Occurrence and Genetic Characterization of Grapevine Pinot Gris Virus in Russia
Abstract
1. Introduction
2. Results
2.1. Determination of qPCR Parameters
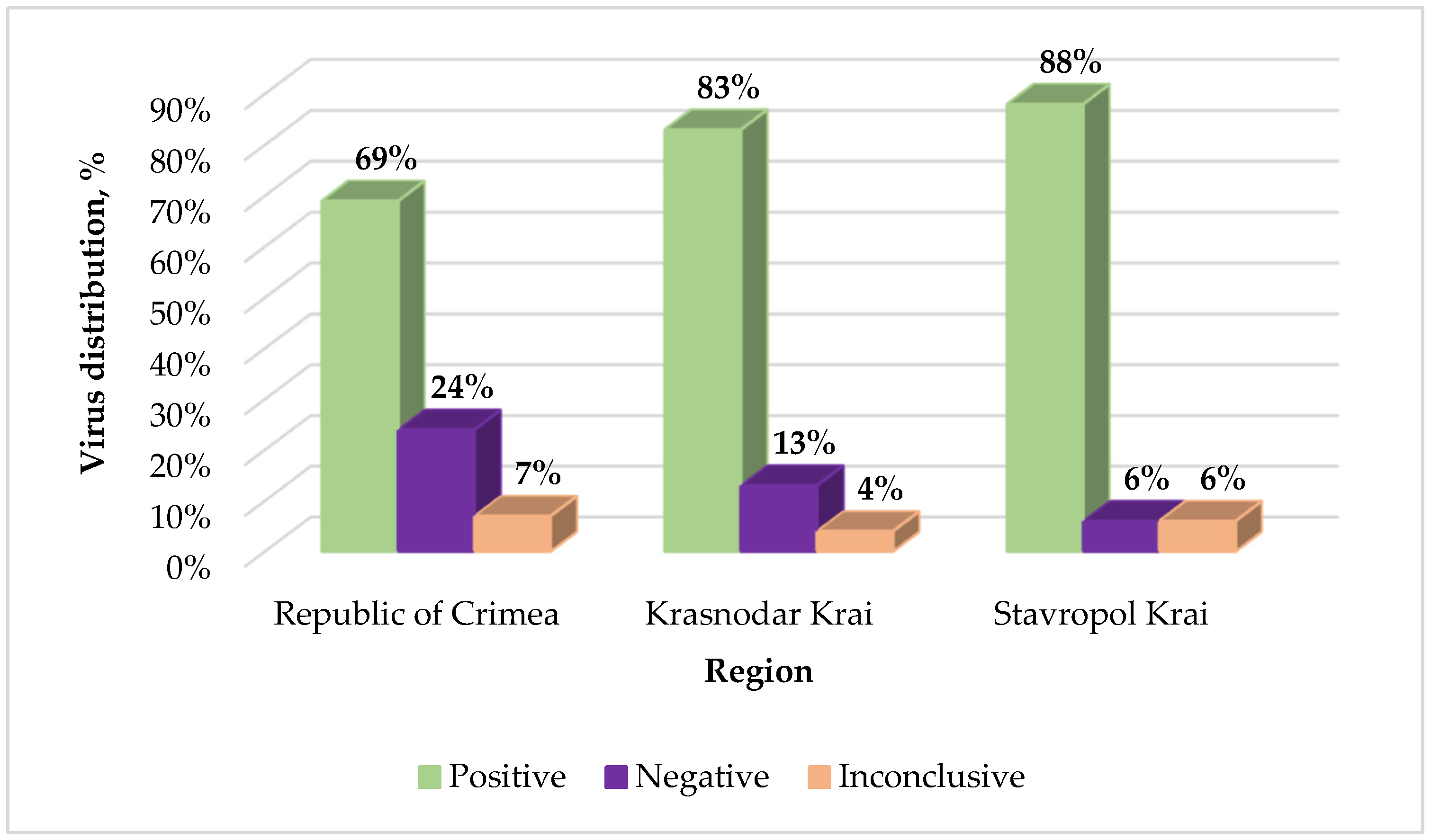
2.2. Monitoring of GPGV in Vineyards of Russia
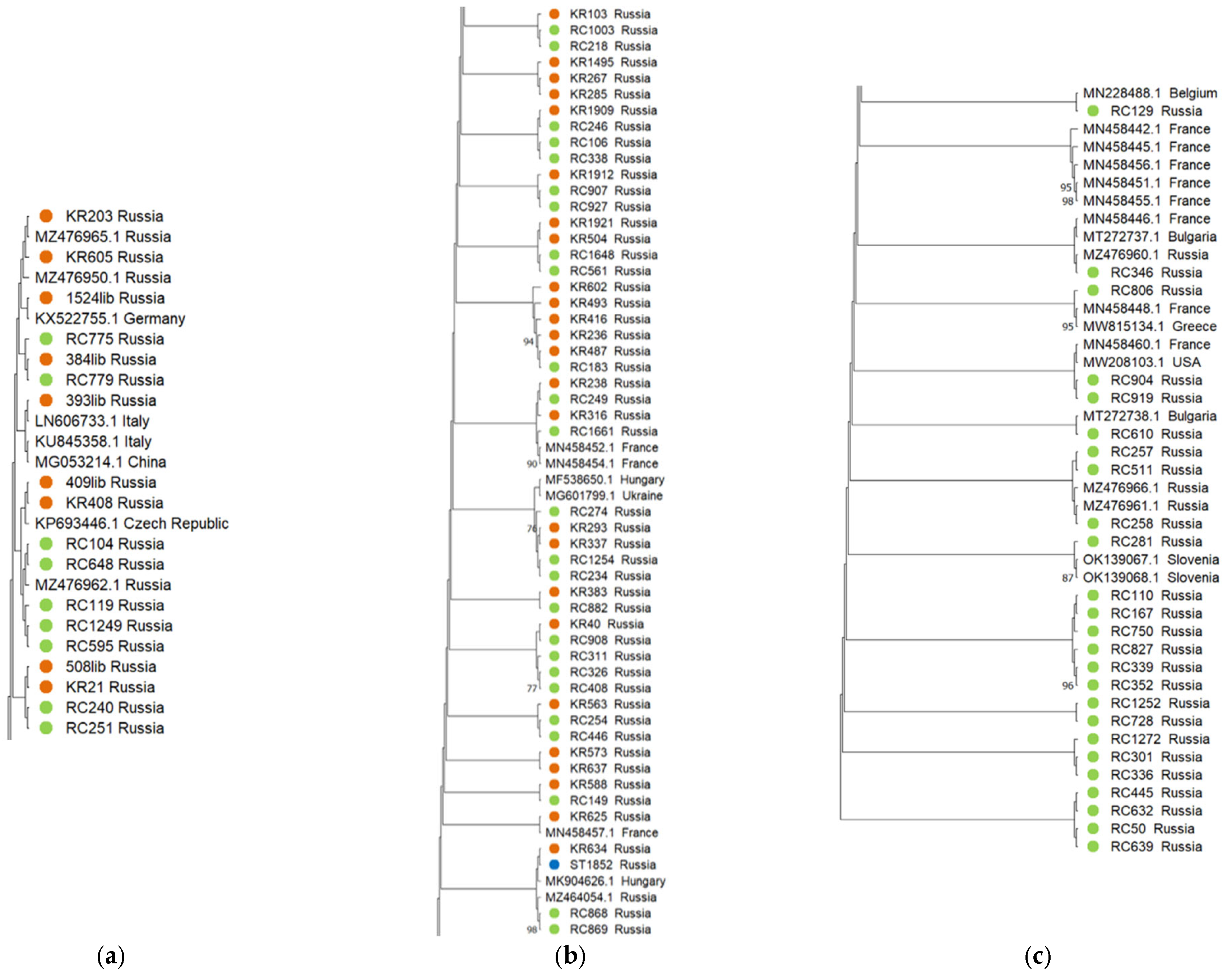
2.3. Phylogenetic Analysis
2.4. Variability of the GPGV Genome
2.5. Genetic Variability of GPGV and Manifestation of Symptoms
3. Discussion
4. Materials and Methods
4.1. Phytosanitary Monitoring of Vineyards
4.2. GPGV Detection
4.3. Variability of GPGV and Phylogenetic Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Giampetruzzi, A.; Roumi, V.; Roberto, R.; Malossini, U.; Yoshikawa, N.; La Notte, P.; Terlizzi, F.; Credi, R.; Saldarelli, P. A new grapevine virus discovered by deep sequencing of virus- and viroid-derived small RNAs in Cv Pinot gris. Virus Res. 2012, 163, 262–268. [Google Scholar] [CrossRef] [PubMed]
- Saldarelli, P.; Gualandri, V.; Malossini, U.; Glasa, M. Grapevine Pinot gris virus. In Grapevine Viruses Molecular Biology, Diagnostics and Management; Springer: Cham, Switzerland, 2017; pp. 351–363. [Google Scholar] [CrossRef]
- Cho, I.S.; Jung, S.M.; Cho, J.D.; Choi, G.S.; Lim, H.S. First report of Grapevine pinot gris virus infecting grapevine in Korea. New Dis. Reports. 2013, 27, 10. [Google Scholar] [CrossRef]
- Glasa, M.; Predajňa, L.; Komínek, P.; Nagyová, A.; Candresse, T.; Olmos, A. Molecular characterization of divergent grapevine Pinot gris virus isolates and their detection in Slovak and Czech grapevines. Arch. Virol. 2014, 159, 2103–2107. [Google Scholar] [CrossRef] [PubMed]
- Mavrič Pleško, I.; Viršček Marn, M.; Seljak, G.; Žežlina, I. First report of grapevine Pinot gris virus infecting grapevine in Slovenia. Plant Dis. 2014, 98, 1014. [Google Scholar] [CrossRef]
- Jo, Y.; Choi, H.; Kyong Cho, J.; Yoon, J.Y.; Choi, S.K.; Kyong Cho, W. In silico approach to reveal viral populations in grapevine cultivar Tannat using transcriptome data. Sci. Rep. 2015, 5, 15841. [Google Scholar] [CrossRef]
- Casati, P.; Maghradze, D.; Quaglino, F.; Ravasio, A.; Failla, O.; Bianco, P.A. First report of Grapevine Pinot gris virus in Georgia. J. Plant Pathol. 2016, 97, 67. [Google Scholar] [CrossRef]
- Beuve, M.; Candresse, T.; Tannières, M.; Lemaire, O. First report of grapevine pinot gris virus (GPGV) in grapevine in France. Plant Dis. 2015, 99, 293–294. [Google Scholar] [CrossRef]
- Reynard, J. Survey of emerging viruses in Switzerland. In Proceedings of the 18th Congress of the International Council for the Study of Virus and Virus-like Diseases of the Grapevine (ICVG), Ankara, Turkey, 7–11 September 2015. [Google Scholar]
- Poojari, S.; Lowery, T.; Rott, M.; Schmidt, A.M.; Úrbez-Torres, J.R. First report of grapevine pinot gris virus in British Columbia, Canada. Plant Dis. 2016, 100, 1513. [Google Scholar] [CrossRef]
