Stability, the Last Frontier: Forage Yield Dynamics of Peas under Two Cultivation Systems
Abstract
:1. Introduction
2. Results
2.1. ANOVA and Descriptive Statistics on the Stability Index
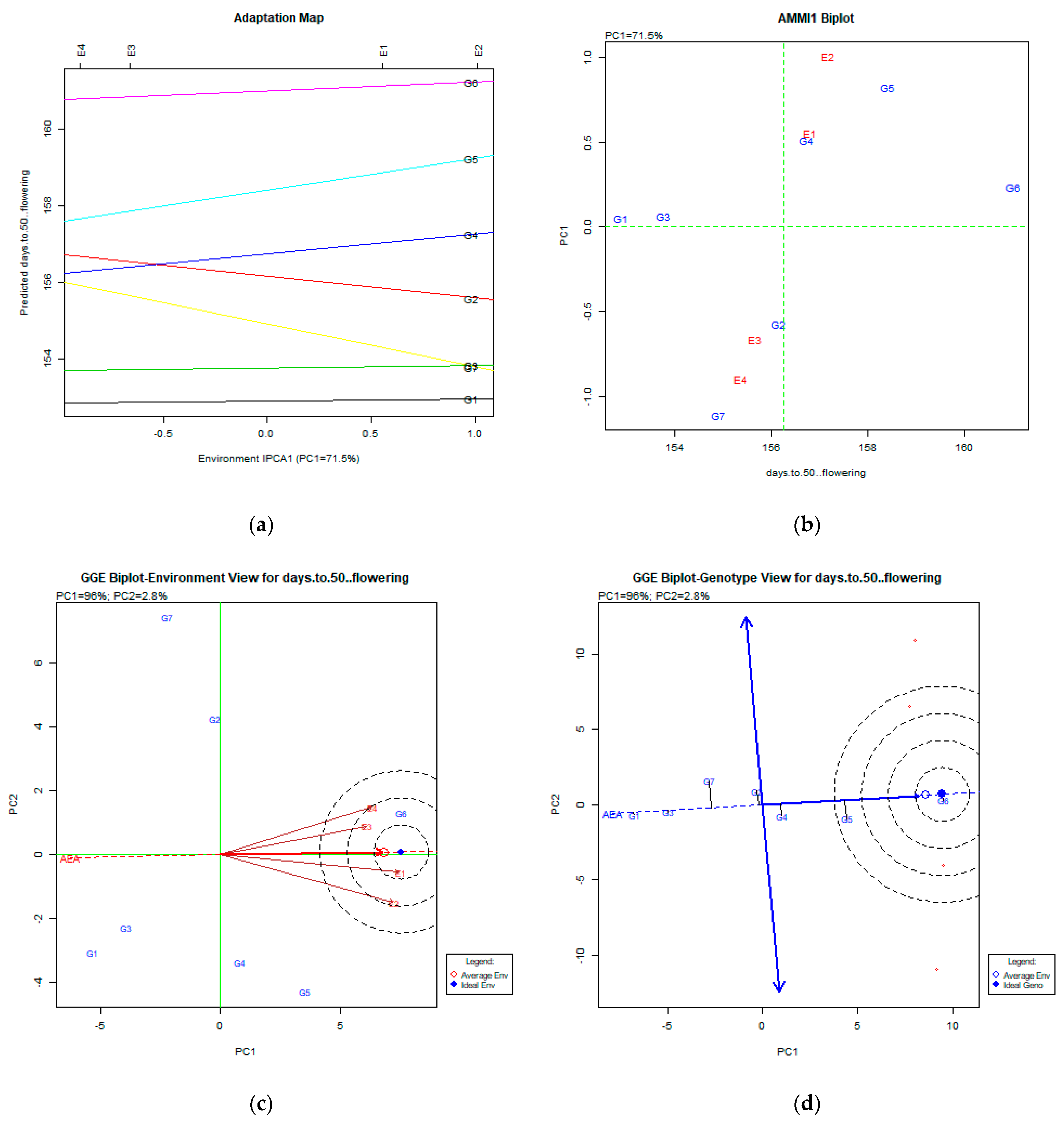
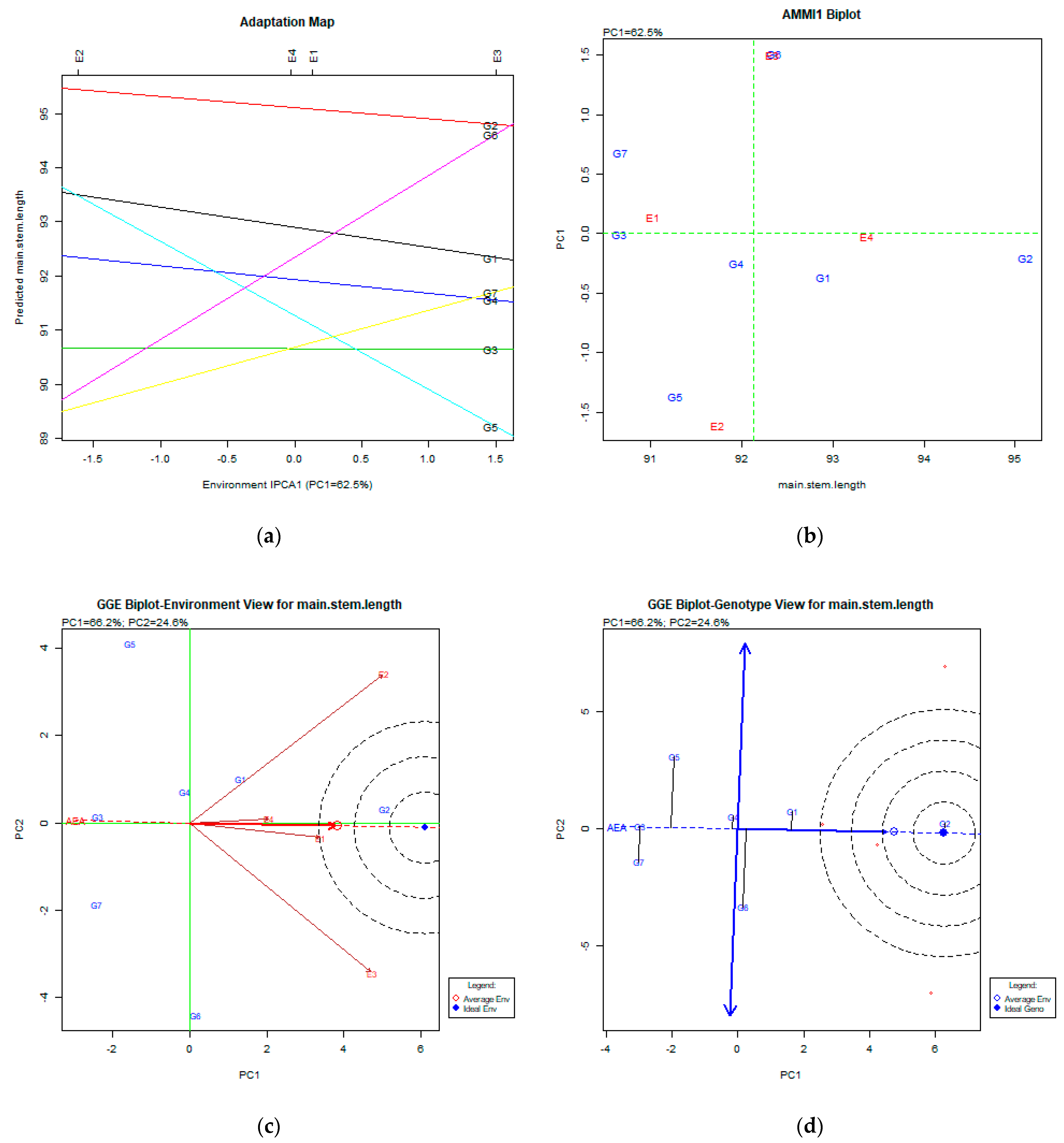
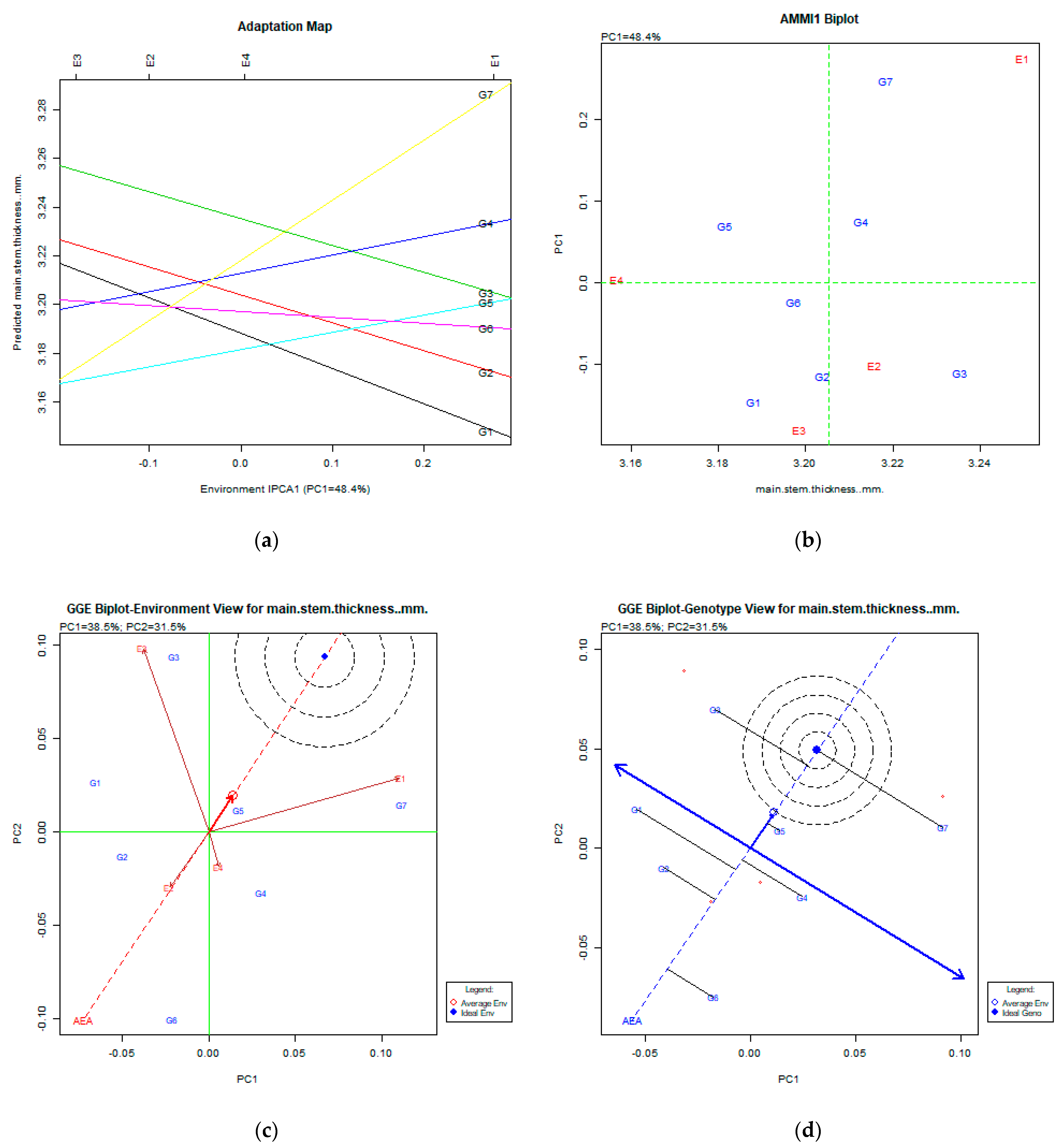
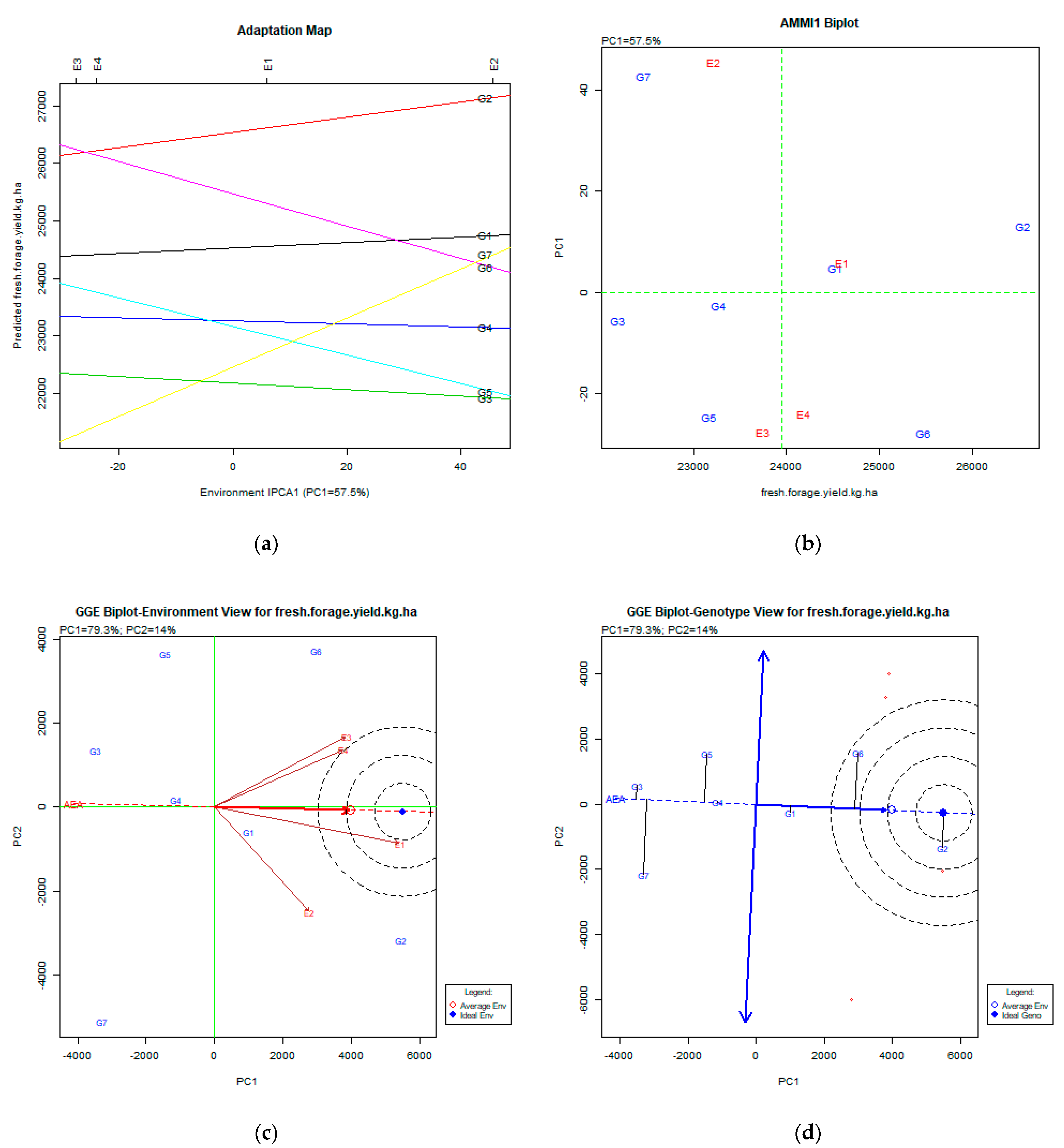
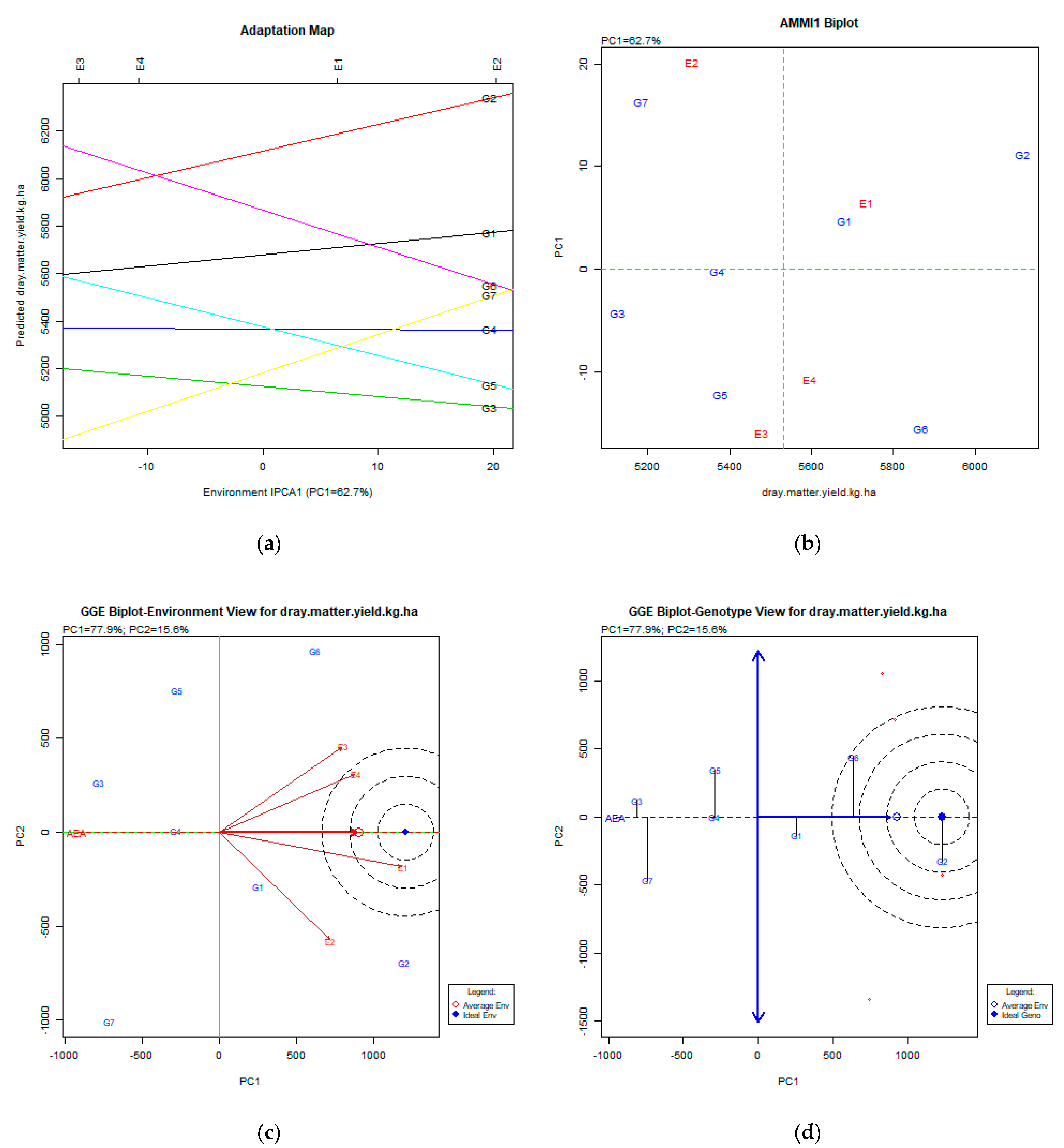
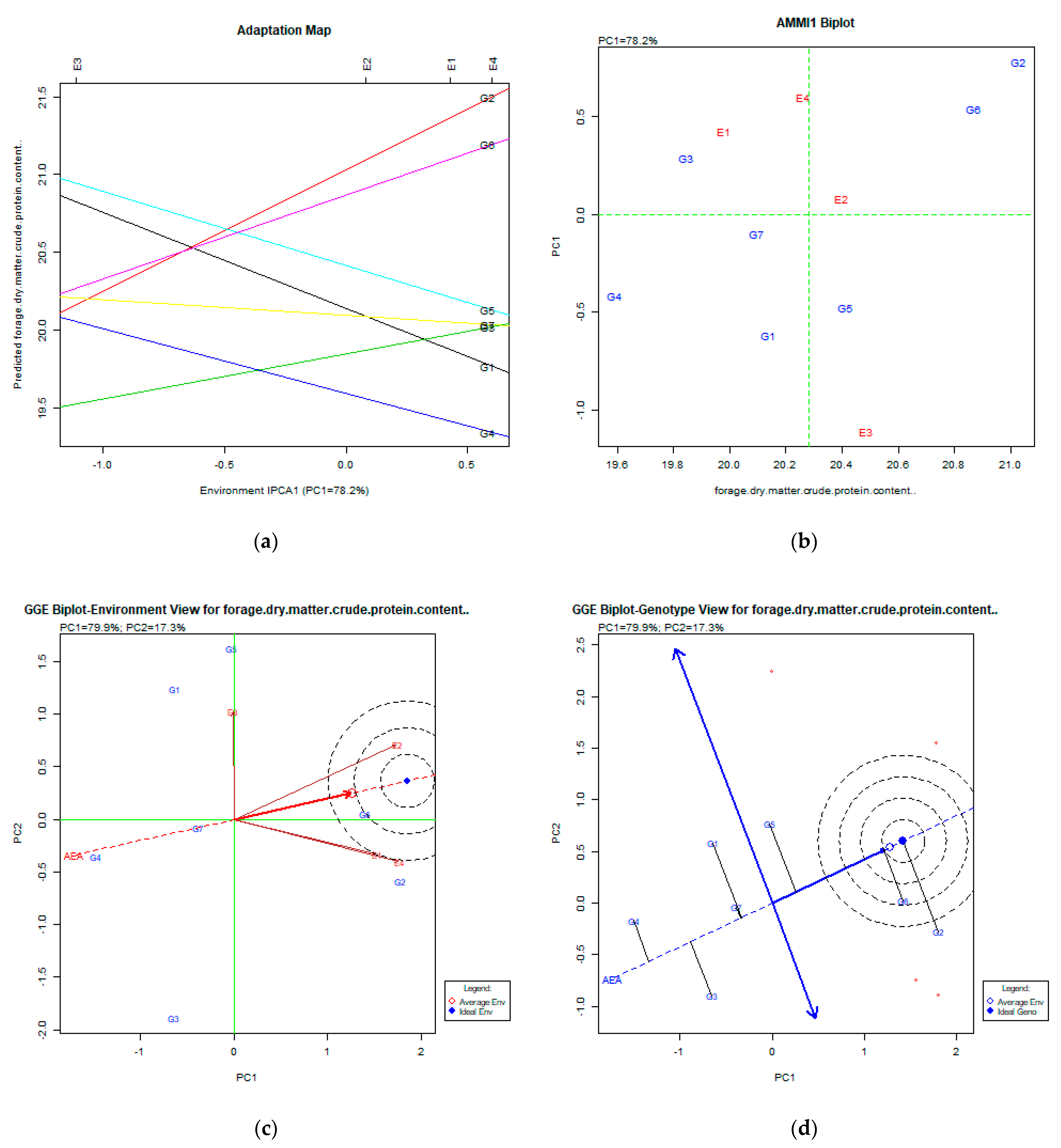
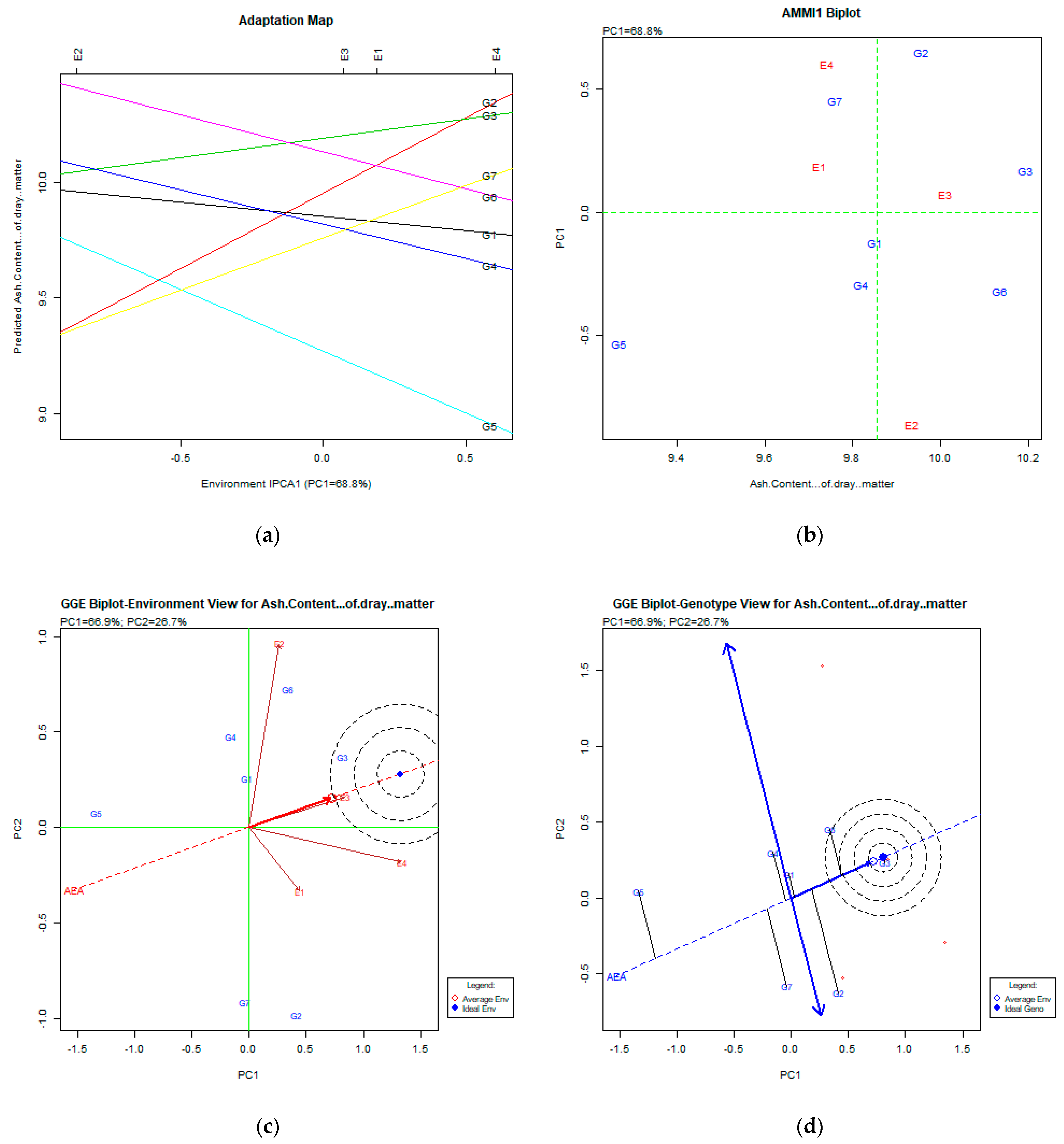
2.2. The AMMI Tool for Multi-Environment Evaluations
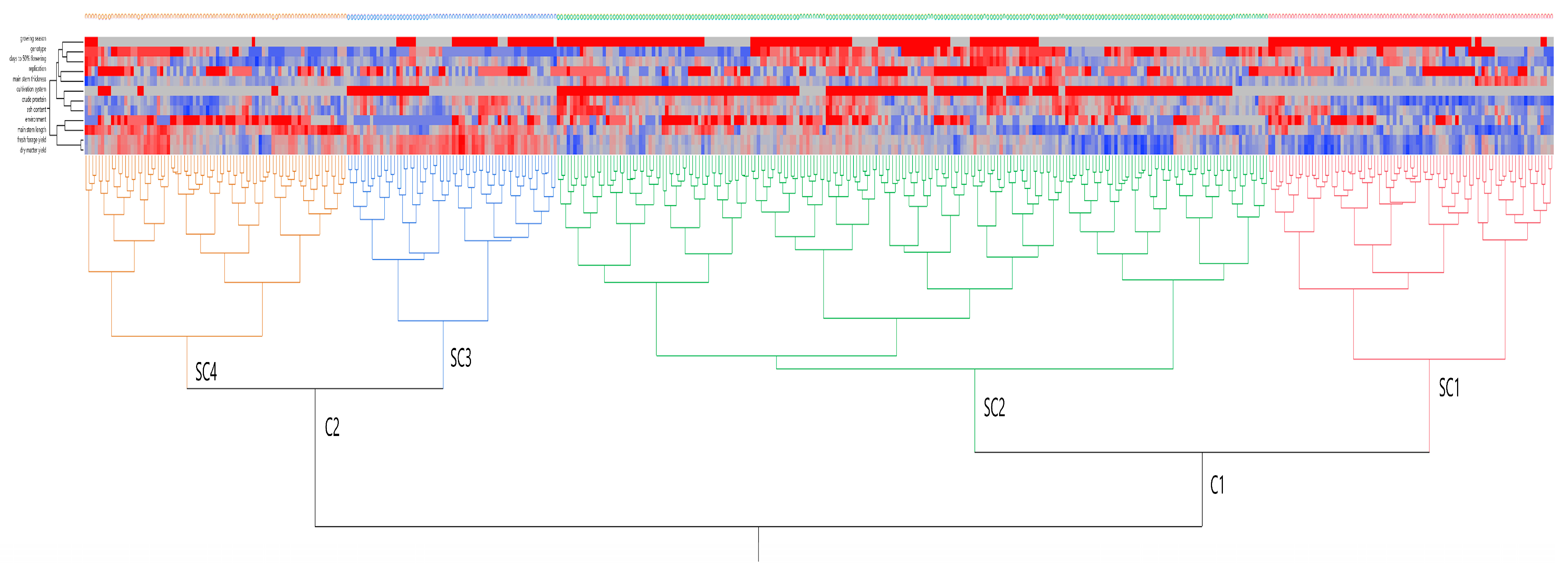
2.3. Exploratory Data Analysis of Peas
2.4. Genotypic and Phenotypic Coefficients of Variation and Heritability
2.5. Correlations between All Characteristics
3. Discussion
3.1. Days to 50% Flowering
3.2. Main Stem Length (cm)
3.3. Main Stem Thickness (mm)
3.4. Fresh Forage Yield (kg ha−1)
3.5. Dry Matter Yield (kg ha−1)
3.6. Forage Dry Matter Crude Protein Content %
3.7. Ash Content % of Dry Matter
3.8. Genotypic and Phenotypic Coefficients of Variation and Heritability
3.9. Correlations between Traits
3.10. Exploratory Data Analysis of Peas
4. Materials and Methods
4.1. Crop Establishment and Experimental Procedures
4.2. Climatic Conditions
4.3. Measurements
4.4. Data Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cacan, E.; Kokten, K.; Bakoglu, A.; Kaplan, M.; Bozkurt, A. Evaluation of some forage pea (Pisum arvense L.) lines and cultivars in terms of herbage yield and quality. Harran Tarım Ve Gıda Bilimleri Derg. 2019, 23, 254–262. [Google Scholar]
- Greveniotis, V.; Bouloumpasi, E.; Zotis, S.; Korkovelos, A.; Ipsilandis, C.G. Yield components stability assessment of peas in conventional and low-input cultivation systems. Agriculture 2021, 11, 805. [Google Scholar] [CrossRef]
- Elzebroek, T.; Wind, K. Guide to Cultivated Plants; CAB International: Oxfordshire, UK, 2008; p. 496. [Google Scholar]
- Tan, M.; Koc, A.; Dumlu, G.Z. Morphological characteristics and seed yield of East Anatolian local forage pea (Pisum sativum ssp. arvense L.) ecotypes. Turk. J. Field Crop. 2012, 17, 24–30. [Google Scholar]
- Arif, U.; Ahmed, M.J.; Rabbani, M.A.; Arif, A.A. Assessment of genetic diversity in pea (Pisum sativum L.) landraces based on physic-chemical and nutritive quality using cluster and principal component analysis. Pak. J. Bot. 2020, 52, 575–580. [Google Scholar] [CrossRef]
- Vafias, B.; Goulas, C.; Lolas, G.; Ipsilandis, C.G. A triple stress effect on monogenotypic and multigenotypic maize populations. Asian J. Plant Sci. 2007, 6, 29–35. [Google Scholar] [CrossRef] [Green Version]
- Fasoulas, A.C. The Honeycomb Methodology of Plant Breeding; Department of Genetics and Plant Breeding, Aristotle University of Thessaloniki: Thessaloniki, Greece, 1988; p. 168. [Google Scholar]
- Fasoula, V.A. Prognostic Breeding: A new paradigm for crop improvement. Plant Breed. Rev. 2013, 37, 297–347. [Google Scholar]
- Yan, W.; Kang, M.S. GGE Biplot Analysis: A Graphical Tool for Breeders, Geneticists and Agronomists; CRC Press: Boca Raton, FL, USA, 2003. [Google Scholar]
- Karimizadeh, R.; Mohammadi, M.; Shefazadeh, M.K. A review on parametric stability analysis methods: Set up by Matlab program. Int. J. Agric. 2012, 2, 433–442. [Google Scholar]
- Kang, M.S.; Gauch, H.G. Genotype-by-Environment Interaction; CRC Press: Boca Raton, FL, USA, 1996. [Google Scholar]
