Genetic Characteristics and Metabolic Interactions between Pseudocercospora fijiensis and Banana: Progress toward Controlling Black Sigatoka
Abstract
1. Introduction
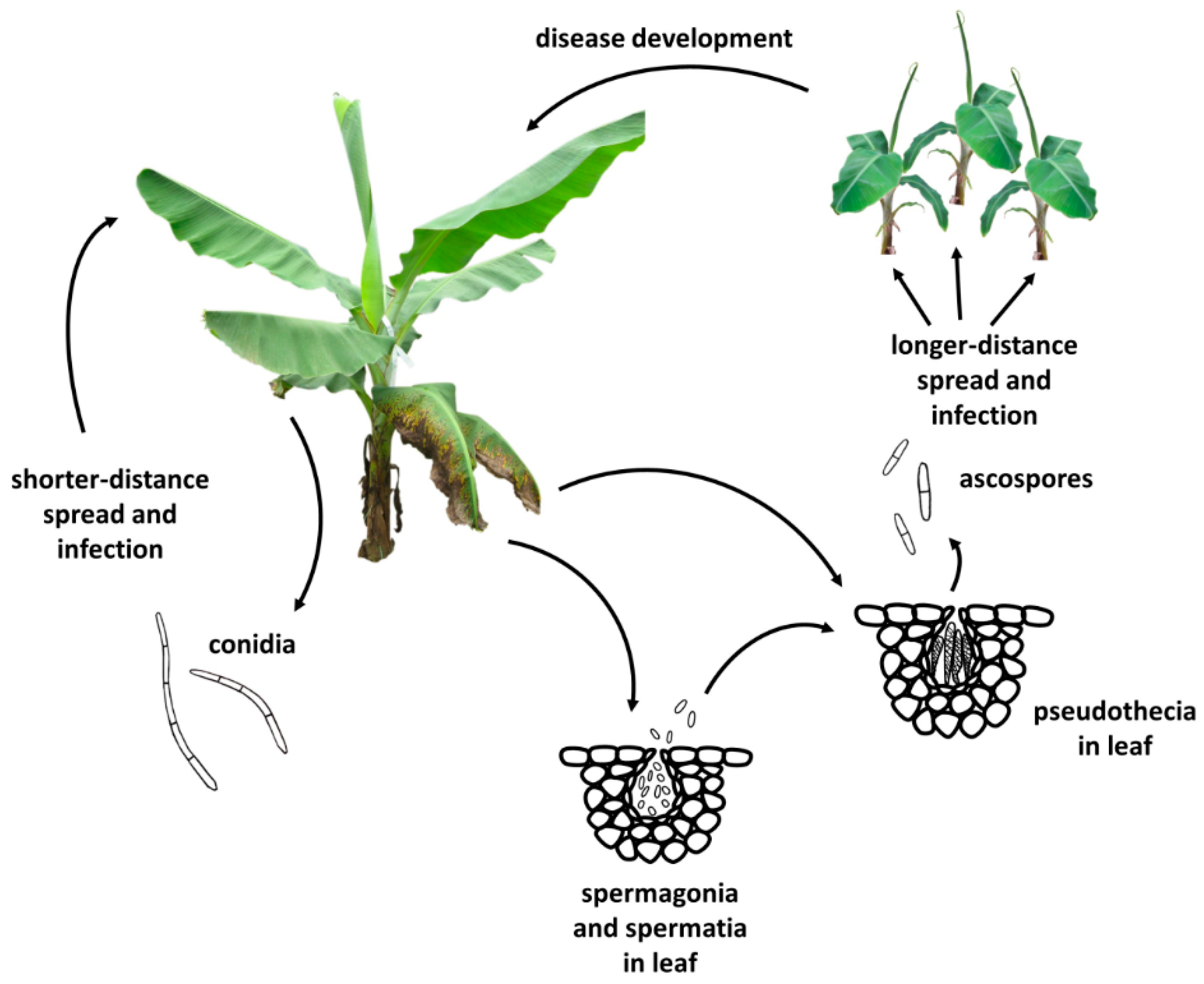
2. Disease Management
3. Breeding for Resistance
4. Genomic Structure of Pseudocercospora fijiensis
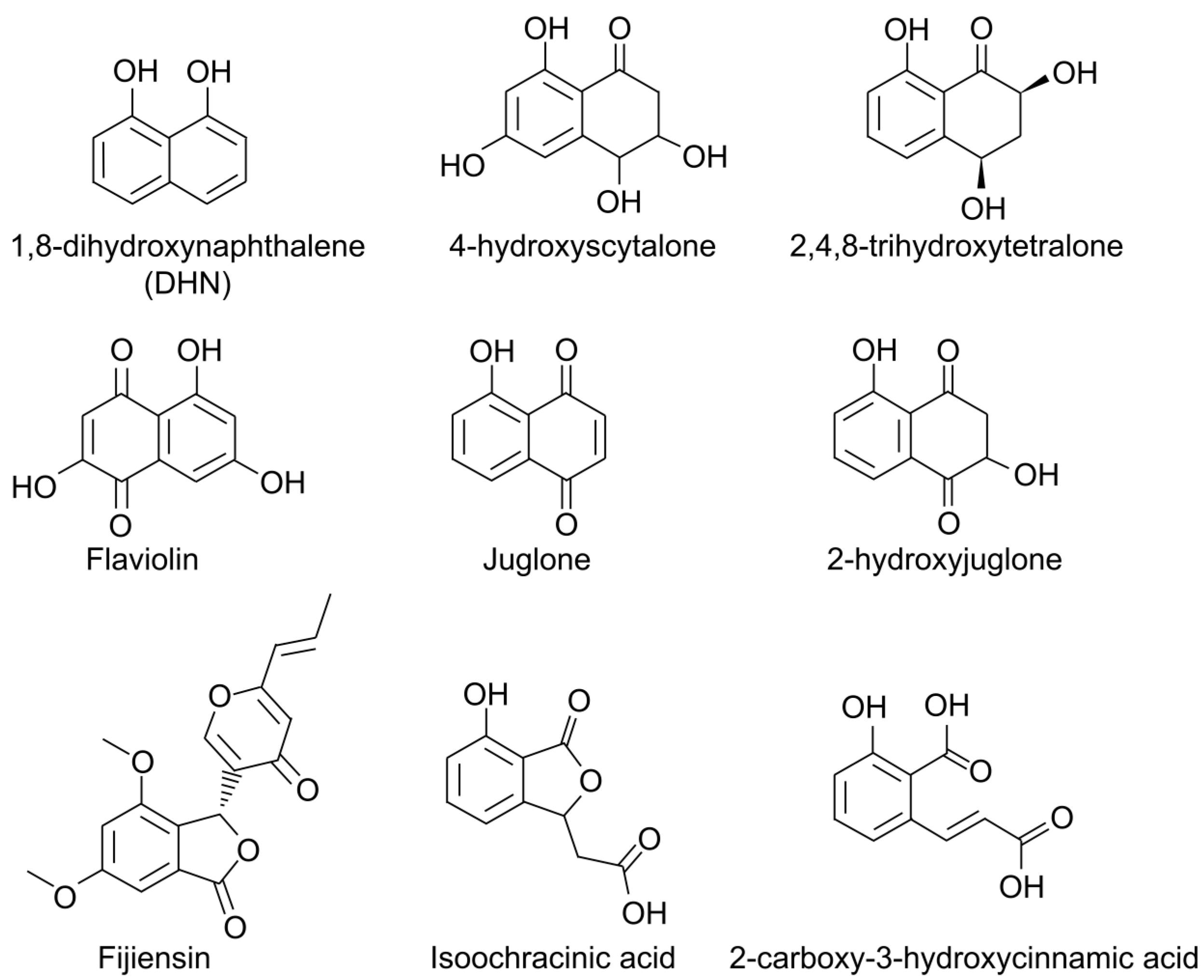
5. Secondary Metabolism
6. Effectors
7. Other Pathogenicity Genes
8. Host Defense Genes
9. Potential for Transgenic Banana Plants Resistant to Black Sigatoka
10. Conclusions and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Frison, E.; Escalant, J.V.; Sharrock, S.L. The Global Musa Genomic Consortium: A boost for banana improvement. In Banana Improvement: Cellular and Molecular Biology, and Induced Mutations; Swennen, M.J.R., Ed.; Science Publishers: Enfield, NH, USA, 2004; pp. 341–349. [Google Scholar]
- Soares, J.M.S.; Rocha, A.J.; Nascimento, F.S.; Santos, A.S.; Miller, R.N.G.; Ferreira, C.F.; Haddad, F.; Amorim, V.B.O.; Amorim, E.P. Genetic improvement for resistance to black Sigatoka in bananas: A systematic review. Front. Plant Sci. 2021, 12, 657916. [Google Scholar] [CrossRef] [PubMed]
- FAOSTAT. FAOSTAT Online Database. 2022. Available online: https://www.fao.org/faostat/en/ (accessed on 22 January 2022).
