in Zea mays and Solanum lycopersicum: A Genome-Wide Comprehensive Analysis. Front. Genet. 2021, 12, 657970. [Google Scholar] [CrossRef]
- Abdullah; Faraji, S.; Mehmood, F.; Malik, H.M.T.; Ahmed, I.; Heidari, P.; Poczai, P. The GASA Gene Family in Cacao (Theobroma cacao, Malvaceae): Genome Wide Identification and Expression Analysis. Agronomy 2021, 11, 1425. [Google Scholar] [CrossRef]
- Musavizadeh, Z.; Najafi-Zarrini, H.; Kazemitabar, S.K.; Hashemi, S.H.; Faraji, S.; Barcaccia, G.; Heidari, P. Genome-Wide Analysis of Potassium Channel Genes in Rice: Expression of the OsAKT and OsKAT Genes under Salt Stress. Genes 2021, 12, 784. [Google Scholar] [CrossRef]
- Faraji, S.; Heidari, P.; Amouei, H.; Filiz, E.; Abdullah; Poczai, P. Investigation and Computational Analysis of the Sulfotransferase (SOT) Gene Family in Potato (Solanum tuberosum): Insights into Sulfur Adjustment for Proper Development and Stimuli Responses. Plants 2021, 10, 2597. [Google Scholar] [CrossRef] [PubMed]
- Vandesteene, L.; López-Galvis, L.; Vanneste, K.; Feil, R.; Maere, S.; Lammens, W.; Rolland, F.; Lunn, J.E.; Avonce, N.; Beeckman, T.; et al. Expansive Evolution of the TREHALOSE-6-PHOSPHATE PHOSPHATASE Gene Family in Arabidopsis. Plant Physiol. 2012, 160, 884–896. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Houtte, H.; López-Galvis, L.; Vandesteene, L.; Beeckman, T.; Van Dijck, P. Redundant and non-redundant roles of the trehalose-6-phosphate phosphatases in leaf growth, root hair specification and energy-responses in Arabidopsis. Plant Signal. Behav. 2013, 8, e23209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wingler, A.; Delatte, T.L.; O’Hara, L.E.; Primavesi, L.F.; Deveraj, J.; Paul, M.J.; Schluepmann, H. Trehalose 6-Phosphate Is Required for the Onset of Leaf Senescence Associated with High Carbon Availability. Plant Physiol. 2012, 158, 1241–1251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schluepmann, H.; van Dijken, A.; Aghdasi, M.; Wobbes, B.; Paul, M.; Smeekens, S. Trehalose Mediated Growth Inhibition of Arabidopsis Seedlings Is Due to Trehalose-6-Phosphate Accumulation. Plant Physiol. 2004, 135, 879–890. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strom, A.R.; Kaasen, I. Trehalose metabolism in Escherichia coli: Stress protection and stress regulation of gene expression. Mol. Microbiol. 1993, 8, 205–210. [Google Scholar] [CrossRef]
- Zhang, Y.; Primavesi, L.; Jhurreea, D.; Andralojc, P.J.; Mitchell, R.; Powers, S.J.; Schluepmann, H.; Delatte, T.; Wingler, A.; Paul, M.J. Inhibition of SNF1-Related Protein Kinase1 Activity and Regulation of Metabolic Pathways by Trehalose-6-Phosphate. Plant Physiol. 2009, 149, 1860–1871. [Google Scholar] [CrossRef] [Green Version]
- Nunes, C.; Primavesi, L.; Patel, M.K.; Martinez-Barajas, E.; Powers, S.J.; Sagar, R.; Fevereiro, P.; Davis, B.G.; Paul, M.J. Inhibition of SnRK1 by metabolites: Tissue-dependent effects and cooperative inhibition by glucose 1-phosphate in combination with trehalose 6-phosphate. Plant Physiol. Biochem. 2013, 63, 89–98. [Google Scholar] [CrossRef]