- Reynard, J.S.; Schumacher, S.; Menzel, W.; Fuchs, J.; Bohnert, P.; Glasa, M.; Wetzel, T.; Fuchs, R. First report of Grapevine pinot gris virus in German vineyards. Plant Dis. 2016, 100, 2545. [Google Scholar] [CrossRef]
- Gazel, M.; Caglayan, K.; Elçi, E.; Öztürk, L. First report of Grapevine pinot gris virus in grapevine in Turkey. Plant Dis. 2016, 100, 657. [Google Scholar] [CrossRef]
- Al Rwahnih, M.; Golino, D.; Rowhani, A. First report of Grapevine pinot gris virus infecting grapevine in the United States. Plant Dis. 2016, 100, 1030. [Google Scholar] [CrossRef]
- Fan, X.D.; Dong, Y.F.; Zhang, Z.P.; Ren, F.; Hu, G.J.; Li, Z.N.; Zhou, J. First report of Grapevine pinot gris virus in Grapevines in China. Plant Dis. 2016, 100, 540. [Google Scholar] [CrossRef]
- Bertazzon, N.; Filippin, L.; Forte, V.; Angelini, E. Grapevine Pinot gris virus seems to have recently been introduced to vineyards in Veneto, Italy. Arch. Virol. 2016, 161, 711–714. [Google Scholar] [CrossRef]
- Fajardo, T.V.M.; Eiras, M.; Nickel, O. First report of Grapevine Pinot gris virus infecting grapevine in Brazil. Australas. Plant Dis. Notes 2017, 12, 45. [Google Scholar] [CrossRef]
- Wu, Q.; Habili, N. The recent importation of Grapevine Pinot gris virus into Australia. Virus Genes 2017, 53, 935–938. [Google Scholar] [CrossRef]
- Eichmeier, A.; Pieczonka, K.; Peňázová, E.; Pečenka, J.; Gajewski, Z. Occurrence of Grapevine Pinot gris virus in Poland and description of asymptomatic exhibitions in grapevines. J. Plant Dis. Prot. 2017, 124, 407–411. [Google Scholar] [CrossRef]
- Rasool, S.; Naz, S.; Rowhani, A.; Diaz-Lara, A.; Golino, D.A.; Farrar, K.D.; Al Rwahnih, M. Survey of grapevine pathogens in Pakistan. J. Plant Pathol. 2019, 101, 725–732. [Google Scholar] [CrossRef]
- Silva, G.; Lecourt, J.; Clover, G.R.G.; Seal, S.E. First record of Grapevine Pinot gris virus infecting Vitis vinifera in the United Kingdom. New Dis. Reports 2018, 38, 7. [Google Scholar] [CrossRef]
- Czotter, N.; Molnar, J.; Szabó, E.; Demian, E.; Kontra, L.; Baksa, I.; Szittya, G.; Kocsis, L.; Deak, T.; Bisztray, G.; et al. NGS of virus-derived small RNAs as a diagnostic method used to determine viromes of Hungarian Vineyards. Front. Microbiol. 2018, 9, 122. [Google Scholar] [CrossRef]
- Abou Kubaa, R.; Choueiri, E.; Jreijiri, F.; El Khoury, Y.; Saldarelli, P. First report of grapevine Pinot gris virus in Lebanon and the Middle East. J. Plant Pathol. 2020, 102, 565. [Google Scholar] [CrossRef]
- Abou Kubaa, R.; Lanotte, P.; Saldarelli, P. First report of grapevine Pinot gris virus in grapevine in Moldavia. J. Plant Pathol. 2019, 101, 441. [Google Scholar] [CrossRef]
- Zamorano, A.; Medina, G.; Fernández, C.; Cui, W.; Quiroga, N.; Fiore, N. First Report of Grapevine Pinot gris virus in Grapevine in Chile. Plant Dis. 2019, 103, 1438. [Google Scholar] [CrossRef]