- Alakonya, A.E.; Kimunye, J.; Mahuku, G.; Amah, D.; Uwimana, B.; Brown, A.; Swennen, R. Progress in understanding Pseudocercospora banana pathogens and the development of resistant Musa germplasm. Plant Pathol. 2018, 67, 759–770. [Google Scholar] [CrossRef]
- FAO. Banana Statistical Compendium 2020; FAO: Rome, Italy, 2021. [Google Scholar]
- FAO. World Banana Forum. Available online: https://www.fao.org/world-banana-forum/about-the-forum/en/ (accessed on 13 February 2022).
- Shepherd, K. Banana breeding—Past and present. Acta Hortic. 1987, 196, 37–44. [Google Scholar] [CrossRef]
- Daniells, J.; O’Keefe, V.; Smyth, H.; Gething, K.; Fanning, K.; Telford, P. Planet of the Cavendish—Understanding the domination. Acta Hortic. 2013, 986, 219–224. [Google Scholar] [CrossRef]
- Churchill, A.C.L. Mycosphaerella fijiensis, the black leaf streak pathogen of banana: Progress towards understanding pathogen biology and detection, disease development, and the challenges of control. Mol. Plant Pathol. 2011, 12, 307–328. [Google Scholar] [CrossRef]
- Ploetz, R.C. Black Sigatoka of banana: The most important disease of a most important fruit. Plant Health Instr. 2001. [Google Scholar] [CrossRef]
- Marín, D.H.; Romero, R.A.; Guzmán, M.; Sutton, T.B. Black Sigatoka: An increasing threat to banana cultivation. Plant Dis. 2003, 87, 208–222. [Google Scholar] [CrossRef]
- Hermanto, C.; Opina, O.S.; Natural, M.P. Assessment of fungicide resistance of a population of Mycosphaerella spp. on Señorita banana variety (Sucrier group). Tree For. Sci. Biotechnol. 2010, 4, 85–90. [Google Scholar]
- Stover, R.H. Distribution and probable origin of Mycosphaerella fijiensis in southeast Asia. Trop. Agric. 1978, 55, 65–68. [Google Scholar]
- Strobl, E.; Mohan, P. Climate and the global spread and impact of bananas’ black leaf Sigatoka disease. Atmosphere 2020, 11, 947. [Google Scholar] [CrossRef]
- Amil, A.F.; Heaney, S.P.; Stanger, C.; Shaw, M.W. Dynamics of QoI sensitivity in Mycosphaerella fijiensis in Costa Rica during 2000 to 2003. Phytopathology 2007, 97, 1451–1457. [Google Scholar] [CrossRef] [PubMed]
- Jacome, L.H.; Schuh, W.; Stevenson, R.E. Effect of temperature and relative humidity on germination and germ tube development of Mycosphaerella fijiensis var. difformis. Phytopathology 1991, 81, 1480–1485. [Google Scholar] [CrossRef]
- Balint-Kurti, P.J.; Churchill, A.C. Towards a molecular understanding of Mycosphaerella/banana interactions. In Banana Improvement: Cellular, Molecular Biology, and Induced Mutations; Jain, S.M., Swennen, R., Eds.; Science Publishers, Inc.: Enfield, NH, USA; Plymouth, UK, 2004; pp. 147–160. [Google Scholar]
- Balint-Kurti, P.J.; May, G.D.; Churchill, A.C. Development of a transformation system for Mycosphaerella pathogens of banana: A tool for the study of host/pathogen interactions. FEMS Microbiol. Lett. 2001, 195, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Beveraggi, A.; Mourichon, X.; Sallé, G. Étude comparée des premières étapes de l’infection chez des bananiers sensibles et résistants infectés par le Cercospora fijiensis (Mycosphaerella fijiensis) agent responsable de la maladie des raies noires. Can. J. Bot. 1995, 73, 1328–1337. [Google Scholar] [CrossRef]
- Liberato, J.R.; Peterson, R.A.; Gasparotto, L.; Ferrari, J.T.; Grice, K.; Porchun, S.C.; Shivas, R.G. Black Sigatoka of Banana. Available online: http://www.padil.gov.au/pests-and-diseases/pest/main/136601 (accessed on 7 May 2021).
- Washington, J.R.; Cruz, J.; Lopez, F.; Fajardo, M. Infection studies of Mycosphaerella fijiensis on banana and the control of black Sigatoka with chlorothalonil. Plant Dis. 1998, 82, 1185–1190. [Google Scholar] [CrossRef]
- Meredith, D.S.; Lawrence, J.S.; Firman, I.D. Ascospore release and dispersal in black leaf streak disease of bananas (Mycosphaerella fijiensis). Trans. Br. Mycol. Soc. 1973, 60, 547–554. [Google Scholar] [CrossRef]
- FRAC. Banana working group meeting minutes. In Proceedings of the Fungicide Resistance Action Committee, Miami, FL, USA, 30 April–1 May 2018. [Google Scholar]
- Chong, P.; Essoh, J.N.; Arango Isaza, R.E.; Keizer, P.; Stergiopoulos, I.; Seidl, M.F.; Guzman, M.; Sandoval, J.; Verweij, P.E.; Scalliet, G.; et al. A world-wide analysis of reduced sensitivity to DMI fungicides in the banana pathogen Pseudocercospora fijiensis. Pest Manag. Sci. 2021, 77, 3273–3288. [Google Scholar] [CrossRef]
- Cañas-Gutiérrez, G.P.; Angarita-Velásquez, M.J.; Restrepo-Flórez, J.M.; Rodríguez, P.; Moreno, C.X.; Arango, R. Analysis of the CYP51 gene and encoded protein in propiconazole-resistant isolates of Mycosphaerella fijiensis. Pest Manag. Sci. 2009, 65, 892–899. [Google Scholar] [CrossRef]
- Becker, P.; Esker, P.; Umaña, G. Incorporation of microorganisms to reduce chemical fungicide usage in black sigatoka control programs in Costa Rica by use of biological fungicides. Crop Prot. 2021, 146, 105657. [Google Scholar] [CrossRef]
- Craenen, K. Black Sigatoka Disease of Banana and Plantain: A Reference Manual; IITA: Ibadan, Nigeria, 1998. [Google Scholar]
- Kablan, L.; Lagauche, A.; Delvaux, B.; Legrève, A. Silicon reduces black Sigatoka development in banana. Plant Dis. 2012, 96, 273–278. [Google Scholar] [CrossRef]
- Mobambo, K.N.; Zuofa, K.; Gauhl, F.; Adeniji, M.O.; Pasberg-Gauhl, C. Effect of soil fertility on host response to black leaf streak of plantain (Musa spp., AAB group) under traditional farming systems in southeastern Nigeria. Int. J. Pest Manag. 1994, 40, 75–80. [Google Scholar] [CrossRef]
- Dubois, T.; Coyne, D.; Kahangi, E.; Turoop, L.; Nsubuga, E.W.N. Endophyte-enhanced banana tissue culture: Technology transfer through public-private partnerships in Kenya and Uganda. ATDF J. 2012, 3, 18. [Google Scholar]
- Vézina, A.; Dubois, T. Planting Material. Available online: https://www.promusa.org/Planting+material (accessed on 7 May 2021).