- Chen, S.L.; Peng, Y.; Zhou, H.; Yu, B.; Teng, S. Research advances in trehalose metabolism and trehalose-6-phosphate signaling in plants. Plant Physiol. J. 2014, 50, 233–242. [Google Scholar]
- Baena-González, E.; Rolland, F.; Thevelein, J.; Sheen, J. A central integrator of transcription networks in plant stress and energy signalling. Nature 2007, 448, 938–942. [Google Scholar] [CrossRef]
- Martínez-Barajas, E.; Delatte, T.; Schluepmann, H.; de Jong, G.J.; Somsen, G.W.; Nunes, C.; Primavesi, L.; Coello, P.; Mitchell, R.; Paul, M.J. Wheat Grain Development Is Characterized by Remarkable Trehalose 6-Phosphate Accumulation Pregrain Filling: Tissue Distribution and Relationship to SNF1-Related Protein Kinase1 Activity. Plant Physiol. 2011, 156, 373–381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, L.; Birch, R.G. Physiological basis for enhanced sucrose accumulation in an engineered sugarcane cell line. Funct. Plant Biol. 2010, 37, 1161–1174. [Google Scholar] [CrossRef]
- Debast, S.; Nunes-Nesi, A.; Hajirezaei, M.R.; Hofmann, J.; Sonnewald, U.; Fernie, A.R.; Börnke, F. Altering Trehalose-6-Phosphate Content in Transgenic Potato Tubers Affects Tuber Growth and Alters Responsiveness to Hormones during Sprouting. Plant Physiol. 2011, 156, 1754–1771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Deng, Y.; Song, X.; Miao, M. Trehalose-6-phosphate and SNF1-related protein kinase 1 are involved in the first-fruit inhibition of cucumber. J. Plant Physiol. 2014, 177, 110–120. [Google Scholar] [CrossRef] [PubMed]
- Chary, S.N.; Hicks, G.R.; Choi, Y.G.; Carter, D.; Raikhel, N.V. Trehalose-6-Phosphate Synthase/Phosphatase Regulates Cell Shape and Plant Architecture in Arabidopsis. Plant Physiol. 2007, 146, 97–107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Abubakar, Y.S.; Yang, C.; Wang, X.; Miao, P.; Lin, M.; Wen, Y.; Wu, Q.; Zhong, H.; Fan, Y.; et al. Trehalose Phosphate Synthase Complex-Mediated Regulation of Trehalose 6-Phosphate Homeostasis Is Critical for Development and Pathogenesis in Magnaporthe oryzae. mSystems 2021, 6, e00462-21. [Google Scholar] [CrossRef]
- Wang, W.; Chen, Q.-B.; Xu, S.; Liu, W.; Zhu, X.; Song, C. Trehalose-6-phosphate phosphatase E modulates ABA-controlled root growth and stomatal movement in Arabidopsis. J. Integr. Plant Biol. 2020, 62, 1518–1534. [Google Scholar] [CrossRef] [Green Version]
- Abdelraheem, A.; Esmaeili, N.; O’Connell, M.; Zhang, J. Progress and perspective on drought and salt stress tolerance in cotton. Ind. Crop. Prod. 2018, 130, 118–129. [Google Scholar] [CrossRef]
- Gao, J.; Wang, N.; Xu, S.S.; Li, Y.; Wang, Y.; Wang, G.X. Exogenous application of trehalose induced H2O2 production and stomatal closure in Vicia faba. Biol. Plant. 2013, 57, 380–384. [Google Scholar] [CrossRef]
