Phylogeographical Analyses of a Relict Fern of Palaeotropical Flora (Vandenboschia speciosa): Distribution and Diversity Model in Relation to the Geological and Climate Events of the Late Miocene and Early Pliocene
Abstract
1. Introduction
2. Materials and Methods
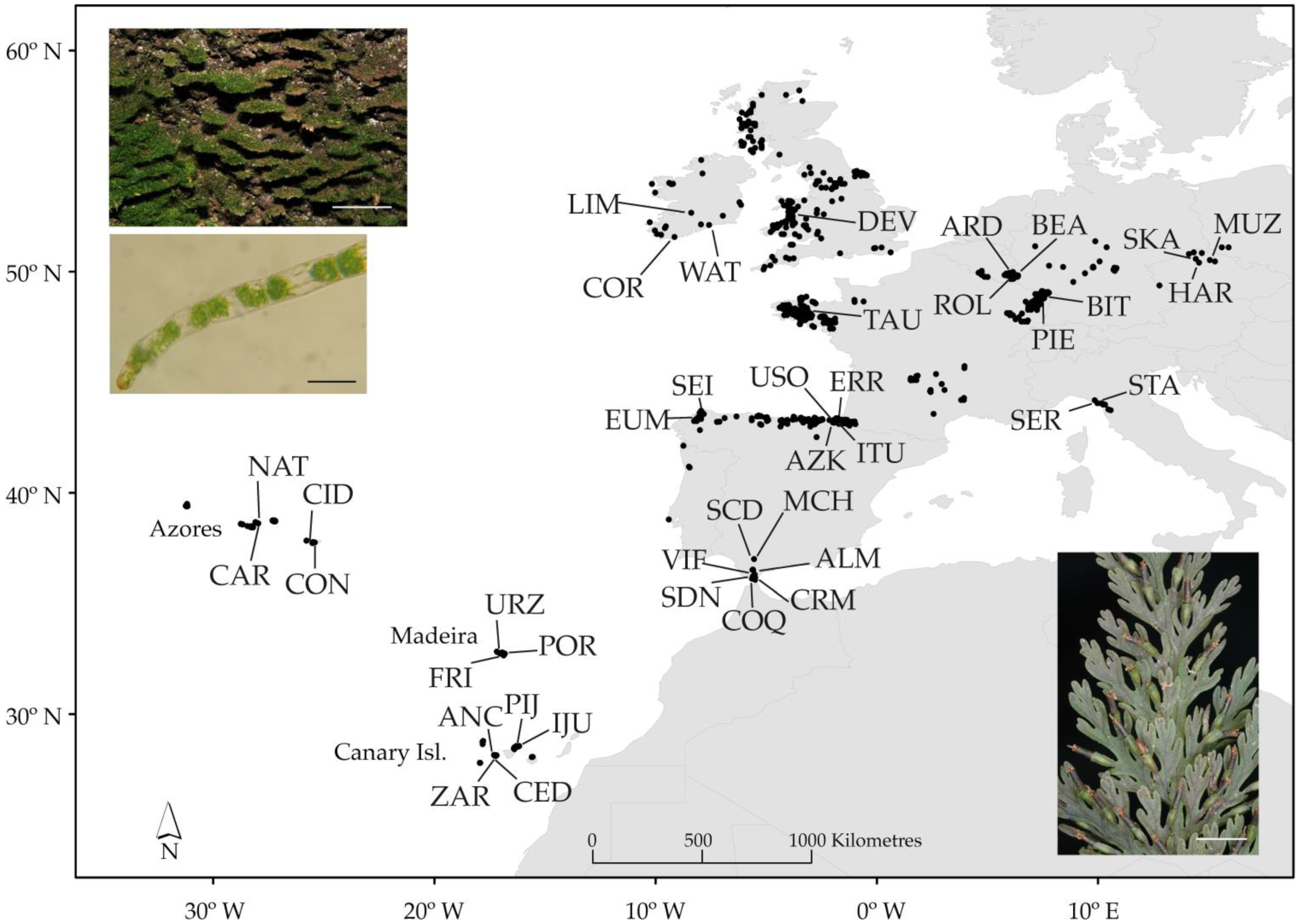
2.1. Plant Material
2.2. DNA Extraction, PCR Amplification, and Sequencing
2.3. Establishment of gapCp Variant Homology by Phylogenetic Analysis
2.4. Genetic Diversity and Structure
2.5. Haplotype Phylogeny and Dating
2.6. Demographic Analyses
2.7. Species Distribution Modelling
3. Results
3.1. PtDNA and Nuclear Marker Characteristics
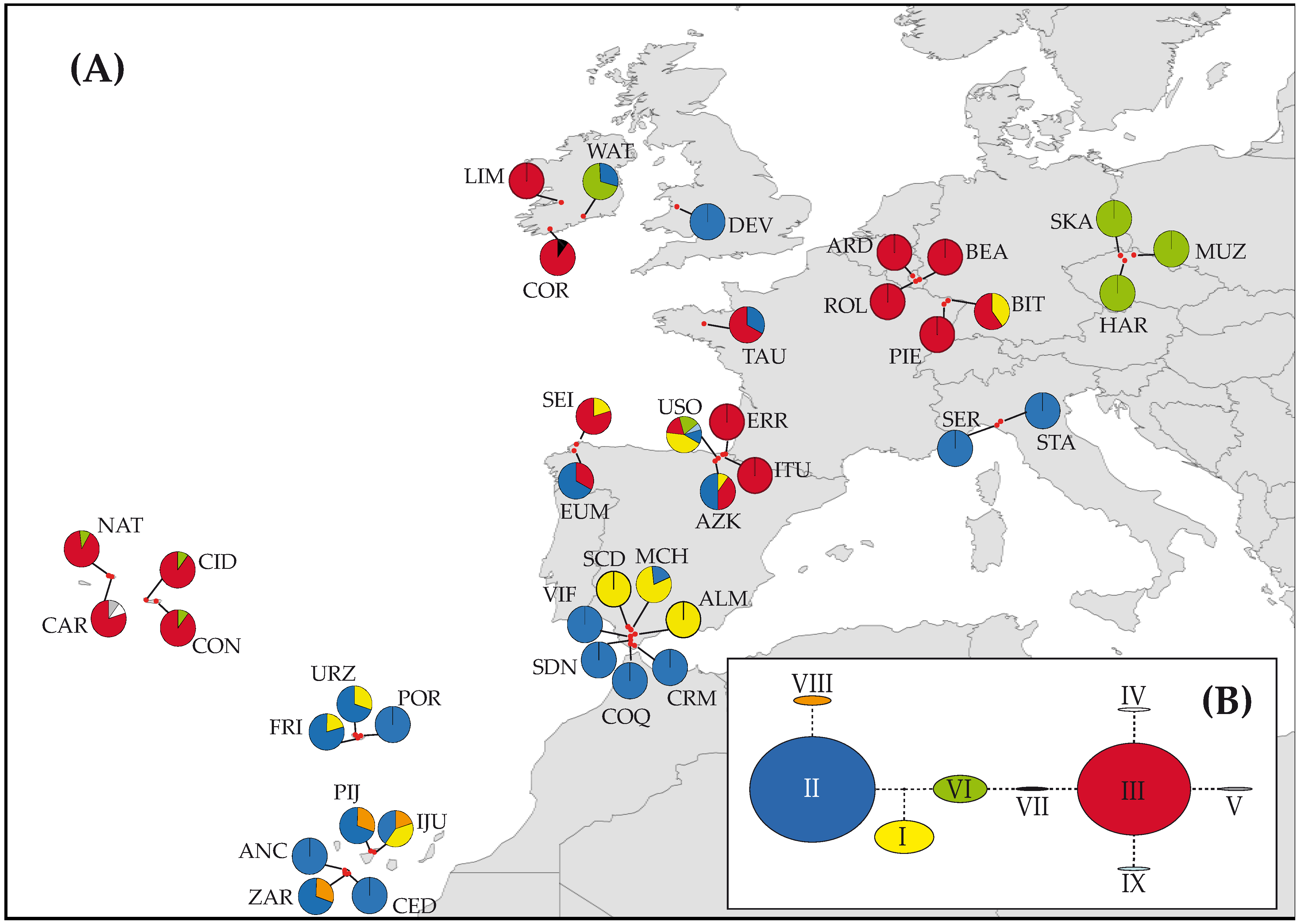