- Debat, H.; Luna, F.; Moyano, S.; Zavallo, D.; Asurmendi, S.; Gomez-Talquenca, S. First report of grapevine Pinot gris virus infecting grapevine in Argentina. J. Plant Pathol. 2020, 102, 1321. [Google Scholar] [CrossRef]
- Eichmeier, A.; Penazova, E.; Nebish, A. First Report of Grapevine Pinot gris Virus on Grapevines in Armenia. Plant Dis. 2020, 104, 1000. [Google Scholar] [CrossRef]
- Tokhmechi, K.; Koolivand, D. First report of grapevine Pinot gris virus infecting grapevine in Iran. J. Plant Pathol. 2020, 102, 549. [Google Scholar] [CrossRef]
- Massart, S.; Vankerkoven, L.; Blouin, A.G.; Nourinejhad Zarghani, S.; Wetzel, T. First Report of Grapevine Pinot Gris Virus and Grapevine Rupestris Stem Pitting-Associated Virus in Grapevine in Belgium. Plant Dis. 2020, 104, 1879. [Google Scholar] [CrossRef]
- Bertazzon, N.; Rahali, M.; Angelini, E.; Crespan, M.; Migliaro, D. First report of grapevine pinot gris virus infecting grapevine in Algeria. Plant Dis. 2021, 105, 234. [Google Scholar] [CrossRef]
- Abe, J.; Nabeshima, T. First report of grapevine Pinot gris virus in wild grapevines (Vitis coignetiae) in Japan. J. Plant Pathol. 2021, 103, 725. [Google Scholar] [CrossRef]
- Bertazzon, N.; Angelini, E.; Signorotto, M.; Genov, N. First report of grapevine Pinot gris virus and grapevine leafroll-associated virus 2 in Bulgarian vineyards. J. Plant Dis. Prot. 2021, 128, 597–599. [Google Scholar] [CrossRef]
- Navrotskaya, E.; Porotikova, E.; Yurchenko, E.; Galbacs, Z.N.; Varallyay, E.; Vinogradova, S. High-Throughput Sequencing of Small RNAs for Diagnostics of Grapevine Viruses and Viroids in Russia. Viruses 2021, 13, 2432. [Google Scholar] [CrossRef]
- Constable, F.E.; Tassie, E.; McLoughlin, S. A Comprehensive Review of Grapevine Pinot Gris Virus (GPGV); Vinehealth Australia: Adelaide, SA, Australia, 2019; pp. 1–120. [Google Scholar]
- Malossini, U.; Bianchedi, P.; Beber, R.; Credi, R.; Saldarelli, P.; Gualandri, V. Spread of GPGV-associated disease in two vineyards in Trentino (Italy). In Proceedings of the 18th Congress of the International Council for the Study of Virus and Virus-like Diseases of the Grapevine (ICVG), Ankara, Turkey, 7–11 September 2015. [Google Scholar]
- Tarquini, G.; Ermacora, P.; Bianchi, G.L.; De Amicis, F.; Pagliari, L.; Martini, M.; Loschi, A.; Saldarelli, P.; Loi, N.; Musetti, R. Localization and subcellular association of Grapevine Pinot Gris Virus in grapevine leaf tissues. Protoplasma 2018, 255, 923–935. [Google Scholar] [CrossRef] [PubMed]
- Saldarelli, P.; Giampetruzzi, A.; Morelli, M.; Malossini, U.; Pirolo, C.; Bianchedi, P.; Gualandri, V. Genetic variability of Grapevine Pinot gris virus and its association with Grapevine leaf mottling and deformation. Phytopathology 2015, 105, 555–563. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, G.L.; De Amicis, F.; De Sabbata, L.; Di Bernardo, N.; Governatori, G.; Nonino, F.; Prete, G.; Marrazzo, T.; Versolatto, S.; Frausin, C. Occurrence of grapevine pinot gris virus in friuli venezia giulia (Italy): Field monitoring and virus quantification by real-time RT-PCR. EPPO Bull. 2015, 45, 22–32. [Google Scholar] [CrossRef]