- Lin, C.S.; Binns, M.R.; Lefkovitch, L.P. Stability analysis: Where do we stand? Crop Sci. 1986, 26, 894–900. [Google Scholar] [CrossRef] [Green Version]
- Jinks, J.; Pooni, H. The genetic basis of environmental sensitivity. In Proceedings of the 2nd International Conference on Quantitative Genetics, Raleigh, NC, USA, May–June 1987; Weir, B.S., Eisen, E.J., Goodman, M.M., Namkoong, G., Eds.; Sinauer: Sunderland, MA, USA, 1988; pp. 505–522. [Google Scholar]
- Acikgoz, E.; Ustun, A.; Gul, İ.; Anlarsal, A.E.; Tekeli, A.S.; Nizam, İ.; Avcioglu, R.; Geren, H.; Cakmakcı, S.; Aydinoglu, B.; et al. Genotype × environment interaction and stability analysis for dry matter and seed yield in field pea (Pisum sativum L.). Span. J. Agric. Res. 2009, 7, 96–106. [Google Scholar] [CrossRef] [Green Version]
- Ceyhan, E.; Kahraman, A.; Ates, M.K.; Karadas, S. Stability analysis on seed yield and its components in peas. Bulg. J. Agric. Sci. 2012, 18, 905–911. [Google Scholar]
- Bocianowski, J.; Ksiezak, J.; Nowosad, K. Genotype by environment interaction for seeds yield in pea (Pisum sativum L.) using additive main effects and multiplicative interaction model. Euphytica 2019, 215, 191. [Google Scholar] [CrossRef] [Green Version]
- Rana, C.; Sharma, A.; Sharma, K.C.; Mittal, P.; Sinha, B.N.; Sharma, V.K.; Chandel, A.; Thakur, H.; Kaila, V.; Sharma, P.; et al. Stability analysis of garden pea (Pisum sativum L.) genotypes under North Western Himalayas using joint regression analysis and GGE biplots. Genet. Resour. Crop. Evol. 2021, 68, 999–1010. [Google Scholar] [CrossRef]
- Popović, V.; Vučković, S.; Jovović, Z.; Rakaščan, N.; Kostić, M.; Ljubičić, N.; Mladenović-Glamočlija, M.; Ikanović, J. Genotype by year interaction effects on soybean morphoproductive traits and biogas production. Genetika 2020, 52, 1055–1073. [Google Scholar] [CrossRef]
- Lakić, Ž.; Stanković, S.; Pavlović, S.; Krnjajic, S.; Popović, V. Genetic variability in quantitative traits of field pea (Pisum sativum L.) genotypes. Czech J. Genet. Plant Breed. 2018, 54, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Amin, M.; Mohammad, T.; Khan, A.J.; Irfaq, M.; Ali, A.; Tahir, G.R. Yield stability of spring wheat (Triticum aestivum L.) in the North West Frontier Province, Pakistan. Songklanakarin J. Sci. Technol. 2005, 27, 1147–1150. [Google Scholar]
- Gauch, H.G. A simple protocol for AMMI analysis of yield trials. Crop Sci. 2013, 53, 1860–1869. [Google Scholar] [CrossRef]
- Ebdon, J.S.; Gauch, H.G. Direct validation of AMMI predictions in turfgrass trials. Crop Sci. 2011, 51, 862–869. [Google Scholar] [CrossRef]
- Greveniotis, V.; Bouloumpasi, E.; Zotis, S.; Korkovelos, A.; Ipsilandis, C.G. Estimations on Trait Stability of Maize Genotypes. Agriculture 2021, 11, 952. [Google Scholar] [CrossRef]
- Tsenov, N.; Atanasova, D.; Nankova, M.; Ivanova, A.; Tsenova, E.; Chamurliiski, P.; Raykov, G. Approaches for grading breeding evaluation of winter wheat varieties for grain yield. Sci. Work. Instiute Agric.-Karnobat 2014, 3, 21–35. [Google Scholar]
- Georgieva, N.; Kosev, V. Model of forage pea (Pisum sativum L.) cultivar in conditions of organic production. Bulg. J. Agric. Sci. 2020, 26, 91–95. [Google Scholar]
- Al-Aysh, F.; Kotmaa, H.; Al-Shareef, A.; Al-Serhan, M. Genotype-environment interact ion and stability analysis in garden pea (Pisum sativum L.) landraces. Agric. Forest. 2013, 59, 183–191. [Google Scholar]
