Comparative Analysis of Heat-Tolerant and Heat-Susceptible Rice Highlights the Role of OsNCED1 Gene in Heat Stress Tolerance
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Heat-Stress Treatment
2.2. RNA Isolation, Microarray Hybridization, Signal Scanning, and Normalization
2.3. Data Filtering, Clustering and Functional Classification
2.4. Quantitative Real-Time PCR
2.5. Cloning of OsNCED1 CDS
2.6. Generation of OsNCED1 Overexpression Lines
2.7. Determination of Expression Pattern and Transcription Levels of OsNCED1 and ABA Signaling Related Genes
2.8. Determination of Pollen Viability, Seed Setting Rate, Cell Membrane Damage, and Oxidative Stress in Transgenic Rice Plants under HS
2.9. Statistical Analysis
3. Results
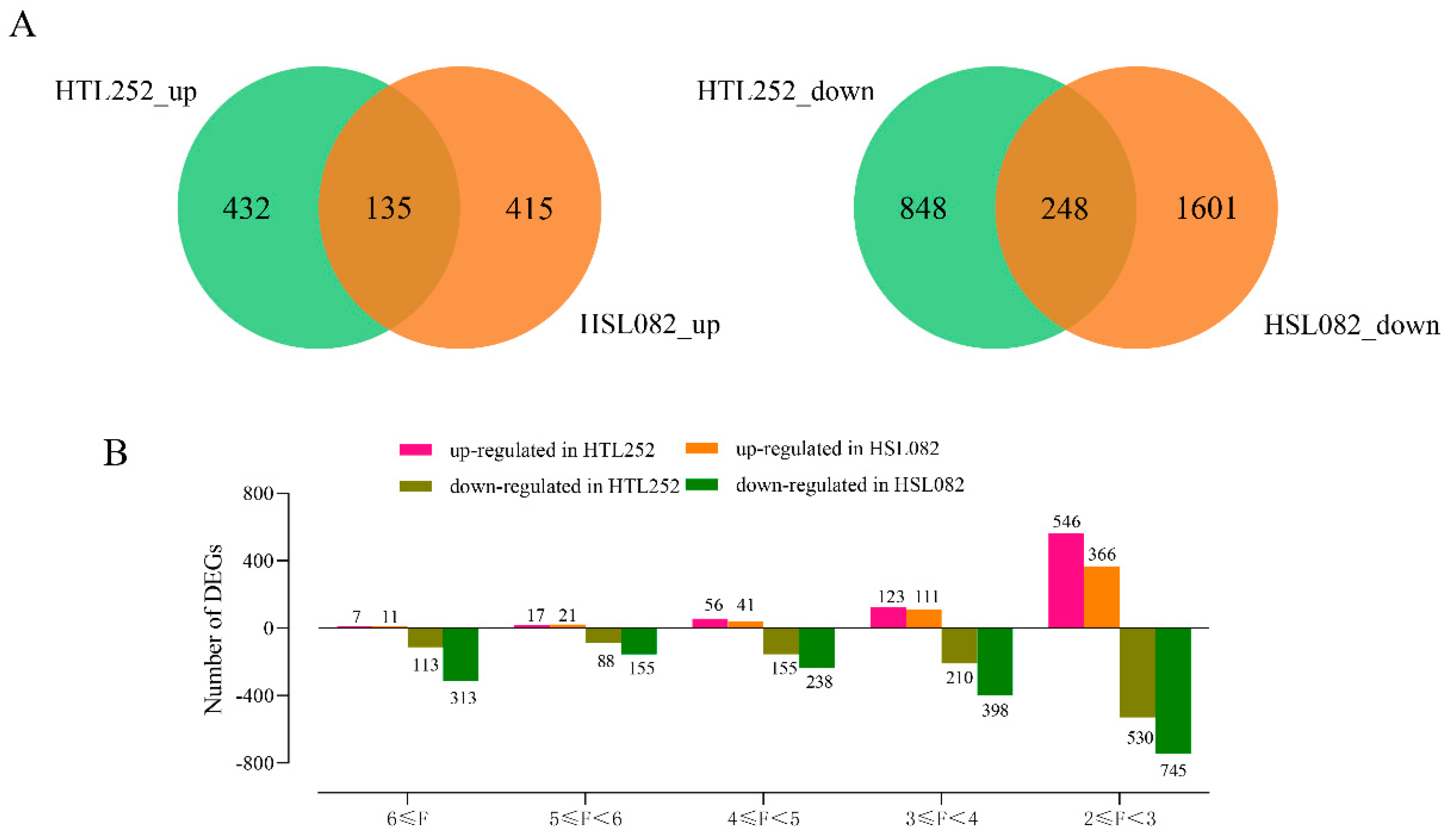
3.1. Differentially Expressed Gene Analysis under HS
3.2. Comparative Analysis between HTL252 and HSL082
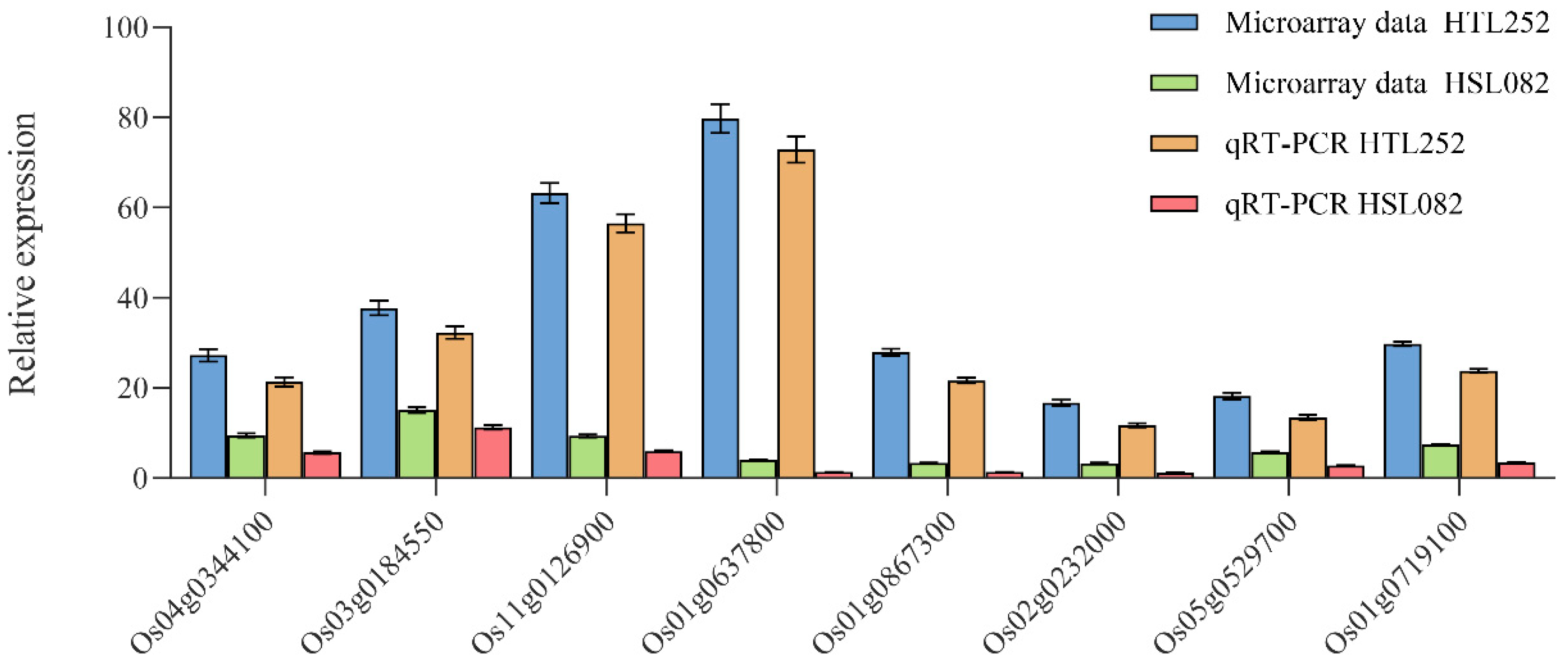
3.3. Quantitative Real-Time PCR (qRT-PCR) Verification
3.4. Expression Pattern of OsNCED1 Gene under HS
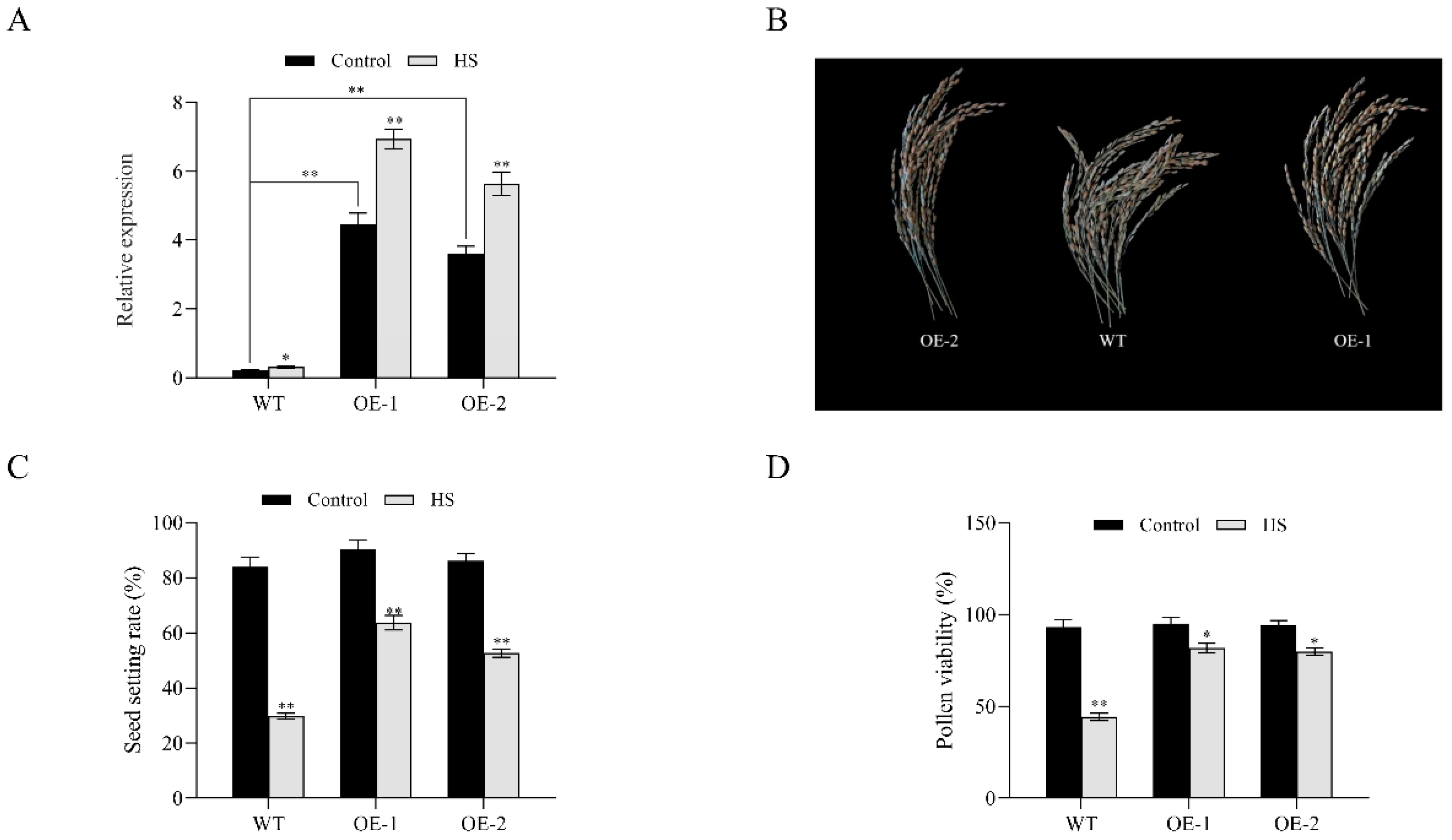
3.5. Analysis of Pollen Viability and Seed Setting Rate in OsNCED1 Transgenic Rice Plants
3.6. Analysis of Membrane Permeability and Antioxidant Enzyme Activity of OsNCED1 Transgenic Rice
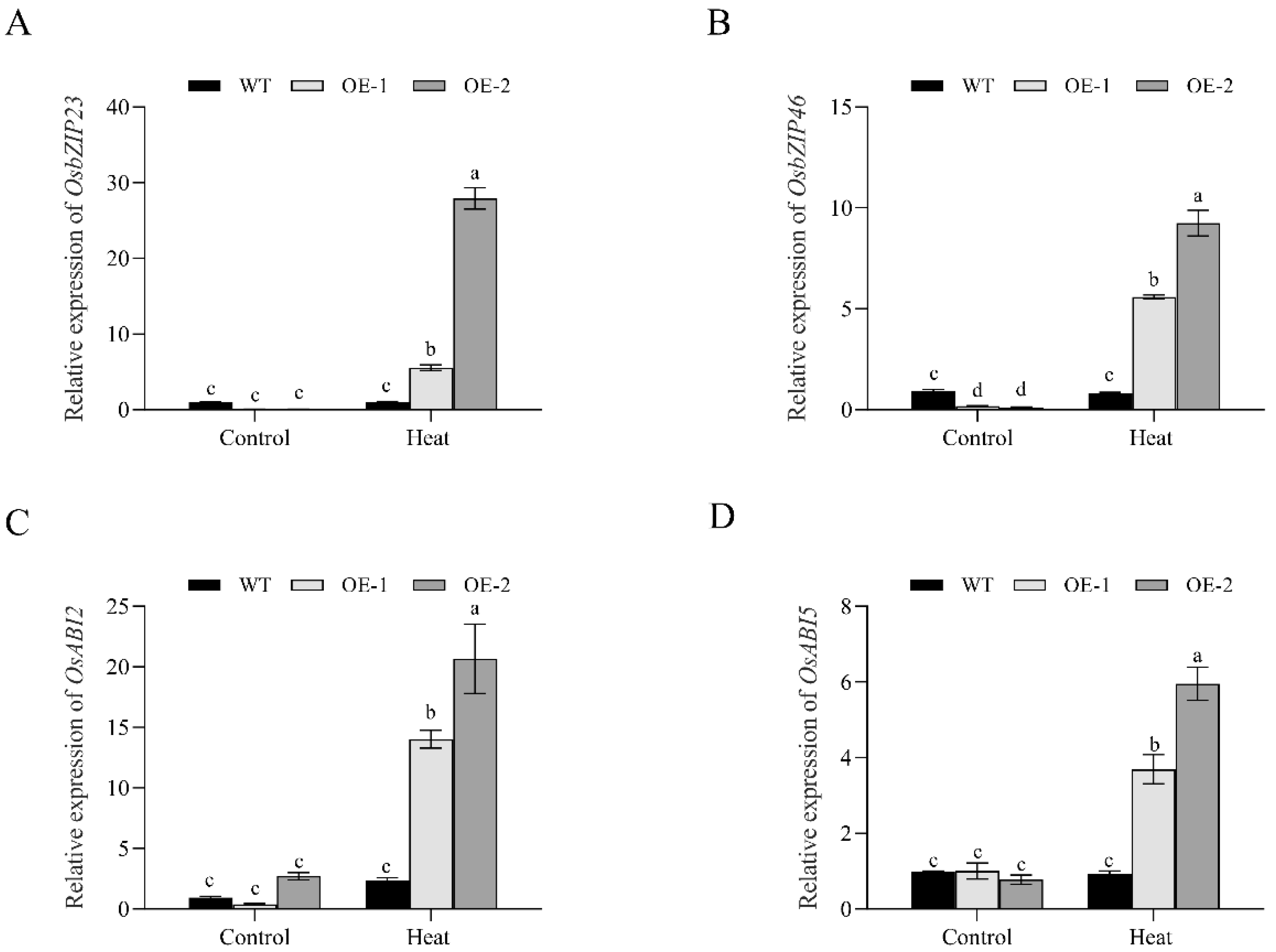
3.7. Changes in the Transcription of ABA-Signaling-Related Genes in OsNCED1 Transgenic Rice Plants
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wahid, A.; Gelani, S.; Ashraf, M.; Foolad, M.R. Heat tolerance in plants: An overview. J. Environ. Exp. Bot. 2007, 61, 199–223. [Google Scholar] [CrossRef]
- Peng, S.; Huang, J.; Sheehy, J.E.; Laza, R.C.; Visperas, R.M.; Zhong, X.; Centeno, G.S.; Khush, G.S.; Cassman, K.G. Rice yields decline with higher night temperature from global warming. Proc. Natl. Acad. Sci. USA 2004, 101, 9971–9975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, S.K.; Chang, M.C.; Tsai, Y.G.; Lur, H.S. Proteomic analysis of the expression of proteins related to rice quality during caryopsis development and the effect of high temperature on expression. J. Proteom. 2005, 5, 2140–2156. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Liu, B.; Piao, S.; Wang, X.; Lobell, D.B.; Huang, Y.; Huang, M.; Yao, Y.; Bassu, S.; Ciais, P. Temperature increase reduces global yields of major crops in four independent estimates. Proc. Natl. Acad. Sci. USA 2017, 114, 9326–9331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, P.; Fan, S.; Gao, P.; Li, S. Gene function and signal pathway review of rice high temperature resistance. J. Sichuan Agric. Univ. 2021, 39, 279–285. [Google Scholar]