- Vicente-Chandler, J.; Abruña, F.; Silva, S. Effect of shade trees on yields of five crops in the humid mountain region of Puerto Rico. J. Agric. Univ. Puerto Rico 1966, 50, 218–225. [Google Scholar] [CrossRef]
- Dold, C.; Staver, C.; Pocasangre, L.; Heller, J. Musa in shaded perennial crops—Response to light interception. In Proceedings of the Conference on International Research on Food Security, Natural Resource Management, and Rural Development University of Hohenheim, Stuttgart, Germany, 7–9 October 2008. [Google Scholar]
- Kimunye, J.; Were, E.; Swennen, R.; Viljoen, A.; Mahuku, G. Sources of resistance to Pseudocercospora fijiensis, the cause of black Sigatoka in banana. Plant Pathol. 2021, 70, 1651–1664. [Google Scholar] [CrossRef] [PubMed]
- Orjeda, G. Evaluating Bananas: A Global Partnership. Results of IMTP Phase II; INIBAP: Montpellier, France, 2000. [Google Scholar]
- Nascimento, F.D.S.; Sousa, Y.M.; Rocha, A.D.J.; Ferreira, C.F.; Haddad, F.; Amorim, E.P. Sources of black Sigatoka resistance in wild banana diploids. Rev. Bras. Frutic. 2020, 42, e038. [Google Scholar] [CrossRef]
- Backiyarani, S.; Sasikala, R.; Sharmiladevi, S.; Uma, S. Decoding the molecular mechanism of parthenocarpy in Musa spp. through protein–protein interaction network. Sci. Rep. 2021, 11, 14592. [Google Scholar] [CrossRef] [PubMed]
- Ortiz, R.; Vuylsteke, D. Inheritance of black Sigatoka disease resistance in plantain-banana (Musa spp.) hybrids. Theor. Appl. Genet. 1994, 89, 146–152. [Google Scholar] [CrossRef]
- Dadzie, B.K.; Orjeda, G. Post-Harvest Characteristics of Black Sigatoka Resistant Banana, Cooking Banana and Plantain Hybrids; Technical Guidelines Inibap 4; International Plant Genetic Resources Institute: Rome, Italy, 1998. [Google Scholar]
- Daniells, J.W. Global banana disease management—Getting serious with sustainability and food security. Acta Hortic. 2009, 828, 411–416. [Google Scholar] [CrossRef]
- Arango Isaza, R.E.; Diaz-Trujillo, C.; Dhillon, B.; Aerts, A.; Carlier, J.; Crane, C.F.; de Jong, T.V.; de Vries, I.; Dietrich, R.; Farmer, A.D.; et al. Combating a global threat to a clonal crop: Banana black Sigatoka pathogen Pseudocercospora fijiensis (synonym Mycosphaerella fijiensis) genomes reveal clues for disease control. PLoS Genet. 2016, 12, e1005876. [Google Scholar] [CrossRef]
- Amarillas, L.; Estrada-Acosta, M.; León-Chan, R.G.; López-Orona, C.; Lightbourn, L. First draft genome sequence resource of a strain of Pseudocercospora fijiensis isolated in North America. Phytopathology 2020, 110, 1620–1622. [Google Scholar] [CrossRef]
- Ohm, R.A.; Feau, N.; Henrissat, B.; Schoch, C.L.; Horwitz, B.A.; Barry, K.W.; Condon, B.J.; Copeland, A.C.; Dhillon, B.; Glaser, F.; et al. Diverse lifestyles and strategies of plant pathogenesis encoded in the genomes of eighteen Dothideomycetes fungi. PLOS Pathog. 2012, 8, e1003037. [Google Scholar] [CrossRef] [PubMed]
- Manzo-Sánchez, G.; Zapater, M.-F.; Luna-Martínez, F.; Conde-Ferráez, L.; Carlier, J.; James-Kay, A.; Simpson, J. Construction of a genetic linkage map of the fungal pathogen of banana Mycosphaerella fijiensis, causal agent of black leaf streak disease. Curr. Genet. 2008, 53, 299–311. [Google Scholar] [CrossRef] [PubMed]
- Gladyshev, E. Repeat-induced point mutation and other genome defense mechanisms in fungi. Eukaryotes Fungi Parasitol. 2017, 4, 687–699. [Google Scholar] [CrossRef]
- Rodríguez-García, C.M.; Raigosa-Flores, N.; Conde-Ferráez, L.; Peraza-Echeverría, L.; Canto-Canché, B.; James-Kay, A. Variation in electrophoretic karyotype among Mexican isolates of Mycosphaerella fijiensis. Can. J. Plant Pathol. 2006, 28, 236–241. [Google Scholar] [CrossRef]
- Mehrabi, R.; Gohari, A.M.; Kema, G.H.J. Karytype variability in plant pathogenic fungi. Annu. Rev. Phytopathol. 2017, 55, 483–503. [Google Scholar] [CrossRef]
- Covert, S.F. Supernumerary chromosomes in filamentous fungi. Curr. Genet. 1998, 33, 311–319. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.H.; Walton, J.D. Chromosomal organization of TOX2, a complex locus controlling host-selective toxin biosynthesis in Cochliobolus carbonum. Plant Cell 1996, 8, 887. [Google Scholar] [CrossRef]
- Han, Y.; Liu, X.; Benny, U.; Kistler, H.C.; VanEtten, H.D. Genes determining pathogenicity to pea are clustered on a supernumerary chromosome in the fungal plant pathogen Nectria haematococca. Plant J. 2001, 25, 305–314. [Google Scholar] [CrossRef]
- Johnson, L.J.; Johnson, R.D.; Akamatsu, H.; Salamiah, A.; Otani, H.; Kohmoto, K.; Kodama, M. Spontaneous loss of a conditionally dispensable chromosome from the Alternaria alternata apple pathotype leads to loss of toxin production and pathogenicity. Curr. Genet. 2001, 40, 65–72. [Google Scholar] [CrossRef]
- Temporini, E.D.; VanEtten, H.D. An analysis of the phylogenetic distribution of the pea pathogenicity genes of Nectria haematococca MPVI supports the hypothesis of their origin by horizontal transfer and uncovers a potentially new pathogen of garden pea: Neocosmospora boniensis. Curr. Genet. 2004, 46, 29–36. [Google Scholar] [CrossRef]
- Wittenberg, A.H.J.; van der Lee, T.A.J.; Ben M’barek, S.; Ware, S.B.; Goodwin, S.B.; Kilian, A.; Visser, R.G.F.; Kema, G.H.J.; Schouten, H.J. Meiosis drives extraordinary genome plasticity in the haploid fungal plant pathogen Mycosphaerella graminicola. PLoS ONE 2009, 4, e5863. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, S.B.; Kema, G.H.J. The genomes of Mycosphaerella graminicola and M. fijiensis. In Genomics of Plant-Associated Fungi: Monocot Pathogens; Dean, R., Lichens-Park, A., Kole, C., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 123–140. [Google Scholar]
- Noar, R.D.; Daub, M.E. Transcriptome sequencing of Mycosphaerella fijiensis during association with Musa acuminata reveals candidate pathogenicity genes. BMC Genom. 2016, 17, 690. [Google Scholar] [CrossRef] [PubMed]
- Heiser, I.; Sachs, E.; Liebermann, B. Photodynamic oxygen activation by rubellin D, a phytotoxin produced by Ramularia collo-cygni (Sutton et Waller). Physiol. Mol. Plant Pathol. 2003, 62, 29–36. [Google Scholar] [CrossRef]
- Choquer, M.; Dekkers, K.L.; Chen, H.Q.; Cao, L.; Ueng, P.P.; Daub, M.E.; Chung, K.R. The CTB1 gene encoding a fungal polyketide synthase is required for cercosporin biosynthesis and fungal virulence of Cercospora nicotianae. Mol. Plant Microbe Interact. 2005, 18, 468–476. [Google Scholar] [CrossRef]