- Sayar, S.M. Additive Main Efects and Multiplicative Interactions (AMMI) analysis for fresh forage yield in common vetch (Vicia Sativa L.) genotypes. Agric. For. 2017, 63, 119–127. [Google Scholar]
- Gauch, H.G.; Zobel, R.W. AMMI analysis of yield trials. In Genotype-by-Environment Interaction; Kang, M.S., Gauch, H.G., Eds.; CRC Press: Boca Raton, FL, USA, 1996; pp. 85–122. [Google Scholar]
- Islam, M.R.; Anisuzzaman, M.; Khatun, H.; Sharma, N.; Islam, M.Z.; Akter, A.; Biswas, P.S. AMMI analysis of yield performance and stability of rice genotypes across different haor areas. Eco-Friendly Agril. J. 2014, 7, 20–24. [Google Scholar]
- Gabriel, K.R. The biplot graphic display of matrices with application to principal component analysis. Biometrika 1971, 58, 453–467. [Google Scholar] [CrossRef]
- Yan, W.; Tinker, N.A. Biplot Analysis of Multi-Environment Trial Data: Principles and Applications. Can. J. Plant Sci. 2006, 86, 623–645. [Google Scholar] [CrossRef] [Green Version]
- Yan, W. Singular Value Partitioning for Biplot Analysis of Multi-environment Trial Data. Argon. J. 2002, 94, 990–996. [Google Scholar]
- Kaya, Y.; Akcura, M.; Taner, S. GGE Biplot Analysis of Multi Environment Yield Trials in Bread Wheat. Turk. J. Agric. For. 2006, 30, 325–337. [Google Scholar]
- Ilker, E.; AykutTonk, F.; Caylak, O.; Tosun, M.; Ozmen, I. Assessment of Genotype × Environment İnteractions for Grain Yield in Maize Hybrids Using AMMI and GGE Biplot Analyses. Turk. J. Field Crop. 2009, 14, 123–135. [Google Scholar]
- Ahmadi, J.; Vaezi, B.; Shaabani, A.; Khademi, K. Multi-environment Yield Trials of Grass Pea (Lathyrus sativus L.) in Iran Using AMMI and SREG GGE. J. AgrIC. Sci. Technol. 2012, 14, 1075–1085. [Google Scholar]
- Mortazavian, S.M.M.; Nikkhah, H.R.; Hassani, F.A.; Sharif-al-Hosseini, M.; Taheri, M.; Mahlooji, M. GGE Biplot and AMMI Analysis of Yield Performance of Barley Genotypes Across Different Environments in Iran. J. AgrIC. Sci. Technol. 2014, 16, 609–622. [Google Scholar]
- Macák, M.; Candráková, E.; Ðalovic, I.; Prasad, P.V.V.; Farooq, M.; Korczyk-Szabó, J.; Kovácik, P.; Šimanský, V. The Influence of Different Fertilization Strategies on the Grain Yield of Field Peas (Pisum sativum L.) under Conventional and Conservation Tillage. Agronomy 2020, 10, 1728. [Google Scholar] [CrossRef]
- Hanáčková, E.; Candráková, E. The influence of soil cultivation and fertilization on the yield and protein content in seeds of common pea (Pisum sativum L.). Agriculture 2014, 60, 105–114. [Google Scholar] [CrossRef]
- Greveniotis, V.; Sioki, E.; Ipsilandis, C.G. Estimations of fibre trait stability and type of inheritance in cotton. Czech J. Genet. Plant Breed. 2018, 54, 190–192. [Google Scholar] [CrossRef] [Green Version]
- Koundinya, A.V.V.; Ajeesh, B.R.; Hegde, V.; Sheela, M.N.; Mohan, C.; Asha, K.I. Genetic parameters, stability and selection of cassava genotypes between rainy and water stress conditions using AMMI, WAAS, BLUP and MTSI. Sci. Hortic. 2021, 281, 109949. [Google Scholar]
- Sayar, M.S.; Han, Y. Forage Yield Performance of Forage Pea (Pisum sativum spp. arvense L.) Genotypes and Assessments Using GGE Biplot Analysis. J. Agric. Sci. Technol. 2016, 18, 1621–1634. [Google Scholar]
- Johnson, H.W.; Robinson, H.E.; Comstock, R.E. Estimate of genetic and environmental variability in soybean. Agron. J. 1955, 47, 314–318. [Google Scholar] [CrossRef]