- Tian, X.H.; Tsutomu, M.; Li, S.H.; Lin, J.C. Research progress and prospect of high temperature stress in rice at flowering stage. Ying Yong Sheng Tai Xue Bao J. Appl. Ecol. 2007, 18, 2632–2636. [Google Scholar]
- Jacob, P.; Hirt, H.; Bendahmane, A. The heat-shock protein/chaperone network and multiple stress resistance. Plant Biotechnol. J. 2017, 15, 405–414. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.H.; An, G. Application of MapMan and RiceNet drives systematic analyses of the early heat stress transcriptome in rice seedlings. J. Plant Biol. 2012, 55, 436–449. [Google Scholar] [CrossRef]
- Jung, K.H.; Gho, H.J.; Nguyen, M.X.; Kim, S.R.; An, G. Genome-wide expression analysis of HSP70 family genes in rice and identification of a cytosolic HSP70 gene highly induced under heat stress. Funct. Integr. Genom. 2013, 13, 391–402. [Google Scholar] [CrossRef]
- Chen, K.; Guo, T.; Li, X.M.; Zhang, Y.M.; Yang, Y.B.; Ye, W.W.; Dong, N.Q.; Shi, C.L.; Kan, Y.; Xiang, Y.H. Translational regulation of plant response to high temperature by a dual-function tRNAHis guanylyltransferase in rice. Mol. Plant. 2019, 12, 1123–1142. [Google Scholar] [CrossRef] [PubMed]
- Dhatt, B.K.; Paul, P.; Sandhu, J.; Hussain, W.; Irvin, L.; Zhu, F.; Adviento-Borbe, M.A.; Lorence, A.; Staswick, P.; Yu, H.; et al. Allelic variation in rice Fertilization Independent Endosperm 1 contributes to grain width under high night temperature stress. New Phytol. 2021, 229, 335–350. [Google Scholar] [CrossRef]
- Jung, K.H.; Ko, H.J.; Nguyen, M.X.; Kim, S.R.; Ronald, P.; An, G. Genome-wide identification and analysis of early heat stress responsive genes in rice. J. Plant Biol. 2012, 55, 458–468. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, X.; Wang, X.; Gao, K.; Qi, W.; Ren, H.; Hu, H.; Sun, D.; Bai, J.; Zheng, S. Heterologous expression of heat stress-responsive AtPLC9 confers heat tolerance in transgenic rice. BMC Plant Biol. 2020, 20, 514. [Google Scholar] [CrossRef] [PubMed]
- Mittal, D.; Madhyastha, D.A.; Grover, A. Gene expression analysis in response to low and high temperature and oxidative stresses in rice: Combination of stresses evokes different transcriptional changes as against stresses applied individually. J. Plant Sci. 2012, 197, 102–113. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, N.K.; Kim, Y.K.; Grover, A. Coexpression network analysis associated with call of rice seedlings for encountering heat stress. Plant Mol. Biol. 2014, 84, 125–143. [Google Scholar] [CrossRef]