3.2. Genetic Diversity and Structure
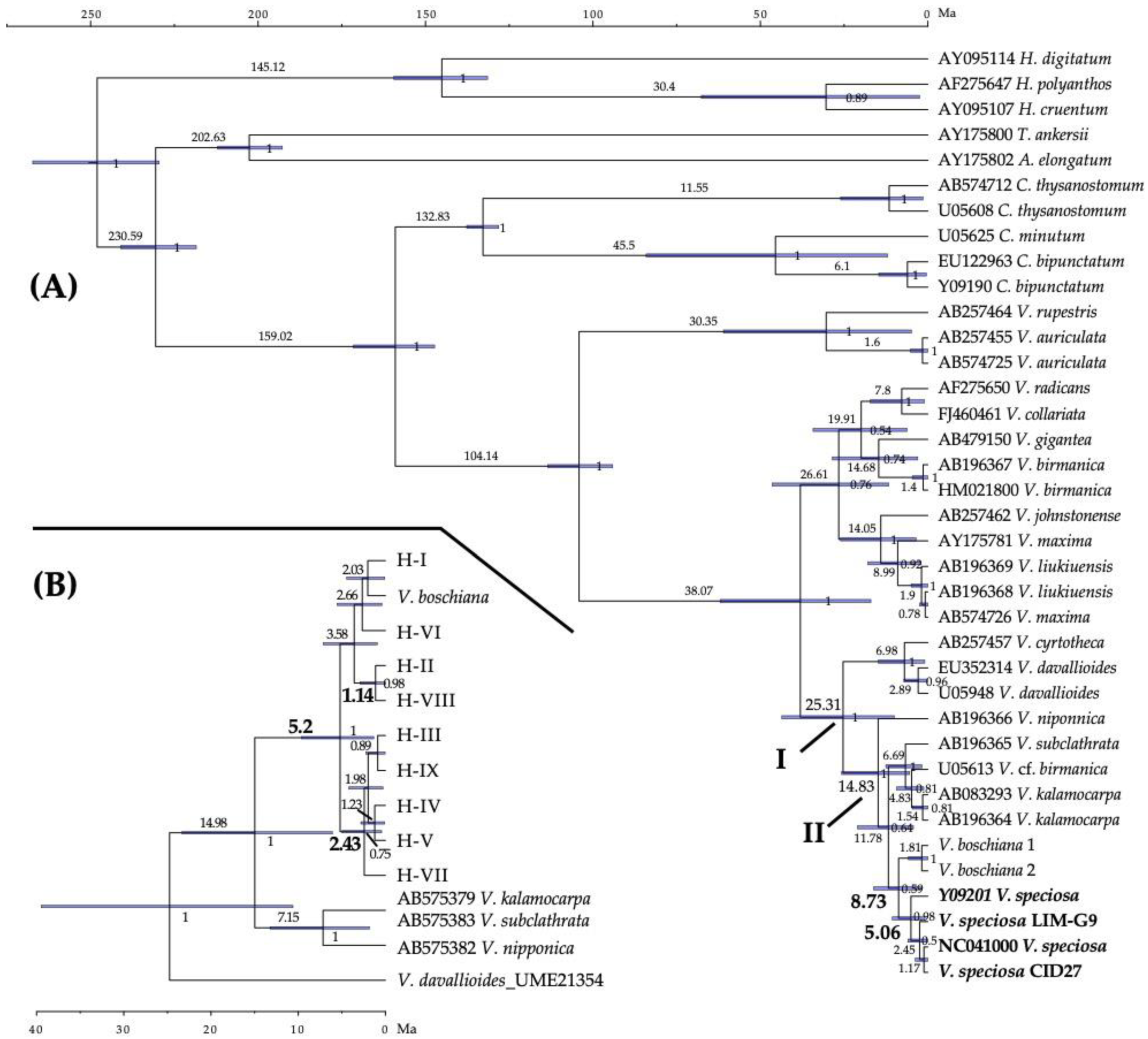
3.3. PtDNA Haplotype Phylogeny and Dating
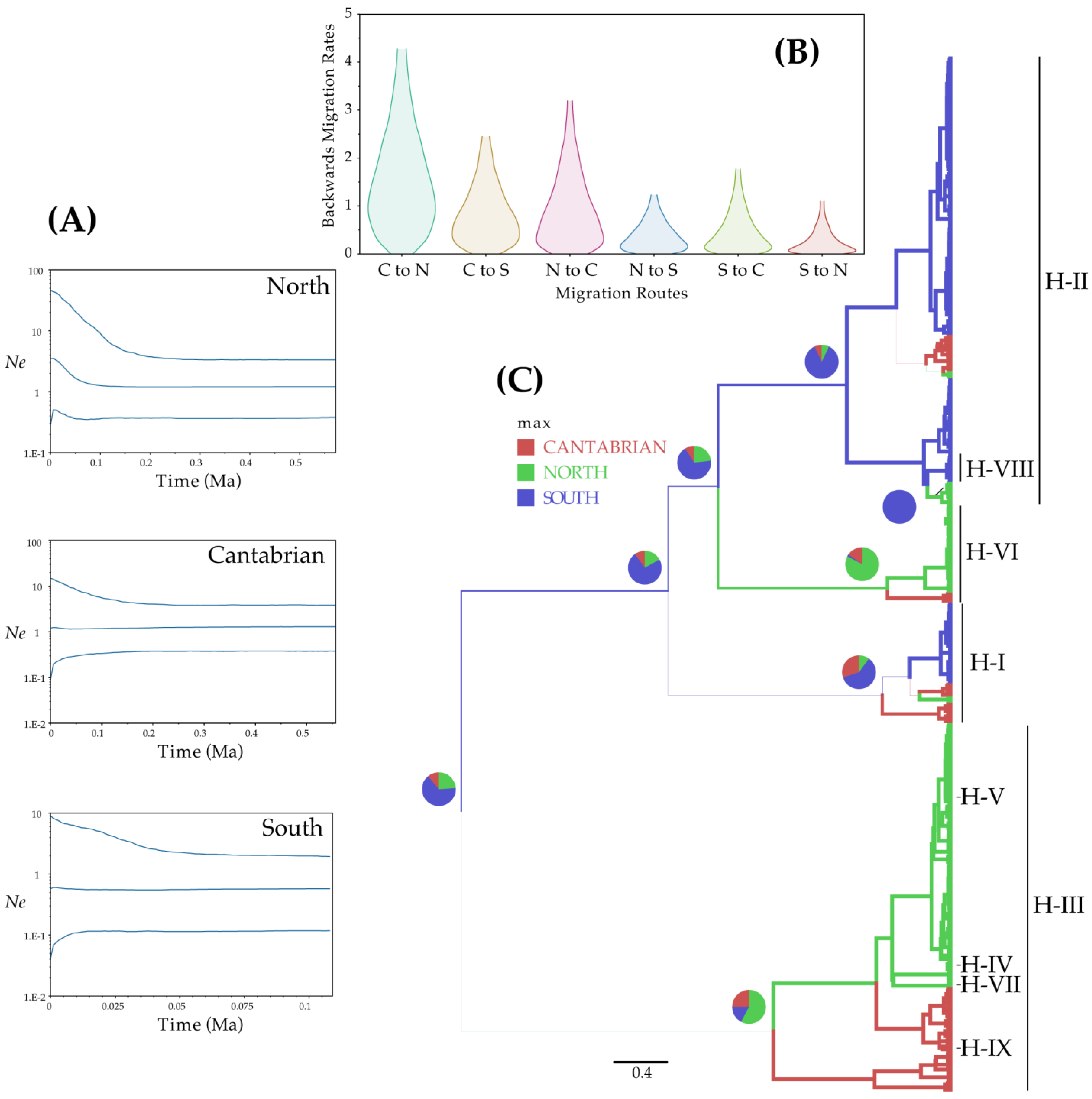
3.4. Demographic Analysis of V. speciosa
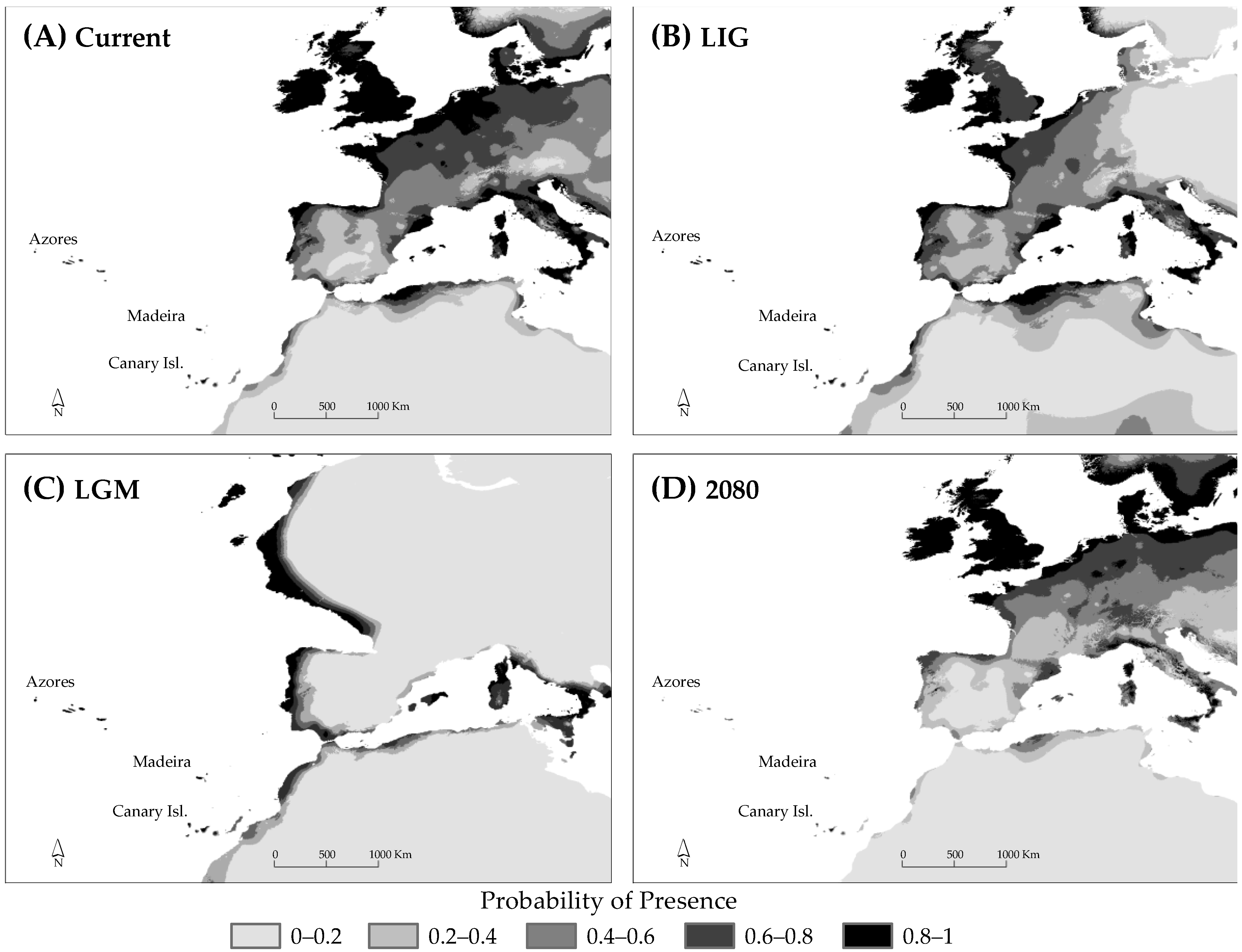
3.5. Species Distribution Modelling
4. Discussion
4.1. Absence of Genetic Structure between the Sporophytic and Gametophytic Phases of V. speciosa
4.2. Tertiary Origin of V. speciosa
4.3. Climate-Change-Driven Phylogeographical History of V. speciosa
4.4. The Cantabrian Cornice Could Act as a Tertiary Refugium and as a Suture Zone during the Quaternary
4.5. Central and Northern Europe
4.6. Macaronesian Archipelagos