- Delorge, I.; Figueroa, C.M.; Feil, R.; Lunn, J.E.; Van Dijck, P. Trehalose-6-phosphate synthase 1 is not the only active TPS in Arabidopsis thaliana. Biochem. J. 2015, 466, 283–290. [Google Scholar] [CrossRef]
- Jin, Q.; Hu, X.; Li, X.; Wang, B.; Wang, Y.; Jiang, H.; Mattson, N.; Xu, Y. Genome-Wide Identification and Evolution Analysis of Trehalose-6-Phosphate Synthase Gene Family in Nelumbo nucifera. Front. Plant Sci. 2016, 7, 1445. [Google Scholar] [CrossRef] [PubMed]
- Li, H.-W.; Zang, B.-S.; Deng, X.-W.; Wang, X.-P. Overexpression of the trehalose-6-phosphate synthase gene OsTPS1 enhances abiotic stress tolerance in rice. Planta 2011, 234, 1007–1018. [Google Scholar] [CrossRef] [PubMed]
- Karim, S.; Aronsson, H.; Ericson, H.; Pirhonen, M.; Leyman, B.; Welin, B.; Mäntylä, E.; Palva, E.T.; Van Dijck, P.; Holmström, K.-O. Improved drought tolerance without undesired side effects in transgenic plants producing trehalose. Plant Mol. Biol. 2007, 64, 371–386. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.; Liang, C.; Meng, Z.; Sun, G.; Meng, Z.; Guo, S.; Zhang, R. CottonFGD: An integrated functional genomics database for cotton. BMC Plant Biol. 2017, 17, 101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef] [Green Version]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; López, R.; Finn, R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef] [Green Version]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein Identification and Analysis Tools in the ExPASy Server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar]
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [Green Version]
- Yu, C.S.; Lin, C.J.; Hwang, J.K. Predicting subcellular localization of proteins for Gram-negative bacteria by support vector machines based on n-peptide compositions. Protein Sci. 2004, 13, 1402–1406. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, w202–w208. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.-Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2014, 31, 1296–1297. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [Green Version]
- Dai, X.; Zhuang, Z.; Zhao, P.X. psRNATarget: A plant small RNA target analysis server (2017 release). Nucleic Acids Res. 2018, 46, W49–W54. [Google Scholar] [CrossRef] [Green Version]
- Otasek, D.; Morris, J.H.; Bouças, J.; Pico, A.R.; Demchak, B. Cytoscape Automation: Empowering workflow-based network analysis. Genome Biol. 2019, 20, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
| Gene ID | Gene Name | Amino Acid Length | PI | MW (Da) | Subcellular Location | Location |
|---|---|---|---|---|---|---|
| Ga01G0805 | GaTPP1 | 197 | 8.79 | 22,151.11 | chloroplast | Ga01:12056674−12058006(−) |
| Ga01G2073 | GaTPP2 | 415 | 5.53 | 46,497.55 | chloroplast | Ga01:98691188−98693520(+) |
| Ga01G2696 | GaTPP3 | 387 | 8.79 | 43,611.07 | cytoplasm, mitochondrion | Ga01:111288363−111290680(+) |