- Hobo, T.; Suwabe, K.; Aya, K.; Suzuki, G.; Yano, K.; Ishimizu, T.; Fujita, M.; Kikuchi, S.; Hamada, K.; Miyano, M.; et al. Various spatiotemporal expression profiles of anther expressed genes in rice. Plant Cell Physiol. 2008, 49, 1417–1428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Usadel, B.; Nagel, A.; Thimm, O.; Redestig, H.; Blaesing, O.E.; Palacios-Rojas, N.; Selbig, J.; Hannemann, J.; Conceição Piques, M.; Steinhauser, D. Extension of the visualization tool MapMan to allow statistical analysis of arrays, display of coresponding genes, and comparison with known responses. J. Plant Physiol. 2005, 138, 1195–1204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rerksiri, W.; Zhang, X.; Xiong, H.; Chen, X. Expression and promoter analysis of six heat stress-inducible genes in rice. Sci. World J. 2013, 11, 1652–1656. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. J. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Counce, P.A.; Keisling, T.C.; Mitchell, A.J. A uniform, objective, and adaptive system for expressing rice development. Crop Sci. 2020, 40, 436–443. [Google Scholar] [CrossRef] [Green Version]
- Zhang, G.L.; Deng, H.B. Experimental Technology and Practice of Seed Science; Hunan Science and Technology Press: Changsha, China, 2018; pp. 142–147. ISBN 978-7-5710-0180-3. [Google Scholar]
- Li, H.S.; Sun, Q.; Zhao, S.J. Principles and Techniques of Plant Physiological and Biochemical Experiments; Higher Education Press: Beijing, China, 2000; pp. 234–239. ISBN 978-7-0400-8076-6. [Google Scholar]
- De, S.N.; Geelen, D. The impact of environmental stress on male reproductive development in plants: Biological processes and molecular mechanisms. J. Plant Cell Environ. 2014, 37, 1–18. [Google Scholar]
- Kan, Y.; Lin, H.X. Molecular regulation and genetic control of rice thermal response. Crop J. 2021, 9, 497–505. [Google Scholar] [CrossRef]
- Sreenivasulu, N.; Butardo, V.; Misra, G.; Cuevas, R.P.; Anacleto, R.; Kavi Kishor, P.B. Designing climate-resilient rice with ideal grain quality suited for high-temperature stress. J. Exp. Bot. 2015, 66, 1737–1748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ozturk, Z.N.; Valentina, T.; Deyholos, M.; Michalowski, C.B.; Galbraith, D.W.; Gozukirmizi, N.; Tuberosa, R.; Bohnert, H.J. Monitoring large-scale changes in transcript abundance in drought-and salt-stressed barley. J. Plant Mol. Biol. 2002, 48, 551–573. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, H.; Gao, F.; Li, J.; Li, Z. Comparison of gene expression between upland and lowland rice cultivars under water stress using cDNA microarray. J. Theor. Appl. Genet. 