- Al-Ashkar, I.; Al-Suhaibani, N.; Abdella, K.; Sallam, M.; Alotaibi, M.; Seleiman, M.F. CombiningGenetic and MultidimensionalAnalyses to Identify InterpretiveTraits Related to Water ShortageTolerance as an Indirect Selection Toolfor Detecting Genotypes of DroughtTolerance in Wheat Breeding. Plants 2021, 10, 931. [Google Scholar] [CrossRef]
- Abebe, T.; Alamerew, S.; Tulu, L. Genetic variability, heritability and genetic advance for yield and its related traits in rainfed lowland rice (Oryza sativa L.) genotypes at Fogera and Pawe, Ethiopia. Adv. Crop Sci. Technol. 2017, 5, 272. [Google Scholar] [CrossRef] [Green Version]
- Greveniotis, V.; Bouloumpasi, E.; Zotis, S.; Korkovelos, A.; Ipsilandis, C.G. Assessment of interactions between yield components of common vetch cultivars in both conventional and low-input cultivation systems. Agriculture 2021, 11, 369. [Google Scholar] [CrossRef]
- Greveniotis, V.; Bouloumpasi, E.; Zotis, S.; Korkovelos, A.; Ipsilandis, C.G. A Stability Analysis Using AMMI and GGE Biplot Approach on Forage Yield Assessment of Common Vetch in Both Conventional and Low-Input Cultivation Systems. Agriculture 2021, 11, 567. [Google Scholar] [CrossRef]
- Georgieva, N.; Nikolova, I.; Kosev, V. Association study of yield and its components in pea (Pisum sativum L.). Int. J. Pharmacogn. 2015, 2, 536–542. [Google Scholar]
- Sanwal, S.K.; Singh, B.; Singh, V.; Mann, A. Multivariateanalysis and its implication in breeding of desired planttype in garden pea (Pisum sativum). Indian J. Agric. Sci. 2015, 85, 1298–1302. [Google Scholar]
- Kumar, A.V.R.; Sharma, R.R. Character association studies in garden pea. Indian J. Hortal. 2006, 63, 185–187. [Google Scholar]
- Singh, S.K.; Singh, V.P.; Srivastava, S.; Singh, A.K.; Chaubey, B.K.; Srivastava, R.K. Estimation of correlation coefficient among yield and attributing traits of field pea (Pisum sativum L.). Legume Res. 2018, 41, 20–26. [Google Scholar] [CrossRef] [Green Version]
- Kosev, V.; Mikić, A. Assessing relationships between seed yield components in spring-sown field pea (Pisum sativum L.) cultivars in Bulgaria by correlation and path analysis. Span. J. Agric. Res. 2012, 10, 1075–1080. [Google Scholar] [CrossRef] [Green Version]
- Yihunie, T.A.; Gesesse, C.A. GGE Biplot analysis of genotype by environment interaction in field pea (Pisum sativum L.) genotypes in North Western Ethiopia. J. Crop Sci. Biotechnol. 2018, 21, 67–74. [Google Scholar] [CrossRef]
- Uzun, A.; Bilgili, U.; Sincik, M.; Acikgoz, E. Yield and quality performances of forage type pea strains contrasting leaf types. Eur. J. Agric. 2005, 22, 85–94. [Google Scholar] [CrossRef]
- Greveniotis, V.A.; Giourieva, V.S.; Bouloumpasi, E.C.; Sioki, E.J.; Mitlianga, P.G. Morpho-physiological Characteristics and Molecular Markers of Maize Crosses Under Multi-location Evaluation. J. Agric. Sci. 2018, 10, 79–90. [Google Scholar] [CrossRef] [Green Version]
- Greveniotis, V.; Bouloumpasi, E.; Tsakiris, I.; Sioki, E.; Ipsilandis, C. Evaluation of Elite Open-Pollinated Maize Lines in Two Contrasting Environments. J. Agric. Sci. 2018, 10, 85–101. [Google Scholar] [CrossRef] [Green Version]
- Ganopoulos, I.; Moysiadis, T.; Xanthopoulou, A.; Ganopoulou, M.; Avramidou, E.; Aravanopoulos, F.A.; Kazantzis, K. Diversity of morpho-physiological traits in worldwide sweet cherry cultivars of GeneBank collection using multivariate analysis. Sci. Hortic. 2015, 197, 381–391. [Google Scholar] [CrossRef]
- AOAC. Official Methods of Analysis, 18th ed.; Association of Official Analytical Chemists: Gaithersburg, MD, USA, 2005. [Google Scholar]