- Tarquini, G.; De Amicis, F.; Martini, M.; Ermacora, P.; Loi, N.; Musetti, R.; Bianchi, G.L.; Firrao, G. Analysis of new grapevine Pinot gris virus (GPGV) isolates from Northeast Italy provides clues to track the evolution of a newly emerging clade. Arch. Virol. 2019, 164, 1655–1660. [Google Scholar] [CrossRef] [PubMed]
- Bertazzon, N.; Forte, V.; Filippin, L.; Causin, R.; Maixner, M.; Angelini, E. Association between genetic variability and titre of Grapevine Pinot gris virus with disease symptoms. Plant Pathol. 2017, 66, 949–959. [Google Scholar] [CrossRef]
- Spilmont, A.-S.; Sevin, A.-F.; Guinard, J.; Beuve, M.; Alliaume, A.; Marais, A.; Faure, C.; Candresse, T.; Lemaire, O. Occurrence of Grapevine Pinot gris virus (GPGV) and Grapevine Leaf Mottling and Deformation (GLMD) syndrome in France: Genetic diversity and field monitoring in diverse viticulture areas. In Proceedings of the 19th Congress of the International Council for the Study of Virus and Virus-Like Diseases of the Grapevine (ICVG), Santiago, Chile, 9–12 April 2018. [Google Scholar]
- Morán, F.; Olmos, A.; Lotos, L.; Predajňa, L.; Katis, N.; Glasa, M.; Maliogka, V.; Ruiz-García, A.B. A novel specific duplex real-time RT-PCR method for absolute quantitation of Grapevine Pinot gris virus in plant material and single mites. PLoS ONE 2018, 13, e0197237. [Google Scholar] [CrossRef] [PubMed]
- Eichmeier, A.; Peňázová, E.; Muljukina, N. Survey of Grapevine Pinot gris virus in certified grapevine stocks in Ukraine. Eur. J. Plant Pathol. 2018, 152, 555–560. [Google Scholar] [CrossRef]
- Tarquini, G.; Ermacora, P.; Firrao, G. Polymorphisms at the 3′end of the movement protein (MP) gene of grapevine Pinot gris virus (GPGV) affect virus titre and small interfering RNA accumulation in GLMD disease. Virus Res. 2021, 302, 198482. [Google Scholar] [CrossRef]
- Tarquini, G.; Zaina, G.; Ermacora, P.; De Amicis, F.; Franco-Orozco, B.; Loi, N.; Martini, M.; Bianchi, G.L.; Pagliari, L.; Firrao, G.; et al. Agroinoculation of Grapevine Pinot Gris Virus in tobacco and grapevine provides insights on viral pathogenesis. PLoS ONE 2019, 14, e0214010. [Google Scholar] [CrossRef]
- Bustin, S.; Huggett, J. qPCR primer design revisited. Biomol. Detect. Quantif. 2017, 14, 19–28. [Google Scholar] [CrossRef]
- Taylor, S.; Wakem, M.; Dijkman, G.; Alsarraj, M.; Nguyen, M. A practical approach to RT-qPCR—Publishing data that conform to the MIQE guidelines. Methods 2010, 50, S1–S5. [Google Scholar] [CrossRef] [PubMed]
- Bivins, A.; Kaya, D.; Bibby, K.; Simpson, S.L.; Bustin, S.A.; Shanks, O.C.; Ahmed, W. Variability in RT-qPCR assay parameters indicates unreliable SARS-CoV-2 RNA quantification for wastewater surveillance. Water Res. 2021, 203, 117516. [Google Scholar] [CrossRef] [PubMed]