2007, 115, 1109–1126. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Xu, X.; Ying, L.; Wang, H.; Li, Z. Comparative transcriptome analysis reveals differential transcription in heat-susceptible and heat-tolerant pepper (Capsicum annum L.) cultivars under heat stress. J. Plant Biol. 2015, 58, 411–424. [Google Scholar] [CrossRef]
- Liu, G.T.; Wang, J.F.; Cramer, G.; Dai, Z.W.; Duan, W.; Xu, H.G.; Wu, B.H.; Fan, P.G.; Wang, L.J.; Li, S.H. Transcriptomic analysis of grape (Vitis vinifera L.) leaves during and after recovery from heat stress. BMC Plant Biol. 2012, 12, 174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharoni, A.M.; Nuruzzaman, M.; Satoh, K.; Shimizu, T.; Kondoh, H.; Sasaya, T.; Choi, I.R.; Omura, T.; Kikuchi, S. Gene structures, classification and expression models of the AP2/EREBP transcription factor family in rice. J. Plant Cell Physiol. 2011, 52, 344–360. [Google Scholar] [CrossRef] [PubMed]
- Ray, S.; Dansana, P.K.; Giri, J.; Deveshwar, P.; Arora, R.; Agarwal, P.; Khurana, J.P.; Kapoor, S.; Tyagi, A.K. Modulation of transcription factor and metabolic pathway genes in response to water-deficit stress in rice. Funct. Integr. Genom. 2011, 11, 157–178. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Shiroto, Y.; Kishitani, S.; Ito, Y.; Toriyama, K. Enhanced heat and drought tolerance in transgenic rice seedlings overexpressing OsWRKY11 under the control of HSP101 promoter. J. Plant Cell Rep. 2009, 28, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, K.; Takasaki, H.; Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. NAC transcription factors in plant abiotic stress responses. J. BBA Gene Regul. Mech. 2012, 1819, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Takasaki, H.; Maruyama, K.; Kidokoro, S.; Ito, Y.; Fujita, Y.; Shinozaki, K.; Yamaguchi-Shinozaki, K.; Nakashima, K. The abiotic stress-responsive NAC-type transcription factor OsNAC5 regulates stress-inducible genes and stress tolerance in rice. J. Mol. Genet. Genom. 2010, 284, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Xiong, H.; Liu, A.; Zhou, X.; Peng, Y.; Li, Z.; Luo, G.; Tian, X.; Chen, X. Microarray data uncover the genome-wide gene expression patterns in response to heat stress in rice post-meiosis panicle. J. Plant Biol. 2014, 57, 327–336. [Google Scholar] [CrossRef]