- Miethbauer, S.; Günther, W.; Schmidtke, K.-U.; Heiser, I.; Gräfe, S.; Gitter, B.; Liebermann, B. Uredinorubellins and caeruleoramularin, photodynamically active anthraquinone derivatives produced by two species of the genus Ramularia. J. Nat. Prod. 2008, 71, 1371–1375. [Google Scholar] [CrossRef]
- Upadhyay, R.K.; Strobel, G.A.; Coval, S.J.; Clardy, J. Fijiensin, the first phytotoxin from Mycosphaerella fijiensis, the causative agent of Black Sigatoka disease. Experientia 1990, 46, 982–984. [Google Scholar] [CrossRef]
- Stierle, A.A.; Upadhyay, R.; Hershenhorn, J.; Strobel, G.A.; Molina, G. The phytotoxins of Mycosphaerella fijiensis, the causative agent of black Sigatoka disease of bananas and plantains. Experientia 1991, 47, 853–859. [Google Scholar] [CrossRef]
- Arai, M.; Tomoda, H.; Okuda, T.; Wang, H.; Tabata, N.; Masuma, R.; Yamaguchi, Y.; Omura, S. Funicone-related compounds, potentiators of antifungal miconazole activity, produced by Talaromyces flavus FKI-0076. J. Antibiot. 2002, 55, 172–180. [Google Scholar] [CrossRef]
- Dethoup, T.; Manoch, L.; Kijjoa, A.; Pinto, M.; Gales, L.; Damas, A.M.; Silva, A.M.S.; Eaton, G.; Herz, W. Merodrimanes and other constituents from Talaromyces thailandiasis. J. Nat. Prod. 2007, 70, 1200–1202. [Google Scholar] [CrossRef]
- Fuska, J.; Fuskova, A.; Nemec, P. Vermistatin, an antibiotic with cytotoxic effects produced from Penicillium vermiculatum. Biologia 1979, 34, 735–739. [Google Scholar]
- Fuska, J.; Uhrín, D.; Proksa, B.; Votický, Z.; Ruppeldt, J. The structure of vermistatin, a new metabolite from Penicillium vermiculatum. J. Antibiot. 1986, 39, 1605–1608. [Google Scholar] [CrossRef] [PubMed]
- Murtaza, N.; Husain, S.A.; Sarfaraz, T.B.; Sultana, N.; Faizi, S. Isolation and identification of vermistatin, ergosterol, stearic acid and mannitol, metabolic products of Penicillium verruculosum. Planta Med. 1997, 63, 191. [Google Scholar] [CrossRef] [PubMed]
- Komai, S.-I.; Hosoe, T.; Itabashi, T.; Nozawa, K.; Yaguchi, T.; Fukushima, K.; Kawai, K.-I. New vermistatin derivatives isolated from Penicillium simplicissimum. Heterocycles 2005, 65, 2771–2776. [Google Scholar] [CrossRef]
- Liu, Z.; Xia, G.; Chen, S.; Liu, Y.; Li, H.; She, Z. Eurothiocin A and B, sulfur-containing benzofurans from a soft coral-derived fungus Eurotium rubrum SH-823. Mar. Drugs 2014, 12, 3669–3680. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.-K.; Huang, H.-R.; She, Z.-G.; Cai, J.-W.; Lan, L.; Zhang, J.-Y.; Fu, L.-W.; Vrijmoed, L.L.P.; Lin, Y.-C. Structural and biological properties of vermistatin and two new vermistatin derivatives isolated from the marine-mangrove endophytic fungus Guignardia sp. No. 4382. Helv. Chim. Acta 2007, 90, 1925–1931. [Google Scholar] [CrossRef]
- Sassa, T.; Nukina, M.; Suzuki, Y. Deoxyfunicone, a new γ-pyrone metabolite from a resorcylide-producing fungus (Penicillium sp.). Agric. Biol. Chem. 1991, 55, 2415–2416. [Google Scholar] [CrossRef]
- Hoss, R.; Helbig, J.; Bochow, H. Function of host and fungal metabolites in resistance response of banana and plantain in the black Sigatoka disease pathosystem (Musa spp.—Mycosphaerella fijiensis). J. Phytopathol. 2000, 148, 387–394. [Google Scholar] [CrossRef]
- Geis, P.A.; Wheeler, M.H.; Szaniszlo, P.J. Pentaketide metabolites of melanin synthesis in the dematiaeeous fungus Wangiella dermatitidis. Arch. Microbiol. 1984, 137, 324–328. [Google Scholar] [CrossRef]
- Stipanovic, R.D.; Bell, A.A. Pentaketide metabolites of Verticillium dahliae. II. Accumulation of naphthol derivatives by the aberrant-melanin mutant BRM-2. Mycologia 1977, 69, 164–172. [Google Scholar] [CrossRef]
- Bell, A.A.; Stipanovic, R.D.; Puhalla, J.E. Pentaketide metabolites of Verticillium dahliae: Identification of (+)-scytalone as a natural precursor to melanin. Tetrahedron 1976, 32, 1353–1356. [Google Scholar] [CrossRef]
- Busogoro, J.P.; Etamé, J.J.; Lognay, G.; Messiaen, J.; van Cutsem, P.; Lepoivre, P. Analysis of the mechanisms of action of Mycosphaerella fijiensis toxins during the development of Black leaf streak disease. In Proceedings of the Banana Improvement: Cellular, Molecular Biology, and Induced Mutations, Leuven, Belgium, 24–28 September 2001; pp. 171–181. [Google Scholar]
- Babula, P.; Adam, V.; Havel, L.; Kizek, R. Noteworthy secondary metabolites naphthoquinones—Their occurrence, pharmacological properties and analysis. Curr. Pharm. Anal. 2009, 5, 47–68. [Google Scholar] [CrossRef]
- Howard, R.J.; Ferrari, M.A. Role of melanin in appressorium function. Exp. Mycol. 1989, 13, 403–418. [Google Scholar] [CrossRef]
- Steiner, U.; Oerke, E.-C. Localized melanization of appressoria is required for pathogenicity of Venturia inaequalis. Phytopathology 2007, 97, 1222–1230. [Google Scholar] [CrossRef] [PubMed]
- Brush, L.; Money, N.P. Invasive hyphal growth in Wangiella dermatitidis is induced by stab inoculation and shows dependence upon melanin biosynthesis. Fungal Genet. Biol. 1999, 28, 190–200. [Google Scholar] [CrossRef] [PubMed]
- Nosanchuk, J.D.; Casadevall, A. Impact of melanin on microbial virulence and clinical resistance to antimicrobial compounds. Antimicrob. Agents Chemother. 2006, 50, 3519–3528. [Google Scholar] [CrossRef]
- Wang, Y.; Casadevall, A. Susceptibility of melanized and nonmelanized Cryptococcus neoformans to nitrogen- and oxygen-derived oxidants. Infect. Immun. 1994, 62, 3004–3007. [Google Scholar] [CrossRef]
- Morel, J.-B.; Dangl, J.L. The hypersensitive response and the induction of cell death in plants. Cell Death Differ. 1997, 4, 671–683. [Google Scholar] [CrossRef]
- Freitas, M.; Lima, J.L.; Fernandes, E. Optical probes for detection and quantification of neutrophils’ oxidative burst. A review. Anal. Chim. Acta 2009, 649, 8–23. [Google Scholar] [CrossRef]
- Korytowski, W.; Pilas, B.; Sarna, T.; Kalyanaraman, B. Photoinduced generation of hydrogen peroxide and hydroxyl radicals in melanins. Photochem. Photobiol. 1987, 45, 185–190. [Google Scholar] [CrossRef]
- Chiarelli-Neto, O.; Pavani, C.; Ferreira, A.S.; Uchoa, A.F.; Severino, D.; Baptista, M.S. Generation and suppression of singlet oxygen in hair by photosensitization of melanin. Free Radic. Biol. Med. 2011, 51, 1195–1202. [Google Scholar] [CrossRef]