- Fasoula, V.A. A novel equation paves the way for an everlasting revolution with cultivars characterized by high and stable crop yield and quality. In Proceedings of the 11th National Hellenic Conference in Genetics and Plant Breeding, Orestiada, Greece, 31 October–2 November 2006; Hellenic Scientific Society for Genetics and Plant Breeding: Orestiada, Greece, 2006; pp. 7–14. [Google Scholar]
- Steel, R.G.D.; Torrie, H.; Dickey, D.A. Principles and Procedures of Statistics. A Biometrical Approach, 3rd ed.; McGraw-Hill: New York, NY, USA, 1997; p. 666. [Google Scholar]
- McIntosh, M.S. Analysis of Combined Experiments. Agron. J. 1983, 75, 153–155. [Google Scholar] [CrossRef]
- Hanson, G.; Robinson, H.F.; Comstock, R.E. Biometrical studies on yield in segregating population of Korean Lespedeza. Agron. J. 1956, 48, 268–274. [Google Scholar] [CrossRef]
- Singh, R.K.; Chaudhary, B.D. Biometrical Methods in Quantitative Genetic Analysis; Kalyani Publishers: New Delhi, India, 1977; p. 304. [Google Scholar]
| Source of Variation | Days to 50% Flowering | Main Stem Length (cm) | Main Stem Thickness (mm) | Fresh Forage Yield (kg ha−1) | Dry Matter Yield (kg ha−1) | Forage Dry Matter Crude Protein Content (%) | Ash Content % of Dry Matter |
|---|---|---|---|---|---|---|---|
| m.s. | m.s. | m.s. | m.s. | m.s. | m.s. | m.s. | |
| Environments (E) | 163.29 ** | 68.52 ** | 0.09 ** | 28,290,714 ** | 1,938,835 ** | 3.95 ** | 3.71 ** |
| REPS/Environments | 1.16 ns | 17.02 ns | 0.01 ns | 6,632,777 ** | 316,336 ** | 8.41 ** | 10.31 ** |
| Genotypes (G) | 499.53 ** | 155.90 ** | 0.02 * | 168,425,535 ** | 8,653,394 ** | 17.54 ** | 5.92 ** |
| Genotypes × Cultivation | 2.01 * | 48.89 * | 0.04 ** | 102,070,746 ** | 5,364,623 ** | 1.18 ** | 0.44 ** |
| Genotypes × Environments (G × E) | 5.37 ** | 32.28 * | 0.03 ** | 17,196,116 ** | 828,043 ** | 2.77 ** | 1.44 ** |
| Cultivations | 334.99 ** | 207.37 ** | 0.04 * | 36,668,636 ** | 1,557,247 ** | 98.63 ** | 42.84 ** |
| Cultivation × Environments | 2.31 * | 32.49 ns | 0.08 ** | 31,475,811 ** | 1,614,424 ** | 0.36 ** | 20.09 ** |
| Cultivation × Genotypes × Environments | 1.03 ns | 42.24 ** | 0.04 ** | 26,630,013 ** | 1,270,305 ** | 0.29 ** | 0.18 ** |
| Error | 0.95 | 2.128 | 0.01 | 1,609,523 | 69,682 | 0.020 | 0.03 |
| Environments | Days to 50% Flowering | Main Stem Length (cm) | Main Stem Thickness (mm) | Fresh Forage Yield (kg ha−1) | Dry Matter Yield (kg ha−1) | Forage Dry Matter Crude Protein Content % | Ash Content % of Dry Matter | |
|---|---|---|---|---|---|---|---|---|
| Conventional | Giannitsa | 1735 | 544 | 301 | 41 | 44 | 379 | 117 |
| Florina | 2127 | 675 | 533 | 59 | 57 | 284 | 108 | |
| Trikala | 3391 | 927 | 1012 | 38 | 44 | 402 | 115 | |
| Kalambaka | 3173 | 2726 | 3009 | 57 | 57 | 357 | 83 | |
| Low-input | Giannitsa | 2091 | 1816 | 484 | 60 | 61 | 449 | 119 |
| Florina | 2142 | 1344 | 664 | 94 | 126 | 438 | 123 | |
| Trikala | 2987 | 2575 | 422 | 68 | 79 | 446 | 130 | |
| Kalambaka | 2461 | 1531 | 649 | 120 | 148 | 435 | 109 | |
| Conventional and Low-input | Giannitsa | 1824 | 720 | 367 | 49 | 51 | 343 | 104 |
| Florina | 2074 | 905 | 593 | 73 | 79 | 307 | 107 | |
| Trikala | 2881 | 1348 | 593 | 46 | 53 | 340 | 113 | |
| Kalambaka | 2458 | 1860 | 1065 | 73 | 80 | 315 | 85 |
| Genotypes | Days to 50% Flowering | Main Stem Length (cm) | Main Stem Thickness (mm) | Fresh Forage Yield (kg ha−1) | Dry Matter Yield (kg ha−1) | Forage Dry Matter Crude Protein Content % | Ash Content % of Dry Matter | |