- Mittal, D.; Chakrabarti, S.; Sarkar, A.; Singh, A.; Grover, A. Heat shock factor gene family in rice: Genomic organization and transcript expression profiling in response to high temperature, low temperature and oxidative stresses. J. Plant Physiol. Biochem. 2009, 47, 785–795. [Google Scholar] [CrossRef]
- Matsumoto, T.; Morishige, H.; Tanaka, T.; Kanamori, H.; Komatsuda, T.; Sato, K.; Itoh, T.; Wu, J.Z.; Nakamura, S. Transcriptome analysis of barley identifies heat shock and HD-Zip I transcription factors up-regulated in response to multiple abiotic stresses. J. Mol. Breed. 2014, 34, 761–768. [Google Scholar] [CrossRef]
- Queitsch, C.; Hong, S.W.; Vierling, E.; Lindquist, S. Heat shock protein 101 plays a crucial role in thermotolerance in Arabidopsis. Plant Cell 2000, 12, 479–492. [Google Scholar] [CrossRef] [Green Version]
- Yamada, K.; Fukao, Y.; Hayashi, M.; Fukazawa, M.; Suzuki, I.; Nishimura, M. Cytosolic HSP90 regulates the heat shock response that is responsible for heat acclimation in Arabidopsis thaliana. J. Biol. Chem. 2007, 282, 37794–37804. [Google Scholar] [CrossRef] [Green Version]
- Hu, X.; Li, Y.; Li, C.; Yang, H.; Wang, W.; Lu, M. Characterization of small heat shock proteins associated with maize tolerance to combined drought and heat stress. J. Plant Growth Regul. 2010, 29, 455–464. [Google Scholar] [CrossRef]
- Li, W.B.; Cai, X.; Jiang, M.Y. Functional analysis and gene identification of the OsCaMs involved in ABA-inducing. Acta Bot. Boreali Occident. Sin. 2017, 37, 1474–1485. [Google Scholar]
- De-Souza, T.C.; Magalhães, P.C.; de-Castro, E.M.; Carneiro, N.P.; Padilha, F.A.; Gomes-Júnior, C.C. ABA application to maize hybrids contrasting for drought tolerance: Changes in water parameters and in antioxidant enzyme activity. J. Plant Growth Regul. 2014, 73, 205–217. [Google Scholar] [CrossRef]
- Yang, D.Q.; Wang, Z.L.; Ni, Y.L.; Yin, Y.P.; Cai, T.; Yang, W.B.; Peng, D.L.; Cui, Z.Y.; Jiang, W.W. Effect of high temperature stress and spraying exogenous ABA post-anthesis on grain filling and grain yield in different types of stay-green wheat. J. Sci. Agric. Sin. 2014, 47, 2109–2125. [Google Scholar]
- Tang, R.S.; Zhang, D.D.; Tong, H.Y. Effects of high temperature on some physiological indexes in rice seedling leaves and regulations by ABA and 6-BA. Jiangsu J. Agric. Sci. 2005, 21, 145–149. [Google Scholar]
- Tang, R.S.; Zheng, J.C.; Jin, Z.Q.; Zhang, D.D.; Huang, Y.H.; Chen, L.G. Possible correlation between high temperature-induced floret sterility and endogenous levels of IAA, GAs and ABA in rice (Oryza sativa L.). Plant Growth Regul. 2008, 54, 37–43. [Google Scholar] [CrossRef]