| Ga02G0355 | GaTPP4 | 365 | 9.01 | 41,110.26 | endoplasmic reticulum | Ga02:4283490−4285630(+) |
| Ga02G1338 | GaTPP5 | 349 | 9.37 | 39,003.96 | mitochondrion | Ga02:91984769−91987027(+) |
| Ga02G1426 | GaTPP6 | 307 | 7.13 | 34,427.13 | chloroplast | Ga02:93890804−93892792(+) |
| Ga04G1370 | GaTPP7 | 174 | 9.15 | 19,797.69 | cell membrane | Ga04:82923593−82924930(−) |
| Ga05G2426 | GaTPP8 | 369 | 9.31 | 41,176.46 | chloroplast | Ga05:22943084−22945173(−) |
| Ga06G0361 | GaTPP9 | 364 | 9.03 | 40,884.08 | peroxisome | Ga06:4054349−4056277(−) |
| Ga08G2961 | GaTPP10 | 248 | 9.3 | 28,235.55 | chloroplast | Ga08:129160428−129168443(−) |
| Ga09G2059 | GaTPP11 | 327 | 6.13 | 36,864.31 | chloroplast | Ga09:78465375−78467180(−) |
| Ga10G0429 | GaTPP12 | 181 | 9.05 | 20,501.79 | chloroplast | Ga10:6841862−6843187(+) |
| Ga11G1620 | GaTPP13 | 370 | 9.03 | 41,573.91 | nucleus | Ga11:66146228−66148090(−) |
| Ga12G0284 | GaTPP14 | 340 | 9.01 | 38,454.32 | endoplasmic reticulum | Ga12:2329765−2331423(−) |
| Ga12G0607 | GaTPP15 | 388 | 8.47 | 43,856.03 | mitochondrion | Ga12:5363443−5365648(+) |
| Ga12G0815 | GaTPP16 | 383 | 7.69 | 42,710.69 | chloroplast | Ga12:7602925−7605180(−) |
| Ga12G1996 | GaTPP17 | 365 | 9.17 | 41,099.14 | chloroplast | Ga12:36111089−36113512(+) |
| Gorai.002G219000 | GrTPP1 | 354 | 9.57 | 39,276.39 | mitochondrion | Gr02:57218239−57220778(+) |
| Gorai.003G038300 | GrTPP2 | 373 | 9.11 | 41,994.42 | endoplasmic reticulum | Gr03:4115747−4118444(+) |
| Gorai.003G113900 | GrTPP3 | 419 | 5.62 | 46,886.94 | chloroplast | Gr03:34671718−34674357(+) |
| Gorai.003G169300 | GrTPP4 | 387 | 8.79 | 43,600.99 | cytoplasm, mitochondrion | Gr03:43992274−43996267(−) |
| Gorai.006G200800 | GrTPP5 | 327 | 6.35 | 37,063.49 | chloroplast | Gr06:45780176−45782343(−) |
| Gorai.007G240600 | GrTPP6 | 370 | 9.23 | 41,706.1 | nucleus | Gr07:33289543−33291870(−) |
| Gorai.008G104600 | GrTPP7 | 377 | 9.13 | 42,485.96 | chloroplast | Gr08:32381215−32384204(−) |
| Gorai.008G216700 | GrTPP8 | 422 | 8.4 | 47,239.17 | chloroplast | Gr08:50362362−50368208(+) |
| Gorai.008G236800 | GrTPP9 | 388 | 8.47 | 43,922.1 | mitochondrion | Gr08:52268635−52273612(−) |
| Gorai.008G270100 | GrTPP10 | 340 | 8.87 | 38,339.14 | endoplasmic reticulum | Gr08:54895162−54901806(+) |
| Gorai.009G238600 | GrTPP11 | 369 | 9.31 | 41,190.49 | chloroplast | Gr09:18961184−18963904(−) |
| Gorai.010G042600 | GrTPP12 | 364 | 9.24 | 40,853.11 | peroxisome | Gr10:4054502−4056819(−) |
| Gbar_A01G006620 | GbTPP1 | 313 | 8.54 | 35,142.68 | chloroplast | A01:10624088−10627713(−) |
| Gbar_A01G018380 | GbTPP2 | 365 | 9.55 | 41,026.49 | mitochondrion | A01:108393899−108396649(+) |
| Gbar_A01G019190 | GbTPP3 | 247 | 8.45 | 27,951 | vacuole | A01:110286108−110293672(+) |
| Gbar_A02G015580 | GbTPP4 | 365 | 9.05 | 41,062.2 | endoplasmic reticulum | A02:95555407−95558198(−) |
| Gbar_A03G001550 | GbTPP5 | 387 | 8.79 | 43,611.07 | cytoplasm, mitochondrion | A03:1858338−1862358(+) |