- Beltrán-García, M.J.; Prado, F.M.; Oliveira, M.S.; Ortiz-Mendoza, D.; Scalfo, A.C.; Pessoa, A., Jr.; Medeiros, M.H.G.; White, J.F.; Di Mascio, P. Singlet molecular oxygen generation by light-activated DHN-melanin of the fungal pathogen Mycosphaerella fijiensis in black Sigatoka disease of bananas. PLoS ONE 2014, 9, e91616. [Google Scholar] [CrossRef]
- Daub, M.E.; Ehrenshaft, M. The photoactivated Cercospora toxin cercosporin: Contributions to plant disease and fundamental biology. Annu. Rev. Phytopathol. 2000, 38, 461–490. [Google Scholar] [CrossRef] [PubMed]
- Manning, V.A.; Chu, A.L.; Steeves, J.E.; Wolpert, T.J.; Ciuffetti, L.M. A host-selective toxin of Pyrenophora tritici-repentis, Ptr ToxA, induces photosystem changes and reactive oxygen species accumulation in sensitive wheat. Mol. Plant-Microbe Interact. 2009, 22, 665–676. [Google Scholar] [CrossRef] [PubMed]
- Choquer, M.; Fournier, E.; Kunz, C.; Levis, C.; Pradier, J.-M.; Simon, A.; Viaud, M. Botrytis cinerea virulence factors: New insights into a necrotrophic and polyphageous pathogen. FEMS Microbiol. Lett. 2007, 277, 1–10. [Google Scholar] [CrossRef]
- Cruz-Cruz, C.A.; García-Sosa, K.; Escalante-Erosa, F.; Peña-Rodríguez, L.M. Production of hydrophilic phytotoxins by Mycosphaerella fijiensis. J. Gen. Plant Pathol. 2009, 75, 191–195. [Google Scholar] [CrossRef]
- Cruz-Cruz, C.A.; García-Sosa, K.; Escalante-Erosa, F.; Peña-Rodríguez, L.M. Physiological effects of the hydrophilic phytotoxins produced by Mycosphaerella fijiensis, the causal agent of black sigatoka in banana plants. J. Gen. Plant Pathol. 2011, 77, 93–100. [Google Scholar] [CrossRef]
- Noar, R.D.; Daub, M.E. Bioinformatics prediction of polyketide synthase gene clusters from Mycosphaerella fijiensis. PLoS ONE 2016, 11, e0158471. [Google Scholar] [CrossRef]
- Khaldi, N.; Seifuddin, F.T.; Turner, G.; Haft, D.; Nierman, W.C.; Wolfe, K.H.; Fedorova, N.D. SMURF: Genomic mapping of fungal secondary metabolite clusters. Fungal Genet. Biol. 2010, 47, 736–741. [Google Scholar] [CrossRef]
- Noar, R.D.; Thomas, E.; Xie, D.-Y.; Carter, M.E.; Ma, D.; Daub, M.E. A polyketide synthase gene cluster associated with the sexual reproductive cycle of the banana pathogen, Pseudocercospora fijiensis. PLoS ONE 2019, 14, e0220319. [Google Scholar] [CrossRef]
- Thomas, E.; Noar, R.D.; Daub, M.E. A polyketide synthase gene cluster required for pathogenicity of Pseudocercospora fijiensis on banana. PLoS ONE 2021, 16, e0258981. [Google Scholar] [CrossRef]
- Noar, R.D.; Thomas, E.; Daub, M.E. A novel polyketide synthase gene cluster in the plant pathogenic fungus Pseudocercospora fijiensis. PLoS ONE 2019, 14, e0212229. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.-Y.; Li, C.-Y.; Yao, N. Fumonisin B1: A tool for exploring the multiple functions of sphingolipids in plants. Front. Plant Sci. 2020, 11, 600458. [Google Scholar] [CrossRef] [PubMed]
- Schindler, D.; Nowrousian, M. The polyketide synthase gene pks4 is essential for sexual development and regulates fruiting body morphology in Sordaria macrospora. Fungal Genet. Biol. 2014, 68, 48–59. [Google Scholar] [CrossRef] [PubMed]
- Nowrousian, M. A novel polyketide biosynthesis gene cluster is involved in fruiting body morphogenesis in the filamentous fungi Sordaria macrospora and Neurospora crassa. Curr. Genet. 2009, 55, 185–198. [Google Scholar] [CrossRef] [PubMed]
- Krasnoff, S.B.; Gibson, D.M.; Belofsky, G.N.; Gloer, K.B.; Gloer, J.B. New destruxins from the entomopathogenic fungus Aschersonia sp. J. Nat. Prod. 1996, 59, 485–489. [Google Scholar] [CrossRef]
- Lira, S.P.; Vita-Marques, A.M.; Seleghim, M.H.; Bugni, T.S.; LaBarbera, D.V.; Sette, L.D.; Sponchiado, S.R.; Ireland, C.M.; Berlinck, R.G. New destruxins from the marine-derived fungus Beauveria felina. J. Antibiot. 2006, 59, 553–563. [Google Scholar] [CrossRef]
- Wang, B.; Kang, Q.; Lu, Y.; Bai, L.; Wang, C. Unveiling the biosynthetic puzzle of destruxins in Metarhizium species. Proc. Natl. Acad. Sci. USA 2012, 109, 1287–1292. [Google Scholar] [CrossRef]
- Buchwaldt, L.; Jensen, J.S. HPLC purification of destruxins produced by Alternaria brassicae in culture and leaves of Brassica napus. Phytochemistry 1991, 30, 2311–2316. [Google Scholar] [CrossRef]
- Buchwaldt, L.; Green, H. Phytotoxicity of destruxin B and its possible role in the pathogenesis of Alternaria brassicae. Plant Pathol. 1992, 41, 55–63. [Google Scholar] [CrossRef]
- Venkatasubbaiah, P.; Tisserat, N.A.; Chilton, W.S. Metabolites of Ophiosphaerella herpotricha, a cause of spring dead spot of bermudagrass. Mycopathologia 1994, 128, 155–159. [Google Scholar] [CrossRef]
- Parada, R.Y.; Oka, K.; Yamagishi, D.; Kodama, M.; Otani, H. Destruxin B produced by Alternaria brassicae does not induce accessibility of host plants to fungal invasion. Physiol. Mol. Plant Pathol. 2007, 71, 48–54. [Google Scholar] [CrossRef]
- de Boer, A.H.; de Vries-van Leeuwen, I.J. Fusicoccanes: Diterpenes with surprising biological functions. Trends Plant Sci. 2012, 17, 360–368. [Google Scholar] [CrossRef] [PubMed]
- Stergiopoulos, I.; de Wit, P.J.G.M. Fungal effector proteins. Annu. Rev. Phytopathol. 2009, 47, 233–263. [Google Scholar] [CrossRef]
- Tan, K.-C.; Oliver, R.P.; Solomon, P.S.; Moffat, C.S. Proteinaceous necrotrophic effectors in fungal virulence. Funct. Plant Biol. 2010, 37, 907–912. [Google Scholar] [CrossRef]
- van den Burg, H.A.; Westerink, N.; Francoijs, K.J.; Roth, R.; Woestenenk, E.; Boeren, S.; de Wit, P.J.; Joosten, M.H.; Vervoort, J. Natural disulfide bond-disrupted mutants of AVR4 of the tomato pathogen Cladosporium fulvum are sensitive to proteolysis, circumvent Cf-4-mediated resistance, but retain their chitin binding ability. J. Biol. Chem. 2003, 278, 27340–27346. [Google Scholar] [CrossRef]
- Giraldo, M.C.; Valent, B. Filamentous plant pathogen effectors in action. Nat. Rev. Microbiol. 2013, 11, 800–814. [Google Scholar] [CrossRef]
- Chang, T.-C.; Salvucci, A.; Crous, P.W.; Stergiopoulos, I. Comparative genomics of the Sigatoka disease complex on banana suggests a link between parallel evolutionary changes in Pseudocercospora fijiensis and Pseudocercospora eumusae and increased virulence on the banana host. PLoS Genet. 2016, 12, e1005904. [Google Scholar] [CrossRef]
- Carreón-Anguiano, K.G.; Islas-Flores, I.; Vega-Arreguín, J.; Sáenz-Carbonell, L.; Canto-Canché, B. EffHunter: A tool for prediction of effector protein candidates in fungal proteomic databases. Biomolecules 2020, 10, 712. [Google Scholar] [CrossRef]