- Priya, R.; Siva, R. Analysis of phylogenetic and functional diverge in plant nine-cis epoxycarotenoid dioxygenase gene family. J. Plant Res. 2015, 128, 519–534. [Google Scholar] [CrossRef] [PubMed]
- Vishwakarma, K.; Upadhyay, N.; Kumar, N.; Yadav, G.; Singh, J.; Mishra, R.K.; Kumar, V.; Verma, R.; Upadhyay, R.G.; Pandey, M.; et al. Abscisic acid signaling and abiotic stress tolerance in plants: A review on current knowledge and future prospects. Front. Plant Sci. 2017, 8, 161. [Google Scholar] [CrossRef] [Green Version]
- Tong, S.M.; Xi, H.X.; Ai, K.J.; Hou, H.S. Overexpression of wheat TaNCED gene in Arabidopsis enhances tolerance to drought stress and delays seed germination. J. Biol. Plantarum. 2017, 61, 64–72. [Google Scholar] [CrossRef]
- Hwang, S.G.; Chen, H.C.; Huang, W.Y.; Chu, Y.C.; Shii, C.T.; Cheng, W.H. Ectopic expression of rice OsNCED3 in Arabidopsis increases ABA level and alters leaf morphology. J. Plant Sci. 2010, 178, 12–22. [Google Scholar] [CrossRef]
- Hwang, S.G.; Lee, C.Y.; Tseng, C.S. Heterologous expression of rice 9-cis-epoxycarotenoid dioxygenase 4 (OsNCED4) in Arabidopsis confers sugar oversensitivity and drought tolerance. J. Bot. Stud. 2018, 59, 2. [Google Scholar] [CrossRef]
- Zhou, H.; Zhang, Y.J.; Luo, Z.X.; Tang, W.B.; Zhang, G.L. Bioinformatic analysis of NCED gene family in rice. J. Mol. Plant Breed. 2020. Available online: https://kns.cnki.net/kcms/detail/46.1068.s.20201123.1142.004.html (accessed on 23 November 2020).
- Oliver, S.N.; Dennis, E.S.; Dolferus, R. ABA regulates apoplastic sugar transport and is a potential signal for cold-induced pollen sterility in rice. J. Plant Cell Physiol. 2007, 48, 1319–1330. [Google Scholar] [CrossRef] [Green Version]
- Jiang, D.; Zhou, L.; Chen, W.; Ye, N.; Xia, J.; Zhuang, C. Overexpression of a microRNA-targeted NAC transcription factor improves drought and salt tolerance in Rice via ABA-mediated pathways. Rice 2019, 12, 76. [Google Scholar] [CrossRef]
- Kim, Y.H.; Khan, A.L.; Waqas, M.; Shim, J.K.; Lee, I.J. Silicon application to rice root zone influenced the phytohormonal and antioxidant responses under salinity stress. J. Plant Growth Regul. 2014, 33, 137–149. [Google Scholar] [CrossRef]
- Xu, Y.; Chu, C.; Yao, S. The impact of high-temperature stress on rice: Challenges and solutions. Crop J. 2021, 9, 963–976. [Google Scholar] [CrossRef]
- Higashi, Y.; Saito, K. Lipidomic studies of membrane glycerolipids in plant leaves under heat stress. J. Prog. Lipid Res. 2019, 75, 100990. [Google Scholar] [CrossRef]