|---|---|---|---|---|---|---|---|---|
| Conventional | Olympos | 4766 | 888 | 1124 | 97 | 121 | 354 | 133 |
| Pisso | 7190 | 2155 | 1735 | 140 | 148 | 475 | 115 | |
| Livioletta | 7544 | 1661 | 388 | 135 | 135 | 459 | 133 | |
| Vermio | 5127 | 1650 | 738 | 88 | 99 | 533 | 111 | |
| Dodoni | 4208 | 1539 | 881 | 58 | 64 | 453 | 81 | |
| Zt1 | 3881 | 579 | 476 | 68 | 66 | 540 | 123 | |
| Zt2 | 5786 | 847 | 364 | 84 | 86 | 526 | 118 | |
| Low-input | Olympos | 5693 | 1885 | 690 | 87 | 108 | 472 | 146 |
| Pisso | 7613 | 1751 | 1016 | 51 | 52 | 486 | 129 | |
| Livioletta | 8642 | 1286 | 408 | 139 | 204 | 471 | 118 | |
| Vermio | 7799 | 1542 | 362 | 125 | 193 | 438 | 116 | |
| Dodoni | 5006 | 2130 | 476 | 59 | 55 | 623 | 122 | |
| Zt1 | 8035 | 993 | 343 | 151 | 202 | 541 | 153 | |
| Zt2 | 10,911 | 3094 | 638 | 64 | 71 | 544 | 107 | |
| Conventional and Low-input | Olympos | 4487 | 1194 | 846 | 93 | 115 | 325 | 126 |
| Pisso | 6254 | 1731 | 1292 | 46 | 48 | 460 | 119 | |
| Livioletta | 6589 | 1413 | 395 | 114 | 130 | 347 | 118 | |
| Vermio | 5305 | 1462 | 473 | 83 | 98 | 390 | 104 | |
| Dodoni | 4090 | 1648 | 586 | 59 | 60 | 407 | 84 | |
| Zt1 | 4080 | 630 | 386 | 85 | 86 | 461 | 120 | |
| Zt2 | 6836 | 1347 | 468 | 71 | 75 | 337 | 95 |
| Genotypes | Days to 50% Flowering | Main Stem Length (cm) | Main Stem Thickness (mm) | Fresh Forage Yield (kg ha−1) | Dry Matter Yield (kg ha−1) | Forage Dry Matter Crude Protein Content % | Ash Content % of Dry Matter | |
|---|---|---|---|---|---|---|---|---|
| Giannitsa | ||||||||
| Conventional | Olympos | 3420 | 5714 | 591 | 294 | 507 | 436 | 134 |
| Pisso | 4193 | 9510 | 1778 | 569 | 437 | 600 | 130 | |
| Livioletta | 6005 | 800 | 239 | 147 | 247 | 574 | 186 | |
| Vermio | 5724 | 933 | 834 | 71 | 73 | 663 | 75 | |
| Dodoni | 3471 | 2586 | 412 | 30 | 32 | 510 | 71 | |
| Zt1 | 2905 | 8231 | 293 | 44 | 43 | 864 | 147 | |
| Zt2 | 4254 | 2239 | 298 | 98 | 97 | 786 | 143 | |
| Low-input | Olympos | 4118 | 2902 | 760 | 130 | 140 | 402 | 146 |
| Pisso | 6867 | 6588 | 411 | 633 | 720 | 893 | 143 | |
| Livioletta | 7437 | 13,802 | 327 | 439 | 470 | 598 | 100 | |
| Vermio | 6470 | 9720 | 684 | 90 | 132 | 461 | 81 | |
| Dodoni | 5693 | 26,290 | 1184 | 24 | 23 | 921 | 124 | |
| Zt1 | 5386 | 8439 | 430 | 399 | 528 | 704 | 133 | |
| Zt2 | 7046 | 6798 | 1474 | 82 | 59 | 598 | 128 | |
| Conventional and Low-input | Olympos | 3600 | 2443 | 684 | 190 | 233 | 315 | 133 |
| Pisso | 4795 | 4764 | 708 | 538 | 509 | 619 | 124 | |
| Livioletta | 5997 | 1621 | 289 | 88 | 107 | 424 | 123 | |
| Vermio | 5870 | 467 | 406 | 64 | 74 | 475 | 79 | |
| Dodoni | 4228 | 2573 | 577 | 29 | 28 | 464 | 81 | |
| Zt1 | 3668 | 268 | 186 | 56 | 55 | 692 | 130 | |
| Zt2 | 4391 | 2417 | 355 | 81 | 65 | 491 | 101 | |
| Florina | ||||||||
| Conventional | Olympos | 4556 | 1110 | 1957 | 178 | 196 | 590 | 148 |
| Pisso | 4709 | 1626 | 3689 | 791 | 513 | 746 | 96 | |
| Livioletta | 7338 | 11,783 | 350 | 146 | 139 | 603 | 104 | |
| Vermio | 8348 | 3337 | 358 | 269 | 442 | 511 | 186 | |
| Dodoni | 5397 | 3092 | 729 | 174 | 170 | 706 | 87 | |
| Zt1 | 4315 | 4055 | 399 | 215 | 240 | 643 | 145 | |
| Zt2 | 9480 | 4485 | 725 | 98 | 124 | 386 | 90 | |
| Low-input | Olympos | 7142 | 1362 | 2903 | 31 | 39 | 734 | 162 |
| Pisso | 5487 | 1700 | 2236 | 307 | 444 | 375 | 109 | |
| Livioletta | 8024 | 2574 | 941 | 149 | 511 | 470 | 140 | |