4.7. Future Forecasts
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Engler, A. Versuch Einer Entwicklungsgeschichte der Pflanzenwelt, Insbesondere der Florengebiete Seit der Tertiärperiode, Die Extratropischen Gebiete der Nördischen Hemisphäre; Engelmann: Leipzig, Germany, 1879; Volume 1. [Google Scholar]
- Mai, D.H. Development and regional differentiation of the European vegetation during the Tertiary. Plant Syst. Evol. 1989, 162, 79–91. [Google Scholar] [CrossRef]
- Mai, D.H. Palaeofloristic changes in Europe and the confirmation of the Arctotertiary-Palaeotropical geofloral concept. Rev. Palaeobot. Palynol. 1991, 68, 29–36. [Google Scholar] [CrossRef]
- Pichi-Sermolli, R.E.G. A survey of the pteridological flora of the mediterranean region: Rassegna della flora pteridologica della regione mediterranea. Webbia 1979, 34, 175–242. [Google Scholar] [CrossRef]
- Pichi-Sermolli, R.E.G. Considerazioni sull’affinitá ed origine della flora pteridologica della Regione Mediterranea. Acta Botánica Malacit. 1991, 16, 235–280. [Google Scholar] [CrossRef]
- Barrón, E. Evolución de las floras terciarias en la Península Ibérica. Monogr. Jardín Botánico Córdoba 2003, 11, 63–74. [Google Scholar]
- Barrón, E.; Peyrot, D. La vegetación forestal en el Terciario. In Paleoambientes y Cambio