- Stergiopoulos, I.; van den Burg, H.A.; Ökmen, B.; Beenen, H.G.; van Liere, S.; Kema, G.H.J.; de Wit, P.J.G.M. Tomato Cf resistance proteins mediate recognition of cognate homologous effectors from fungi pathogenic on dicots and monocots. Proc. Natl. Acad. Sci. USA 2010, 107, 7610–7615. [Google Scholar] [CrossRef]
- van den Burg, H.A.; Harrison, S.J.; Joosten, M.H.; Vervoort, J.; de Wit, P.J. Cladosporium fulvum Avr4 protects fungal cell walls against hydrolysis by plant chitinases accumulating during infection. Mol. Plant-Microbe Interact. 2006, 19, 1420–1430. [Google Scholar] [CrossRef]
- Sánchez-Vallet, A.; Saleem-Batcha, R.; Kombrink, A.; Hansen, G.; Valkenburg, D.-J.; Thomma, B.P.H.J.; Mesters, J.R. Fungal effector Ecp6 outcompetes host immune receptor for chitin binding through intrachain LysM dimerization. eLife 2013, 2, e00790. [Google Scholar] [CrossRef] [PubMed]
- Bolton, M.D.; van Esse, H.P.; Vossen, J.H.; de Jonge, R.; Stergiopoulos, I.; Stulemeijer, I.J.; van den Berg, G.C.; Borrás-Hidalgo, O.; Dekker, H.L.; de Koster, C.G.; et al. The novel Cladosporium fulvum lysin motif effector Ecp6 is a virulence factor with orthologues in other fungal species. Mol. Microbiol. 2008, 69, 119–136. [Google Scholar] [CrossRef] [PubMed]
- Passos, M.A.N.; de Cruz, V.O.; Emediato, F.L.; de Teixeira, C.C.; Azevedo, V.C.R.; Brasileiro, A.C.M.; Amorim, E.P.; Ferreira, C.F.; Martins, N.F.; Togawa, R.C.; et al. Analysis of the leaf transcriptome of Musa acuminata during interaction with Mycosphaerella musicola: Gene assembly, annotation and marker development. BMC Genom. 2013, 14, 78. [Google Scholar] [CrossRef] [PubMed]
- Laugé, R.; Joosten, M.H.; Van den Ackerveken, G.F.; Van den Broek, H.W.; De Wit, P.J. The in planta-produced extracellular proteins ECP1 and ECP2 of Cladosporium fulvum are virulence factors. Mol. Plant-Microbe Interact. 1997, 10, 725–734. [Google Scholar] [CrossRef]
- Kulkarni, R.D.; Kelkar, H.S.; Dean, R.A. An eight-cysteine-containing CFEM domain unique to a group of fungal membrane proteins. Trends Biochem. Sci. 2003, 28, 118–121. [Google Scholar] [CrossRef]
- Zhang, Z.-N.; Wu, Q.-Y.; Zhang, G.-Z.; Zhu, Y.-Y.; Murphy, R.W.; Liu, Z.; Zou, C.-G. Systematic analyses reveal uniqueness and origin of the CFEM domain in fungi. Sci. Rep. 2015, 5, 13032. [Google Scholar] [CrossRef]
- Ohtaki, S.; Maeda, H.; Takahashi, T.; Yamagata, Y.; Hasegawa, F.; Gomi, K.; Nakajima, T.; Abe, K. Novel hydrophobic surface binding protein, HsbA, produced by Aspergillus oryzae. Appl. Env. Microbiol. 2006, 72, 2407–2413. [Google Scholar] [CrossRef]
- Nagano, N.; Umemura, M.; Izumikawa, M.; Kawano, J.; Ishii, T.; Kikuchi, M.; Tomii, K.; Kumagai, T.; Yoshimi, A.; Machida, M.; et al. Class of cyclic ribosomal peptide synthetic genes in filamentous fungi. Fungal Genet. Biol. 2016, 86, 58–70. [Google Scholar] [CrossRef]
- Ye, Y.; Ozaki, T.; Umemura, M.; Liu, C.; Minami, A.; Oikawa, H. Heterologous production of asperipin-2a: Proposal for sequential oxidative macrocyclization by a fungi-specific DUF3328 oxidase. Org. Biomol. Chem. 2019, 17, 39–43. [Google Scholar] [CrossRef]
- Kessler, S.C.; Zhang, X.; McDonald, M.C.; Gilchrist, C.L.M.; Lin, Z.; Rightmyer, A.; Solomon, P.S.; Turgeon, B.G.; Chooi, Y.-H. Victorin, the host-selective cyclic peptide toxin from the oat pathogen Cochliobolus victoriae, is ribosomally encoded. Proc. Natl. Acad. Sci. USA 2020, 117, 24243–24250. [Google Scholar] [CrossRef]
- Escobar-Tovar, L.; Guzmán-Quesada, M.; Sandoval-Fernández, J.A.; Gómez-Lim, M.A. Comparative analysis of the in vitro and in planta secretomes from Mycosphaerella fijiensis isolates. Fungal Biol. 2015, 119, 447–470. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Fresneda, R.; Martínez-Esparza, M.; Maicas, S.; Argüelles, J.-C.; Valentín, E. In Candida parapsilosis the ATC1 gene encodes for an acid trehalase involved in trehalose hydrolysis, stress resistance and virulence. PLoS ONE 2014, 9, e99113. [Google Scholar] [CrossRef] [PubMed]
- Bowman, S.M.; Free, S.J. The structure and synthesis of the fungal cell wall. BioEssays 2006, 28, 799–808. [Google Scholar] [CrossRef] [PubMed]
- Kantún-Moreno, N.; Vázquez-Euán, R.; Tzec-Simá, M.; Peraza-Echeverría, L.; Grijalva-Arango, R.; Rodríguez-García, C.; James, A.C.; Ramírez-Prado, J.; Islas-Flores, I.; Canto-Canché, B. Genome-wide in silico identification of GPI proteins in Mycosphaerella fijiensis and transcriptional analysis of two GPI-anchored β-1,3-glucanosyltransferases. Mycologia 2013, 105, 285–296. [Google Scholar] [CrossRef]
- Kitagaki, H.; Wu, H.; Shimoi, H.; Ito, K. Two homologous genes, DCW1 (YKL046c) and DFG5, are essential for cell growth and encode glycosylphosphatidylinositol (GPI)-anchored membrane proteins required for cell wall biogenesis in Saccharomyces cerevisiae. Mol. Microbiol. 2002, 46, 1011–1022. [Google Scholar] [CrossRef]
- Caracuel, Z.; Martínez-Rocha, A.L.; Di Pietro, A.; Madrid, M.P.; Roncero, M.I. Fusarium oxysporum gas1 encodes a putative beta-1,3-glucanosyltransferase required for virulence on tomato plants. Mol. Plant Microbe Interact. 2005, 18, 1140–1147. [Google Scholar] [CrossRef]
- Klis, F.M.; Sosinska, G.J.; de Groot, P.W.; Brul, S. Covalently linked cell wall proteins of Candida albicans and their role in fitness and virulence. FEMS Yeast Res. 2009, 9, 1013–1028. [Google Scholar] [CrossRef]
- Sagaram, U.S.; Shaw, B.D.; Shim, W.B. Fusarium verticillioides GAP1, a gene encoding a putative glycolipid-anchored surface protein, participates in conidiation and cell wall structure but not virulence. Microbiology 2007, 153, 2850–2861. [Google Scholar] [CrossRef]
- Sun, Y.; Yi, X.; Peng, M.; Zeng, H.; Wang, D.; Li, B.; Tong, Z.; Chang, L.; Jin, X.; Wang, X. Proteomics of Fusarium oxysporum race 1 and race 4 reveals enzymes involved in carbohydrate metabolism and ion transport that might play important roles in banana Fusarium wilt. PLoS ONE 2014, 9, e113818. [Google Scholar] [CrossRef]
- Burgos-Canul, Y.Y.; Canto-Canché, B.; Berezovski, M.V.; Mironov, G.; Loyola-Vargas, V.M.; Barba de Rosa, A.P.; Tzec-Simá, M.; Brito-Argáez, L.; Carrillo-Pech, M.; Grijalva-Arango, R.; et al. The cell wall proteome from two strains of Pseudocercospora fijiensis with differences in virulence. World J. Microbiol. Biotechnol. 2019, 35, 105. [Google Scholar] [CrossRef]