| Vermio | 11,714 | 3077 | 404 | 113 | 299 | 549 | 151 | |
| Dodoni | 7228 | 3662 | 801 | 55 | 78 | 967 | 127 | |
| Zt1 | 4205 | 9218 | 326 | 291 | 395 | 812 | 166 | |
| Zt2 | 9946 | 4165 | 499 | 284 | 329 | 823 | 116 | |
| Conventional and Low-input | Olympos | 5354 | 849 | 2451 | 57 | 70 | 486 | 144 |
| Pisso | 5101 | 1705 | 1225 | 95 | 93 | 531 | 104 | |
| Livioletta | 7078 | 4333 | 506 | 97 | 122 | 309 | 107 | |
| Vermio | 8853 | 2523 | 381 | 114 | 168 | 469 | 155 | |
| Dodoni | 5516 | 3340 | 746 | 78 | 104 | 596 | 102 | |
| Zt1 | 4151 | 3239 | 384 | 248 | 311 | 632 | 153 | |
| Zt2 | 8990 | 2881 | 592 | 149 | 192 | 397 | 101 | |
| Trikala | ||||||||
| Conventional | Olympos | 8945 | 3180 | 2478 | 32 | 39 | 485 | 183 |
| Pisso | 11,898 | 2917 | 1701 | 79 | 75 | 327 | 178 | |
| Livioletta | 20,733 | 3844 | 496 | 268 | 178 | 387 | 169 | |
| Vermio | 11,755 | 4665 | 1098 | 114 | 172 | 549 | 107 | |
| Dodoni | 11,729 | 1154 | 2544 | 86 | 178 | 558 | 129 | |
| Zt1 | 16,981 | 1810 | 1666 | 114 | 112 | 527 | 92 | |
| Zt2 | 10,124 | 17,502 | 2158 | 75 | 102 | 460 | 150 | |
| Low-input | Olympos | 5348 | 12,101 | 854 | 375 | 486 | 772 | 144 |
| Pisso | 9580 | 908 | 2804 | 256 | 267 | 450 | 128 | |
| Livioletta | 12,243 | 7686 | 1106 | 64 | 109 | 236 | 155 | |
| Vermio | 13,050 | 12,343 | 228 | 569 | 796 | 936 | 97 | |
| Dodoni | 12,591 | 3145 | 201 | 479 | 391 | 796 | 121 | |
| Zt1 | 24,270 | 3716 | 901 | 196 | 339 | 476 | 132 | |
| Zt2 | 22,794 | 8975 | 351 | 109 | 82 | 396 | 139 | |
| Conventional and Low-input | Olympos | 5520 | 4435 | 640 | 63 | 78 | 506 | 145 |
| Pisso | 8503 | 1122 | 2079 | 18 | 20 | 380 | 159 | |
| Livioletta | 10,698 | 2202 | 680 | 110 | 143 | 255 | 169 | |
| Vermio | 8453 | 7203 | 393 | 81 | 102 | 417 | 101 | |
| Dodoni | 9319 | 1216 | 374 | 142 | 225 | 515 | 111 | |
| Zt1 | 9155 | 2563 | 1007 | 155 | 176 | 465 | 103 | |
| Zt2 | 11,099 | 12,133 | 647 | 72 | 76 | 243 | 103 | |
| Kalambaka | ||||||||
| Conventional | Olympos | 11,945 | 1617 | 1364 | 222 | 275 | 885 | 90 |
| Pisso | 38,704 | 2185 | 8494 | 100 | 112 | 902 | 112 | |
| Livioletta | 27,429 | 2785 | 2088 | 190 | 241 | 463 | 176 | |
| Vermio | 8138 | 13,133 | 4768 | 103 | 110 | 531 | 121 | |
| Dodoni | 16,768 | 9926 | 3175 | 121 | 163 | 856 | 162 | |
| Zt1 | 13,542 | 5841 | 3894 | 163 | 188 | 463 | 169 | |
| Zt2 | 3435 | 12,901 | 3572 | 58 | 51 | 507 | 87 | |
| Low-input | Olympos | 9304 | 1443 | 678 | 125 | 164 | 897 | 146 |
| Pisso | 8098 | 4612 | 1367 | 297 | 545 | 781 | 156 | |
| Livioletta | 14,546 | 442 | 189 | 361 | 419 | 844 | 98 | |
| Vermio | 11,685 | 6158 | 777 | 317 | 470 | 518 | 160 | |
| Dodoni | 7719 | 2190 | 958 | 195 | 463 | 873 | 121 | |
| Zt1 | 21,627 | 4766 | 1075 | 177 | 264 | 804 | 152 | |
| Zt2 | 12,166 | 3613 | 1116 | 47 | 54 | 952 | 72 | |
| Conventional and Low-input | Olympos | 6814 | 1591 | 966 | 153 | 203 | 516 | 110 |
| Pisso | 8481 | 2977 | 2505 | 62 | 71 | 681 | 132 | |
| Livioletta | 10,069 | 815 | 372 | 227 | 231 | 496 | 128 | |
| Vermio | 6461 | 7037 | 1340 | 166 | 180 | 452 | 114 | |
| Dodoni | 7493 | 3521 | 1517 | 151 | 188 | 564 | 86 | |
| Zt1 | 4138 | 3336 | 1756 | 80 | 104 | 431 | 117 | |
| Zt2 | 5731 | 3069 | 1694 | 53 | 55 | 334 | 77 | |
| Traits | Min | Max | Mean | sd | GCV (%) | PCV (%) | H2 (%) | ||
|---|---|---|---|---|---|---|---|---|---|
| Days to 50% flowering | 148.1 | 165.7 | 156.3 | 3.38 | 7.76 | 7.81 | 1.78 | 1.79 | 99.4 |