- Bertazzon, N.; Forte, V.; Angelini, E. Fast transmission of grapevine “Pinot gris” virus (GPGV) in vineyard. Vitis 2020, 59, 29–34. [Google Scholar] [CrossRef]
- Hily, J.-M.; Poulicard, N.; Candresse, T.; Vigne, E.; Beuve, M.; Renault, L.; Velt, A.; Spilmont, A.-S.; Lemaire, O. Datamining, Genetic Diversity Analyses, and Phylogeographic Reconstructions Redefine the Worldwide Evolutionary History of Grapevine Pinot gris virus and Grapevine berry inner necrosis virus. Phytobiomes J. 2020, 4, 165–177. [Google Scholar] [CrossRef]
- Angelini, E.; Bazzo, I.; Bertazzon, N.; Filippin, L.; Forte, V. A new disease in the vineyard: Grapevine leaf mottling and deformation. Aust. N. Z. Grapegrow. Winemak. 2015, 622, 56–59. [Google Scholar]
- Yurchenko, E.G.; Savchuk, N.V.; Porotikova, E.V.; Vinogradova, S.V. First Report of Grapevine (Vitis sp.) Cluster Blight Caused by Fusarium proliferatum in Russia. Plant Dis. 2019, 104, 991. [Google Scholar] [CrossRef]
- Porotikova, E.; Yurchenko, E.; Vinogradova, S. Molecular identification of phytoplasmas in Russian vineyards. Phytopathogenic Mollicutes 2019, 9, 25–26. [Google Scholar] [CrossRef]
- Porotikova, E.V.; Yurchenko, E.G.; Vinogradova, S.V. First Report of ‘Candidatus Phytoplasma solani’ Associated with Bois Noir on Grapevine (Vitis vinifera) in Krasnodar Region of Russia. Plant Dis. 2019, 104, 277. [Google Scholar] [CrossRef]
- Porotikova, E.; Terehova, U.; Volodin, V.; Yurchenko, E.; Vinogradova, S. Distribution and Genetic Diversity of Grapevine Viruses in Russia. Plants 2021, 10, 1080. [Google Scholar] [CrossRef] [PubMed]
- Morelli, M.; de Moraes Catarino, A.; Susca, L.; Saldarelli, P.; Gualandri, V.; Martelli, G.P. First Report of Grapevine Pinot Gris Virus From Table Grapes in Southern Italy. J. Plant Pathol. 2014, 96, 439. [Google Scholar]
- Vončina, D.; Al Rwahnih, M.; Rowhani, A.; Gouran, M.; Almeida, R.P.P. Viral diversity in autochthonous croatian grapevine cultivars. Plant Dis. 2017, 101, 1230–1235. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Zou, Q.; Zhao, L. VPTMdb: A viral posttranslational modification database. Brief. Bioinform. 2021, 22, bbaa251. [Google Scholar] [CrossRef] [PubMed]
- Nemes, K.; Gellért, Á.; Almási, A.; Vági, P.; Sáray, R.; Kádár, K.; Salánki, K. Phosphorylation regulates the subcellular localization of Cucumber Mosaic Virus 2b protein. Sci. Rep. 2017, 7, 13444. [Google Scholar] [CrossRef]
- Lee, J.Y.; Lucas, W.J. Phosphorylation of viral movement proteins—Regulation of cell-to-cell trafficking. Trends Microbiol. 2001, 9, 5–8. [Google Scholar] [CrossRef]
- Shapka, N.; Stork, J.; Nagy, P.D. Phosphorylation of the p33 replication protein of Cucumber necrosis tombusvirus adjacent to the RNA binding site affects viral RNA replication. Virology 2005, 343, 65–78. [Google Scholar] [CrossRef] [PubMed]