- Bahuguna, R.N.; Jha, J.; Pal, M.; Shah, D.; Lawas, L.M.; Khetarpal, S.; Jagadish, K.S. Physiological and biochemical characterization of NERICA-L-44: A novel source of heat tolerance at the vegetative and reproductive stages in rice. J. Physiol. Plantarum. 2015, 154, 543–559. [Google Scholar] [CrossRef]
- Qiao, B.; Zhang, Q.; Liu, D.; Wang, H.; Yin, J.; Wang, R.; He, M.; Cui, M.; Shang, Z.; Wang, D.; et al. A calcium-binding protein, rice annexin OsANN1, enhances heat stress tolerance by modulating the production of H2O2. J. Exp. Bot. 2015, 66, 5853–5866. [Google Scholar] [CrossRef] [Green Version]
- Sailaja, B.; Subrahmanyam, D.; Neelamraju, S.; Vishnukiran, T.; Rao, Y.V.; Vijayalakshmi, P.; Voleti, S.R.; Bhadana, V.P.; Mangrauthia, S.K. Integrated physiological, biochemical, and molecular analysis identifies important traits and mechanisms associated with differential response of rice genotypes to elevated temperature. Front. Plant Sci. 2015, 6, 1044. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Q.; Zhou, L.; Liu, J.; Cao, Z.; Du, X.; Huang, F.; Pan, G.; Cheng, F. Involvement of CAT in the detoxification of HT-induced ROS burst in rice anther and its relation to pollen fertility. J. Plant Cell Rep. 2018, 37, 741–757. [Google Scholar] [CrossRef]
- Li, S.; Wan, C.; Kong, J.; Zhang, Z.; Zhu, Y. Programmed cell death during microgenesis in a Honglian CMS line of rice is correlated with oxidative stress in mitochondria. J. Funct. Plant Biol. 2004, 31, 369–376. [Google Scholar] [CrossRef]
- Sharma, E.; Borah, P.; Kaur, A.; Bhatnagar, A.; Mohapatra, T.; Kapoor, S.; Khurana, J.P. A comprehensive transcriptome analysis of contrasting rice cultivars highlights the role of auxin and ABA responsive genes in heat stress response. Genomics 2021, 113, 1247–1261. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Y.; Zhang, Q.; Cui, Y.; Xiang, J.; Chen, H.; Hu, G.; Chen, Y.; Wang, X.; Zhu, D.; et al. Comparative transcriptome analysis of panicle development under heat stress in two rice (Oryza sativa L.) cultivars differing in heat tolerance. PeerJ 2019, 7, e7595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, Z.; He, F.; Feng, X.; Liang, T.; Wang, H.; Ding, S.; Tian, X. Transcriptomic analysis reveals important roles of lignin and flavonoid biosynthetic pathways in rice thermotolerance during reproductive stage. Front. Genet. 2020, 11, 1120. [Google Scholar] [CrossRef]
- Devireddy, A.R.; Zandalinas, S.I.; Fichman, Y.; Mittler, R. Integration of reactive oxygen species and hormone signaling during abiotic stress. Plant J. 2021, 105, 459–476. [Google Scholar] [CrossRef]