- Stergiopoulos, I.; Zwiers, L.-H.; De Waard, M.A. The ABC transporter MgAtr4 is a virulence factor of Mycosphaerella graminicola that affects colonization of substomatal cavities in wheat leaves. Mol. Plant-Microbe Interact. 2003, 16, 689–698. [Google Scholar] [CrossRef] [PubMed]
- Couoh-Uicab, Y.; Islas-Flores, I.; Kantún-Moreno, N.; Zwiers, L.H.; Tzec-Simá, M.; Peraza-Echeverría, S.; Brito-Argáez, L.; Peraza-Echeverría, L.; Grijalva-Arango, R.; James, A.; et al. Cloning, in silico structural characterization and expression analysis of MfAtr4, an ABC transporter from the banana pathogen Mycosphaerella fijiensis. Afr. J. Biotechnol. 2012, 11, 54–79. [Google Scholar] [CrossRef]
- Onyilo, F.; Tusiime, G.; Chen, L.-H.; Falk, B.; Stergiopoulos, I.; Tripathi, J.N.; Tushemereirwe, W.; Kubiriba, J.; Changa, C.; Tripathi, L. Agrobacterium tumefaciens-mediated transformation of Pseudocercospora fijiensis to determine the role of PfHog1 in osmotic stress regulation and virulence modulation. Front. Microbiol. 2017, 8, 830. [Google Scholar] [CrossRef] [PubMed]
- Onyilo, F.; Tusiime, G.; Tripathi, J.N.; Chen, L.-H.; Falk, B.; Stergiopoulos, I.; Tushemereirwe, W.; Kubiriba, J.; Tripathi, L. Silencing of the mitogen-activated protein kinases (MAPK) Fus3 and Slt2 in Pseudocercospora fijiensis reduces growth and virulence on host plants. Front. Plant Sci. 2018, 9, 291. [Google Scholar] [CrossRef]
- Xu, J.-R. MAP kinases in fungal pathogens. Fungal Genet. Biol. 2000, 31, 137–152. [Google Scholar] [CrossRef]
- Kojima, K.; Takano, Y.; Yoshimi, A.; Tanaka, C.; Kikuchi, T.; Okuno, T. Fungicide activity through activation of a fungal signaling pathway. Mol. Microbiol. 2004, 53, 1785–1796. [Google Scholar] [CrossRef]
- Rouard, M.; Droc, G.; Martin, G.; Sardos, J.; Hueber, Y.; Guignon, V.; Cenci, A.; Geigle, B.; Hibbins, M.S.; Yahiaoui, N.; et al. Three new genome assemblies support a rapid radiation in Musa acuminata (wild banana). Genome Biol. Evol. 2018, 10, 3129–3140. [Google Scholar] [CrossRef]
- Davey, M.W.; Gudimella, R.; Harikrishna, J.A.; Sin, L.W.; Khalid, N.; Keulemans, J. A draft Musa balbisiana genome sequence for molecular genetics in polyploid, inter- and intra-specific Musa hybrids. BMC Genom. 2013, 14, 683. [Google Scholar] [CrossRef]
- Wang, Z.; Miao, H.; Liu, J.; Xu, B.; Yao, X.; Xu, C.; Zhao, S.; Fang, X.; Jia, C.; Wang, J.; et al. Musa balbisiana genome reveals subgenome evolution and functional divergence. Nat. Plants 2019, 5, 810–821. [Google Scholar] [CrossRef]
- Wu, W.; Yang, Y.-L.; He, W.-M.; Rouard, M.; Li, W.-M.; Xu, M.; Roux, N.; Ge, X.-J. Whole genome sequencing of a banana wild relative Musa itinerans provides insights into lineage-specific diversification of the Musa genus. Sci. Rep. 2016, 6, 31586. [Google Scholar] [CrossRef]
- Belser, C.; Istace, B.; Denis, E.; Dubarry, M.; Baurens, F.-C.; Falentin, C.; Genete, M.; Berrabah, W.; Chèvre, A.-M.; Delourme, R.; et al. Chromosome-scale assemblies of plant genomes using nanopore long reads and optical maps. Nat. Plants 2018, 4, 879–887. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, H.A.; Rodriguez-Arango, E.; Morales, J.G.; Kema, G.; Arango, R.E. Defense gene expression associated with biotrophic phase of Mycosphaerella fijiensis M. Morelet infection in banana. Plant Dis. 2016, 100, 1170–1175. [Google Scholar] [CrossRef] [PubMed]
- Alejandro, R.H.; Hidalgo, W.F.; Sanchez, J.D.; Menezes, R.C.; Schneider, B.; Arango, R.E.; Morales, J.G. Differential regulation of jasmonic acid pathways in resistant (Calcutta 4) and susceptible (Williams) banana genotypes during the interaction with Pseudocercospora fijiensis. Plant Pathol. 2020, 69, 872–882. [Google Scholar] [CrossRef]
- Timm, E.S.; Pardo, L.H.; Coello, R.P.; Navarrete, T.C.; Villegas, O.N.; Ordóñez, E.S. Identification of differentially-expressed genes in response to Mycosphaerella fijiensis in the resistant Musa accession ‘Calcutta-4’ using suppression subtractive hybridization. PLoS ONE 2016, 11, e0160083. [Google Scholar] [CrossRef][Green Version]
- Portal, O.; Izquierdo, Y.; De Vleesschauwer, D.; Sánchez-Rodríguez, A.; Mendoza-Rodríguez, M.; Acosta-Suárez, M.; Ocaña, B.; Jiménez, E.; Höfte, M. Analysis of expressed sequence tags derived from a compatible Mycosphaerella fijiensis–banana interaction. Plant Cell Rep. 2011, 30, 913–928. [Google Scholar] [CrossRef]
- Uma, S.; Backiyarani, S.; Saravanakumar, A.S.; Chandrasekar, A.; Thangavelu, R.; Saraswathi, M.S. Identification of Mycosphaerella eumusae responsive unique genes/transcripts from a resistant banana cultivar. Acta Hortic. 2016, 1114, 111–118. [Google Scholar] [CrossRef]
- Arinaitwe, G.; Remy, S.; Strosse, H.; Swennen, R.; Sági, L. Agrobacterium- and particle bombardment-mediated transformation of a wide range of banana cultivars. In Banana Improvement: Cellular, Molecular Biology, and Induced Mutations; Jain, S.M., Swennen, R., Eds.; Science Publishers, Inc.: Enfield, NH, USA; Plymouth, UK, 2004; pp. 351–357. [Google Scholar]
- Dong, T.; Bi, F.-C.; Huang, Y.-H.; He, W.-D.; Deng, G.-M.; Gao, H.-J.; Sheng, O.; Li, C.-Y.; Yang, Q.-S.; Yi, G.-J.; et al. Highly efficient biolistic transformation of embryogenic cell suspensions of banana via a liquid medium selection system. HortScience 2020, 55, 703–708. [Google Scholar] [CrossRef]
- Shivani; Tiwari, S. Enhanced Agrobacterium-mediated transformation efficiency of banana cultivar Grand Naine by reducing oxidative stress. Sci. Hortic. 2019, 246, 675–685. [Google Scholar] [CrossRef]
- Chakrabarti, A.; Ganapathi, T.R.; Mukherjee, P.K.; Bapat, V.A. MSI-99, a magainin analogue, imparts enhanced disease resistance in transgenic tobacco and banana. Planta 2003, 216, 587–596. [Google Scholar] [CrossRef]
- Remy, S. Genetic Transformation of Banana (Musa spp.) for Disease Resistance by Expression of Antimicrobial Proteins; Katholieke Universiteit: Leuven, Belgium, 2000. [Google Scholar]
- Remy, S.; Deconinck, I.; Swennen, R.; Sagi, L. Development of a leaf disc assay to assess fungal tolerance in banana. In Proceedings of the International Symposium on the Molecular and Cellular Biology of Banana, Ithaca, NY, USA, 22–25 March 1999. [Google Scholar]
- Remy, S.; Kovács, G.; Swennen, R.; Panis, B. Genetically modified bananas: Past, present and future. Acta Hortic. 2013, 974, 71–80. [Google Scholar] [CrossRef]