- Trutnyeva, K.; Bachmaier, R.; Waigmann, E. Mimicking carboxyterminal phosphorylation differentially effects subcellular distribution and cell-to-cell movement of Tobacco mosaic virus movement protein. Virology 2005, 332, 563–577. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jakubiec, A.; Tournier, V.; Drugeon, G.; Pflieger, S.; Camborde, L.; Vinh, J.; Héricourt, F.; Redeker, V.; Jupin, I. Phosphorylation of Viral RNA-dependent RNA Polymerase and Its Role in Replication of a Plus-strand RNA Virus. J. Biol. Chem. 2006, 281, 21236–21249. [Google Scholar] [CrossRef] [PubMed]
- Kleinow, T.; Nischang, M.; Beck, A.; Kratzer, U.; Tanwir, F.; Preiss, W.; Kepp, G.; Jeske, H. Three C-terminal phosphorylation sites in the Abutilon mosaic virus movement protein affect symptom development and viral DNA accumulation. Virology 2009, 390, 89–101. [Google Scholar] [CrossRef]
- Ivanov, K.I.; Puustinen, P.; Gabrenaite, R.; Vihinen, H.; Rönnstrand, L.; Valmu, L.; Kalkkinen, N.; Mäkinen, K. Phosphorylation of the Potyvirus Capsid Protein by Protein Kinase CK2 and Its Relevance for Virus Infection. Plant Cell 2003, 15, 2124–2139. [Google Scholar] [CrossRef]
- Hung, C.J.; Huang, Y.W.; Liou, M.R.; Lee, Y.C.; Lin, N.S.; Meng, M.; Tsai, C.H.; Hu, C.C.; Hsu, Y.H. Phosphorylation of Coat Protein by Protein Kinase CK2 Regulates Cell-to-Cell Movement of Bamboo mosaic virus Through Modulating RNA Binding. Mol. Plant Microbe Interact. 2014, 27, 1211–1225. [Google Scholar] [CrossRef]
- Kumar, R.; Mehta, D.; Mishra, N.; Nayak, D.; Sunil, S. Role of Host-Mediated Post-Translational Modifications (PTMs) in RNA Virus Pathogenesis. Int. J. Mol. Sci. 2021, 22, 323. [Google Scholar] [CrossRef] [PubMed]
- Resh, M.D. Fatty acylation of proteins: New insights into membrane targeting of myristoylated and palmitoylated proteins. Biochim. Biophys. Acta Mol. Cell Res. 1999, 1451, 1–16. [Google Scholar] [CrossRef]
- Strecker, T.; Maisa, A.; Daffis, S.; Eichler, R.; Lenz, O.; Garten, W. The role of myristoylation in the membrane association of the Lassa virus matrix protein Z. Virol. J. 2006, 3, 93. [Google Scholar] [CrossRef] [PubMed]
- Rott, M.E.; Jelkmann, W. Characterization and Detection of Several Filamentous Viruses of Cherry: Adaptation of an Alternative Cloning Method (DOP-PCR), and Modification of an RNA Extraction Protocol. Eur. J. Plant Pathol. 2001, 107, 411–420. [Google Scholar] [CrossRef]
- Gambino, G.; Bondaz, J.; Gribaudo, I. Detection and Elimination of Viruses in Callus, Somatic Embryos and Regenerated Plantlets of Grapevine. Eur. J. Plant Pathol. 2006, 114, 397–404. [Google Scholar] [CrossRef]
- Angelini, E.; Luca Bianchi, G.; Filippin, L.; Morassutti, C.; Borgo, M. A new TaqMan method for the identification of phytoplasmas associated with grapevine yellows by real-time PCR assay. J. Microbiol. Methods 2007, 68, 613–622. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- LightCycler® 96 System, Operator’s Guide V2.0; Roche Diagnostics GmbH: Mannheim, Germany, 2012.