| Gbar_A03G006910 | GbTPP6 | 377 | 5.39 | 42,113.75 | peroxisome | A03:13211987−13217500(−) |
| Gbar_A05G022530 | GbTPP7 | 369 | 9.31 | 41,220.52 | chloroplast | A05:22046615−22049049(−) |
| Gbar_A06G003690 | GbTPP8 | 364 | 9.03 | 40,884.08 | peroxisome | A06:4817982−4820242(−) |
| Gbar_A06G003820 | GbTPP9 | 239 | 8.56 | 26,686.69 | mitochondrion | A06:5644700−5662178(+) |
| Gbar_A08G027440 | GbTPP10 | 259 | 6.05 | 28,424.14 | nucleus | A08:119673859−119677052(−) |
| Gbar_A09G019400 | GbTPP11 | 327 | 6.13 | 36,909.37 | chloroplast | A09:71628362−71630477(−) |
| Gbar_A11G020620 | GbTPP12 | 370 | 9.03 | 41,588.89 | nucleus | A11:42027092−42029383(−) |
| Gbar_A12G021250 | GbTPP13 | 383 | 7.69 | 42,710.69 | chloroplast | A12:94907154−94913205(+) |
| Gbar_A12G023290 | GbTPP14 | 388 | 8.47 | 43,870.05 | mitochondrion | A12:97098300−97103172(−) |
| Gbar_A12G026200 | GbTPP15 | 340 | 8.92 | 38,427.25 | endoplasmic reticulum | A12:99713201−99720168(+) |
| Gbar_D01G019570 | GbTPP16 | 354 | 9.57 | 39,306.42 | mitochondrion | D01:57326568−57329308(+) |
| Gbar_D03G003620 | GbTPP17 | 373 | 9.12 | 41,978.42 | endoplasmic reticulum | D03:4085281−4087934(+) |
| Gbar_D03G011280 | GbTPP18 | 384 | 5.77 | 42,710.49 | peroxisome | D03:39684077−39689078(+) |
| Gbar_D03G016980 | GbTPP19 | 387 | 8.96 | 43,682.05 | cytoplasm, mitochondrion | D03:49371797−49375803(−) |
| Gbar_D05G023190 | GbTPP20 | 369 | 9.31 | 41,156.47 | chloroplast | D05:20587648−20591044(−) |
| Gbar_D06G003870 | GbTPP21 | 364 | 9.23 | 40,886.18 | chloroplast | D06:4415421−4417669(−) |
| Gbar_D11G023430 | GbTPP22 | 370 | 9.1 | 41,707.04 | nucleus | D11:35062160−35064485(−) |
| Gbar_D12G010160 | GbTPP23 | 360 | 9.25 | 40,613.84 | mitochondrion | D12:33516324−33519721(−) |
| Gbar_D12G021190 | GbTPP24 | 383 | 8.52 | 42,648.71 | chloroplast | D12:51949877−51955716(+) |
| Gbar_D12G022910 | GbTPP25 | 388 | 8.47 | 43,922.1 | mitochondrion | D12:53800044−53805108(−) |
| Gbar_D12G026150 | GbTPP26 | 340 | 8.74 | 38,360.16 | endoplasmic reticulum | D12:56766186−56773133(+) |
| Gh_A01G209200 | GhTPP1 | 365 | 9.55 | 41,026.49 | mitochondrion | A01:108650087−108652331(+) |
| Gh_A02G174500 | GhTPP2 | 365 | 9.05 | 41,062.2 | endoplasmic reticulum | A02:101053973−101056656(−) |
| Gh_A03G016000 | GhTPP3 | 387 | 8.99 | 43,610.13 | cytoplasm, mitochondrion | A03:1987850−1990163(+) |
| Gh_A03G074500 | GhTPP4 | 415 | 5.65 | 46,558.73 | chloroplast | A03:14292959−14295295(−) |
| Gh_A05G212200 | GhTPP5 | 369 | 9.31 | 41,190.49 | chloroplast | A05:22966211−22968675(−) |
| Gh_A06G040600 | GhTPP6 | 364 | 9.03 | 40,884.08 | peroxisome | A06:4961458−4963382(−) |
| Gh_A06G042800 | GhTPP7 | 134 | 5.87 | 14,881.02 | cytoplasm | A06:5763598−5765589(+) |
| Gh_A09G201500 | GhTPP8 | 327 | 6.13 | 36,864.31 | chloroplast | A09:77041091−77042896(−) |
| Gh_A11G215100 | GhTPP9 | 339 | 9.05 | 38,310.18 | endoplasmic reticulum, mitochondrion, plastid | A11:44824072−44825936(−) |