- Kovács, G.; Sági, L.; Jacon, G.; Arinaitwe, G.; Busogoro, J.P.; Thiry, E.; Strosse, H.; Swennen, R.; Remy, S. Expression of a rice chitinase gene in transgenic banana (‘Gros Michel’, AAA genome group) confers resistance to black leaf streak disease. Transgenic Res. 2013, 22, 117–130. [Google Scholar] [CrossRef] [PubMed]
- Keller, H.; Pamboukdjian, N.; Ponchet, M.; Poupet, A.; Delon, R.; Verrier, J.-L.; Roby, D.; Ricci, P. Pathogen-induced elicitin production in transgenic tobacco generates a hypersensitive response and nonspecific disease resistance. Plant Cell 1999, 11, 223. [Google Scholar] [CrossRef] [PubMed]
- Nowara, D.; Gay, A.; Lacomme, C.; Shaw, J.; Ridout, C.; Douchkov, D.; Hensel, G.; Kumlehn, J.; Schweizer, P. HIGS: Host-induced gene silencing in the obligate biotrophic fungal pathogen Blumeria graminis. Plant Cell 2010, 22, 3130. [Google Scholar] [CrossRef] [PubMed]
- Yin, C.; Jurgenson, J.E.; Hulbert, S.H. Development of a host-induced RNAi system in the wheat stripe rust fungus Puccinia striiformis f. sp. tritici. Mol. Plant Microbe Interact. 2011, 24, 554–561. [Google Scholar] [CrossRef]
- Tinoco, M.L.P.; Dias, B.B.A.; Dall’Astta, R.C.; Pamphile, J.A.; Aragão, F.J.L. In vivo trans-specific gene silencing in fungal cells by in planta expression of a double-stranded RNA. BMC Biol. 2010, 8, 27. [Google Scholar] [CrossRef] [PubMed]
- Andrade, C.M.; Tinoco, M.L.P.; Rieth, A.F.; Maia, F.C.O.; Aragão, F.J.L. Host-induced gene silencing in the necrotrophic fungal pathogen Sclerotinia sclerotiorum. Plant Pathol. 2016, 65, 626–632. [Google Scholar] [CrossRef]
- Thomas, E.; Herrero, S.; Eng, H.; Gomaa, N.; Gillikin, J.; Noar, R.; Beseli, A.; Daub, M.E. Engineering Cercospora disease resistance via expression of Cercospora nicotianae cercosporin-resistance genes and silencing of cercosporin production in tobacco. PLoS ONE 2020, 15, e0230362. [Google Scholar] [CrossRef]
- Laurie, J.D.; Linning, R.; Bakkeren, G. Hallmarks of RNA silencing are found in the smut fungus Ustilago hordei but not in its close relative Ustilago maydis. Curr. Genet. 2008, 53, 49–58. [Google Scholar] [CrossRef]
- Drinnenberg, I.A.; Weinberg, D.E.; Xie, K.T.; Mower, J.P.; Wolfe, K.H.; Fink, G.R.; Bartel, D.P. RNAi in budding yeast. Science 2009, 326, 544–550. [Google Scholar] [CrossRef]
- D’Souza, C.A.; Kronstad, J.W.; Taylor, G.; Warren, R.; Yuen, M.; Hu, G.; Jung, W.H.; Sham, A.; Kidd, S.E.; Tangen, K.; et al. Genome variation in Cryptococcus gattii, an emerging pathogen of immunocompetent hosts. mBio 2011, 2, e00342-10. [Google Scholar] [CrossRef]
- Mumbanza, F.M.; Kiggundu, A.; Tusiime, G.; Tushemereirwe, W.K.; Niblett, C.; Bailey, A. In vitro antifungal activity of synthetic dsRNA molecules against two pathogens of banana, Fusarium oxysporum f. sp. cubense and Mycosphaerella fijiensis. Pest Manag. Sci. 2013, 69, 1155–1162. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, T.; Islamovic, E.; Klos, K.; Schwartz, P.; Gillespie, J.; Hunter, S.; Bregitzer, P. Silencing efficiency of dsRNA fragments targeting Fusarium graminearum TRI6 and patterns of small interfering RNA associated with reduced virulence and mycotoxin production. PLoS ONE 2018, 13, e0202798. [Google Scholar] [CrossRef] [PubMed]
- McDougall, P. The Cost and Time Involved in the Discovery, Development and Authorisation of a New Plant Biotechnology Derived Trait; Crop Life International: Midlothian, UK, 2011; pp. 1–24. [Google Scholar]
- Lucht, J.M. Public acceptance of plant biotechnology and GM crops. Viruses 2015, 7, 4254–4281. [Google Scholar] [CrossRef] [PubMed]
- Siegrist, M.; Hartmann, C. Consumer acceptance of novel food technologies. Nat. Food 2020, 1, 343–350. [Google Scholar] [CrossRef]
- San Miguel, K.; Scott, J.G. The next generation of insecticides: dsRNA is stable as a foliar-applied insecticide. Pest Manag. Sci. 2016, 72, 801–809. [Google Scholar] [CrossRef] [PubMed]
- Vetukuri, R.R.; Dubey, M.; Kalyandurg, P.B.; Carlsson, A.S.; Whisson, S.C.; Ortiz, R. Spray-induced gene silencing: An innovative strategy for plant trait improvement and disease control. Crop Breed. Appl. Biotechnol. 2021, 21, e387921S387911. [Google Scholar] [CrossRef]
- Robinson, K.E.; Worrall, E.A.; Mitter, N. Double stranded RNA expression and its topical application for non-transgenic resistance to plant viruses. J. Plant Biochem. Biotechnol. 2014, 23, 231–237. [Google Scholar] [CrossRef]
- Wu, S.; Zhu, H.; Liu, J.; Yang, Q.; Shao, X.; Bi, F.; Hu, C.; Huo, H.; Chen, K.; Yi, G. Establishment of a PEG-mediated protoplast transformation system based on DNA and CRISPR/Cas9 ribonucleoprotein complexes for banana. BMC Plant Biol. 2020, 20, 425. [Google Scholar] [CrossRef]
- Naim, F.; Dugdale, B.; Kleidon, J.; Brinin, A.; Shand, K.; Waterhouse, P.; Dale, J. Gene editing the phytoene desaturase alleles of Cavendish banana using CRISPR/Cas9. Transgenic Res. 2018, 27, 451–460. [Google Scholar] [CrossRef]
- Tripathi, L.; Ntui, V.O.; Tripathi, J.N. CRISPR/Cas9-based genome editing of banana for disease resistance. Curr. Opin. Plant Biol. 2020, 56, 118–126. [Google Scholar] [CrossRef]
- McDonald, B.A.; Linde, C. Pathogen population genetics, evolutionary potential, and durable resistance. Annu. Rev. Phytopathol. 2002, 40, 349–379. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Noar, R.D.; Thomas, E.; Daub, M.E. Genetic Characteristics and Metabolic Interactions between Pseudocercospora fijiensis and Banana: Progress toward Controlling Black Sigatoka. Plants 2022, 11, 948. https://doi.org/10.3390/plants11070948
Noar RD, Thomas E, Daub ME. Genetic Characteristics and Metabolic Interactions between Pseudocercospora fijiensis and Banana: Progress toward Controlling Black Sigatoka. Plants. 2022; 11(7):948. https://doi.org/10.3390/plants11070948
Chicago/Turabian StyleNoar, Roslyn D., Elizabeth Thomas, and Margaret E. Daub. 2022. "Genetic Characteristics and Metabolic Interactions between Pseudocercospora fijiensis and Banana: Progress toward Controlling Black Sigatoka" Plants 11, no. 7: 948. https://doi.org/10.3390/plants11070948
APA StyleNoar, R. D., Thomas, E., & Daub, M. E. (2022). Genetic Characteristics and Metabolic Interactions between Pseudocercospora fijiensis and Banana: Progress toward Controlling Black Sigatoka. Plants, 11(7), 948. https://doi.org/10.3390/plants11070948