- Sayers, E.W.; Cavanaugh, M.; Clark, K.; Pruitt, K.D.; Schoch, C.L.; Sherry, S.T.; Karsch-Mizrachi, I. GenBank. Nucleic Acids Res. 2021, 49, D92–D96. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- FinchTV. Available online: https://digitalworldbiology.com/FinchTV (accessed on 28 January 2022).
- Martin, D.P.; Murrell, B.; Golden, M.; Khoosal, A.; Muhire, B. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015, 1, vev003. [Google Scholar] [CrossRef]
- Muhire, B.M.; Varsani, A.; Martin, D.P. SDT: A Virus Classification Tool Based on Pairwise Sequence Alignment and Identity Calculation. PLoS ONE 2014, 9, e108277. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef] [PubMed]
- Hall, T. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Open Reading Frame Finder. Available online: https://www.ncbi.nlm.nih.gov/orffinder/ (accessed on 2 February 2022).
- Expasy. ScanProsite—SIB Swiss Institute of Bioinformatics. Available online: https://www.expasy.org/resources/scanprosite (accessed on 15 February 2022).

| Assay | Gene | PCR Efficiency, % | Slope | R2 | Y-Intercept |
|---|---|---|---|---|---|
| Simplex | CP | 96 | −3.4114 | 0.97 | 24.75 |
| Cpn21 | 102 | −3.2668 | 0.98 | 21.33 | |
| GAPDH | 74 | −4.1485 | 0.98 | 19.20 | |
| Duplex | CP | 97 | −3.4034 | 0.99 | 24.64 |
| Cpn21 | 94 | −3.4856 | 0.97 | 22.85 | |
| Duplex | CP | 93 | −3.4939 | 0.97 | 22.84 |
| GAPDH | 78 | −4.0060 | 0.98 | 18.63 |
| Gene | Position in Genome | Position in Protein | Codon | Amino acid | Occurrence of aa | Variant Frequency, % | Putative Post-Translational Modification |
|---|---|---|---|---|---|---|---|
| MP | 5669 | 31 | AAT | N | 118 | 99.2 | N-glycosylation |
| AGT | S | 1 | 0.8 | - * | |||
| 5681 | 35 | TCG/TCA | S | 113 | 95.0 | Protein kinase C phosphorylation | |
| TTG | L | 6 | 5.0 | - | |||
| 6335 | 253 | AGC/AGT | S | 80 | 67.2 | Protein kinase C phosphorylation | |
| AAC/AAT | N | 37 | 31.1 | - | |||
| ARC | X | 2 | 1.7 | - | |||
| 6392 | 272 | GGA/GGR | G | 116 | 97.5 | N-myristoylation | |
| GAA | E | 3 | 2.5 | - | |||
| 6404 | 276 | TTC/TTY | F | 118 | 99.2 | - | |
| TCC | S | 1 | 0.8 | Casein kinase II phosphorylation site | |||
| 6407 | 277 | CTT | L | 24 | 20.2 | - | |
| TTT | F | 88 | 73.9 | - | |||
| TCT | S | 6 | 5.0 | Casein kinase II phosphorylation site | |||
| ATT | I | 1 | 0.8 | - | |||
| 6413 | 279 | GAA/GAG/GAR | E | 118 | 99.2 | - | |
| GGA | G | 1 | 0.8 | N-myristoylation | |||
| 6419 | 281 | ACA | T | 115 | 96.6 | Casein kinase II phosphorylation | |
| GCA | A | 3 | 2.5 | - | |||
| ATA | I | 1 | 0.8 | - | |||
| 6446 | 290 | CGC/CGA/AGA/CGT | R | 115 | 96.6 | - | |
| TGC | C | 1 | 0.8 | - | |||
| MGC | X | 2 | 1.7 | - | |||
| AGC | S | 1 | 0.8 | Protein kinase C phosphorylation | |||
| 6449 | 291 | ACT/ACC | T | 112 | 94.1 | Casein kinase II phosphorylation | |
| ATT | I | 2 | 1.7 | - | |||
| AAT | N | 1 | 0.8 | - | |||
| GCC/GCT | A | 3 | 2.5 | - | |||