| Gh_A12G223300 | GhTPP10 | 383 | 7.69 | 42,710.69 | chloroplast | A12:98041663−98047469(+) |
| Gh_A12G243500 | GhTPP11 | 388 | 8.47 | 43,870.05 | mitochondrion | A12:100217966−100222660(−) |
| Gh_A12G275900 | GhTPP12 | 340 | 8.92 | 38,427.25 | endoplasmic reticulum | A12:103292432−103294090(+) |
| Gh_D01G206700 | GhTPP13 | 354 | 9.57 | 39,306.42 | mitochondrion | D01:58776685−58778828(+) |
| Gh_D03G037900 | GhTPP14 | 365 | 9.12 | 41,103.3 | endoplasmic reticulum | D03:4191881−4194554(+) |
| Gh_D03G122800 | GhTPP15 | 422 | 5.79 | 47,156.41 | chloroplast | D03:41577803−41580170(+) |
| Gh_D03G179600 | GhTPP16 | 387 | 8.65 | 43,629.99 | cytoplasm, mitochondrion | D03:51536913−51539437(−) |
| Gh_D05G229100 | GhTPP17 | 369 | 9.19 | 41,157.41 | chloroplast | D05:20569568−20571657(−) |
| Gh_D06G040500 | GhTPP18 | 364 | 9.23 | 40,886.18 | chloroplast | D06:4645870−4648178(−) |
| Gh_D09G194700 | GhTPP19 | 327 | 6.14 | 37,129.59 | chloroplast | D09:46877954−46879754(−) |
| Gh_D11G237500 | GhTPP20 | 370 | 9.11 | 41,657.97 | nucleus | D11:35983647−35985917(−) |
| Gh_D12G107500 | GhTPP21 | 377 | 9.23 | 42,452.94 | chloroplast | D12:36644512−36646944(−) |
| Gh_D12G217800 | GhTPP22 | 383 | 8.52 | 42,648.71 | chloroplast | D12:55343343−55349138(+) |
| Gh_D12G238200 | GhTPP23 | 388 | 8.47 | 43,956.12 | mitochondrion | D12:57317788−57319992(−) |
| Gh_D12G270400 | GhTPP24 | 340 | 8.74 | 38,360.16 | endoplasmic reticulum | D12:60342947−60344621(+) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, W.; Cui, H.; Xiao, X.; Wu, B.; Sun, J.; Zhang, Y.; Yang, Q.; Zhao, Y.; Liu, G.; Qin, T. Genome-Wide Identification of Cotton (Gossypium spp.) Trehalose-6-Phosphate Phosphatase (TPP) Gene Family Members and the Role of GhTPP22 in the Response to Drought Stress. Plants 2022, 11, 1079. https://doi.org/10.3390/plants11081079
Wang W, Cui H, Xiao X, Wu B, Sun J, Zhang Y, Yang Q, Zhao Y, Liu G, Qin T. Genome-Wide Identification of Cotton (Gossypium spp.) Trehalose-6-Phosphate Phosphatase (TPP) Gene Family Members and the Role of GhTPP22 in the Response to Drought Stress. Plants. 2022; 11(8):1079. https://doi.org/10.3390/plants11081079
Chicago/Turabian StyleWang, Weipeng, Hua Cui, Xiangfen Xiao, Bingjie Wu, Jialiang Sun, Yaxin Zhang, Qiuyue Yang, Yuping Zhao, Guoxiang Liu, and Tengfei Qin. 2022. "Genome-Wide Identification of Cotton (Gossypium spp.) Trehalose-6-Phosphate Phosphatase (TPP) Gene Family Members and the Role of GhTPP22 in the Response to Drought Stress" Plants 11, no. 8: 1079. https://doi.org/10.3390/plants11081079
APA StyleWang, W., Cui, H., Xiao, X., Wu, B., Sun, J., Zhang, Y., Yang, Q., Zhao, Y., Liu, G., & Qin, T. (2022). Genome-Wide Identification of Cotton (Gossypium spp.) Trehalose-6-Phosphate Phosphatase (TPP) Gene Family Members and the Role of GhTPP22 in the Response to Drought Stress. Plants, 11(8), 1079. https://doi.org/10.3390/plants11081079