- Kim, T.H. Mechanism of ABA signal transduction: Agricultural highlights for improving drought tolerance. J. Plant Biol. 2014, 57, 1–8. [Google Scholar] [CrossRef]
- Nakashima, K.; Yamaguchi-Shinozaki, K. ABA signaling in stress-response and seed development. J. Plant Cell Rep. 2013, 32, 959–970. [Google Scholar] [CrossRef]
- Wang, W.Q.; Xu, D.Y.; Sui, Y.P.; Ding, X.H.; Song, X.J. A multiomic study uncovers a bZIP23-PER1A–mediated detoxification pathway to enhance seed vigor in rice. Proc. Natl. Acad. Sci. USA 2022, 119, e2026355119. [Google Scholar] [CrossRef]
- Bi, C.; Ma, Y.; Zhen, W.; Yu, Y.T.; Shan, L.; Lu, K.; Wang, X.F. Arabidopsis ABI5 plays a role in regulating ROS homeostasis by activating CATALASE 1 transcription in seed germination. J. Plant Mol. Biol. 2017, 94, 197–213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miao, Y.; Lv, D.; Wang, P.; Wang, X.C.; Chen, J.; Miao, C.; Song, C.P. An Arabidopsis glutathione peroxidase functions as both a redox transducer and a scavenger in abscisic acid and drought stress responses. J. Plant Cell. 2006, 18, 2749–2766. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Yang, J.; Lu, S.; Cai, J.; Guo, Z. Overexpressing SgNCED1 in tobacco increases ABA level, antioxidant enzyme activities, and stress tolerance. J. Plant Growth Regul. 2008, 27, 151–158. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, H.; Wang, Y.; Zhang, Y.; Xiao, Y.; Liu, X.; Deng, H.; Lu, X.; Tang, W.; Zhang, G. Comparative Analysis of Heat-Tolerant and Heat-Susceptible Rice Highlights the Role of OsNCED1 Gene in Heat Stress Tolerance. Plants 2022, 11, 1062. https://doi.org/10.3390/plants11081062
Zhou H, Wang Y, Zhang Y, Xiao Y, Liu X, Deng H, Lu X, Tang W, Zhang G. Comparative Analysis of Heat-Tolerant and Heat-Susceptible Rice Highlights the Role of OsNCED1 Gene in Heat Stress Tolerance. Plants. 2022; 11(8):1062. https://doi.org/10.3390/plants11081062
Chicago/Turabian StyleZhou, Huang, Yingfeng Wang, Yijin Zhang, Yunhua Xiao, Xiong Liu, Huabing Deng, Xuedan Lu, Wenbang Tang, and Guilian Zhang. 2022. "Comparative Analysis of Heat-Tolerant and Heat-Susceptible Rice Highlights the Role of OsNCED1 Gene in Heat Stress Tolerance" Plants 11, no. 8: 1062. https://doi.org/10.3390/plants11081062
APA StyleZhou, H., Wang, Y., Zhang, Y., Xiao, Y., Liu, X., Deng, H., Lu, X., Tang, W., & Zhang, G. (2022). Comparative Analysis of Heat-Tolerant and Heat-Susceptible Rice Highlights the Role of OsNCED1 Gene in Heat Stress Tolerance. Plants, 11(8), 1062. https://doi.org/10.3390/plants11081062