| Main stem length (cm) | 85.2 | 99.8 | 92.3 | 2.98 | 2.04 | 2.31 | 1.55 | 1.65 | 88.4 |
| Main stem thickness (mm) | 2.78 | 3.76 | 3.21 | 0.14 | - | - | - | - | - |
| Fresh forage yield (kg ha−1) | 17,257 | 34,932 | 23,947 | 3201 | 2,205,760 | 2,631,649 | 6.20 | 6.77 | 83.8 |
| Dry matter yield (kg ha−1) | 4080 | 7930 | 5531 | 714.2 | 114,321 | 135,209 | 6.11 | 6.48 | 84.6 |
| Forage dry matter crude protein content (%) | 17.8 | 23.3 | 20.3 | 1.14 | 0.251 | 0.274 | 2.47 | 2.58 | 91.4 |
| Ash content % of dry matter | 7.29 | 12.45 | 9.9 | 0.98 | 0.081 | 0.093 | 2.87 | 3.08 | 87.0 |
| Days to 50% Flowering | Main Stem Length (cm) | Main Stem Thickness (mm) | Fresh Forage Yield (kg ha−1) | Dry Matter Yield (kg ha−1) | Forage Dry Matter Crude Protein Content % | |
|---|---|---|---|---|---|---|
| Main stem length (cm) | −0.061 | |||||
| Main stem thickness (mm) | −0.004 | −0.231 ** | ||||
| Fresh forage yield (kg ha−1) | 0.032 | 0.203 ** | 0.016 | |||
| Dry matter yield (kg ha−1) | 0.028 | 0.210 ** | 0.009 | 0.974 ** | ||
| Forage dry matter crude protein content (%) | 0.289 ** | 0.006 | 0.004 | 0.100 * | 0.084 | |
| Ash content % of dry matter | 0.100 * | −0.050 | 0.048 | −0.084 | −0.091 | 0.676 ** |
| Environments | Longitude | Latitude | Elevation (m) | Soil Texture | Planting Date | Harvesting Date |
|---|---|---|---|---|---|---|
| Giannitsa | 22°39′ E | 40°77′ N | 10 | Clay (C) | Early November 2008 and 2009 | Late May 2009 and 2010 |
| Florina | 21°22′ E | 40°46′ N | 705 | Sandy loam (SL) | Early November 2008 and 2009 | Late May 2009 and 2010 |
| Trikala | 21°64′ E | 39°55′ N | 120 | Sandy clay loam (SCL) | Early November 2008 and 2009 | Late May 2009 and 2010 |
| Kalambaka | 21°65′ E | 39°64′ N | 190 | Silty clay (SiC) | Early November 2008 and 2009 | Late May 2009 and 2010 |
| Year and Environments | Mean Monthly MaximumTemperature (°C) | Mean Monthly Minimum Temperature (°C) | Mean Temperature (°C) | Rainfall (mm) |
|---|---|---|---|---|
| Giannitsa 2008–2009 | 22.9 | −0.1 | 10.5 | 51.5 |
| Giannitsa 2009–2010 | 23.3 | 0.6 | 11.0 | 55.5 |
| Florina 2008–2009 | 21.0 | −5.7 | 7.1 | 44.1 |
| Florina 2009–2010 | 21.3 | −3.8 | 8.0 | 62.3 |
| Trikala 2008–2009 | 23.4 | 1.3 | 11.0 | 55.8 |
| Trikala 2009–2010 | 24.4 | 2.9 | 12.0 | 85.1 |
| Kalambaka 2008–2009 | 21.1 | 0.4 | 10.8 | 68.4 |
| Kalambaka 2009–2010 | 23.5 | 2.2 | 11.7 | 98.8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Greveniotis, V.; Bouloumpasi, E.; Zotis, S.; Korkovelos, A.; Ipsilandis, C.G. Stability, the Last Frontier: Forage Yield Dynamics of Peas under Two Cultivation Systems. Plants 2022, 11, 892. https://doi.org/10.3390/plants11070892
Greveniotis V, Bouloumpasi E, Zotis S, Korkovelos A, Ipsilandis CG. Stability, the Last Frontier: Forage Yield Dynamics of Peas under Two Cultivation Systems. Plants. 2022; 11(7):892. https://doi.org/10.3390/plants11070892
Chicago/Turabian StyleGreveniotis, Vasileios, Elisavet Bouloumpasi, Stylianos Zotis, Athanasios Korkovelos, and Constantinos G. Ipsilandis. 2022. "Stability, the Last Frontier: Forage Yield Dynamics of Peas under Two Cultivation Systems" Plants 11, no. 7: 892. https://doi.org/10.3390/plants11070892
APA StyleGreveniotis, V., Bouloumpasi, E., Zotis, S., Korkovelos, A., & Ipsilandis, C. G. (2022). Stability, the Last Frontier: Forage Yield Dynamics of Peas under Two Cultivation Systems. Plants, 11(7), 892. https://doi.org/10.3390/plants11070892