| RCT | X | 1 | 0.8 | - | |||
| 6452 | 292 | GAA/GAG | E | 113 | 95.0 | - | |
| GGA | G | 1 | 0.8 | N-myristoylation | |||
| RAA | X | 2 | 1.7 | - | |||
| GCA | A | 1 | 0.8 | - | |||
| AAA | K | 2 | 1.7 | - | |||
| 6455 | 293 | AAT | N | 112 | 94.1 | N-glycosylation | |
| AGT/AGC | S | 3 | 2.5 | - | |||
| AAA | K | 1 | 0.8 | - | |||
| GAT | D | 3 | 2.5 | - | |||
| 6461 | 295 | TCA/TCG | S | 116 | 97.5 | Protein kinase C phosphorylation | |
| YCA | X | 1 | 0.8 | - | |||
| CCA | P | 2 | 1.7 | - | |||
| 6476 | 300 | TTC/TTT | F | 106 | 89.1 | - | |
| TCC | S | 8 | 6.7 | Protein kinase C phosphorylation | |||
| TTA/CTC | L | 4 | 3.4 | - | |||
| TTM | X | 1 | 0.8 | - | |||
| 6485 | 303 | GGT | G | 116 | 97.5 | - | |
| RGT | X | 1 | 0.8 | - | |||
| GAT | D | 1 | 0.8 | - | |||
| AGT | S | 1 | 0.8 | Casein kinase II phosphorylation | |||
| 6596 | 340 | GAT | D | 116 | 97.5 | - | |
| AAT | N | 3 | 2.5 | N-glycosylation | |||
| 6602 | 342 | TCA | S | 117 | 98.3 | Casein kinase II phosphorylation | |
| CCA | P | 2 | 1.7 | - | |||
| 6605 | 343 | GGA | G | 118 | 99.2 | N-myristoylation | |
| AGA | R | 1 | 0.8 | - | |||
| 6644 | 356 | GTT | V | 3 | 2.5 | - | |
| GCT | A | 111 | 93.3 | - | |||
| ACT | T | 3 | 2.5 | Protein kinase C phosphorylation | |||
| GVT/RCT | X | 2 | 1.7 | - | |||
| 6665 | 363 | ACT | T | 113 | 95.0 | Protein kinase C phosphorylation | |
| GCT | A | 5 | 4.2 | - | |||
| RCT | X | 1 | 0.8 | - | |||
| 6668 | 364 | TCA | S | 118 | 99.2 | Casein kinase II phosphorylation | |
| CCA | P | 1 | 0.8 | - | |||
| 6674 | 366 | GCT | A | 3 | 2.5 | - | |
| GTT | V | 111 | 93.3 | - | |||
| ATT | I | 2 | 1.7 | - | |||
| ACT | T | 2 | 1.7 | Protein kinase C phosphorylation | |||
| GYT | X | 1 | 0.8 | - | |||
| 6686 | 370 | TAA | Stop codon | 2 | 1.7 | - | |
| CAA | Q | 116 | 97.5 | - | |||
| YAA | X | 1 | 0.8 | - | |||
| 6689 | 371 | TAA | Stop codon | 1 | 0.9 | - | |
| CAA | Q | 115 | 98.3 | - | |||
| YAA | X | 1 | 0.9 | - | |||
| 6701 | 376 | TGA | Stop codon | 116 | 100.0 | - | |
| CP | 6870 | 94 | ACT/ACC/ACY | T | 118 | 99.2 | Casein kinase II phosphorylation |
| ATT | I | 1 | 0.8 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shvets, D.; Vinogradova, S. Occurrence and Genetic Characterization of Grapevine Pinot Gris Virus in Russia. Plants 2022, 11, 1061. https://doi.org/10.3390/plants11081061
Shvets D, Vinogradova S. Occurrence and Genetic Characterization of Grapevine Pinot Gris Virus in Russia. Plants. 2022; 11(8):1061. https://doi.org/10.3390/plants11081061
Chicago/Turabian StyleShvets, Darya, and Svetlana Vinogradova. 2022. "Occurrence and Genetic Characterization of Grapevine Pinot Gris Virus in Russia" Plants 11, no. 8: 1061. https://doi.org/10.3390/plants11081061
APA StyleShvets, D., & Vinogradova, S. (2022). Occurrence and Genetic Characterization of Grapevine Pinot Gris Virus in Russia. Plants, 11(8), 1061. https://doi.org/10.3390/plants11081061