Climático; Carrión, J.S., Fernández, S., Fuentes, N., Eds.; Fudación Séneca: Murcia, Spain, 2006; pp. 55–76. ISBN 84-932456-6-6. [Google Scholar]
- Rodríguez-Sánchez, F.; Arroyo, J. Reconstructing the demise of Tethyan plants: Climate-driven range dynamics of Laurus since the Pliocene. Glob. Ecol. Biogeogr. 2008, 17, 685–695. [Google Scholar] [CrossRef]
- Hewitt, G. Ice ages. In Evolution on Planet Earth; Rothschild, L.J., Lister, A.M., Eds.; Academic Press: London, UK, 2003; pp. 339–361. [Google Scholar]
- Vogel, J.C.; Rumsey, F.J.; Schneller, J.J.; Barrett, J.A.; Gibby, M. Where are the glacial refugia in Europe? Evidence from pteridophytes. Biol. J. Linn. Soc. 1999, 66, 23–37. [Google Scholar] [CrossRef]
- Postigo Mijarra, J.M.; Barrón, E.; Gómez Manzaneque, F.; Morla, C. Floristic changes in the Iberian Peninsula and Balearic Islands (south-west Europe) during the cenozoic. J. Biogeogr. 2009, 36, 2025–2043. [Google Scholar] [CrossRef]
- Ivanov, D.; Utescher, T.; Mosbrugger, V.; Syabryaj, S.; Djordjević-Milutinović, D.; Molchanoff, S. Miocene vegetation and climate dynamics in Eastern and Central Paratethys (Southeastern Europe). Palaeogeogr. Palaeoclimatol. Palaeoecol. 2011, 304, 262–275. [Google Scholar] [CrossRef]
- Stewart, J.R.; Lister, A.M. Cryptic northern refugia and the origins of the modern biota. Trends Ecol. Evol. 2001, 16, 608–613. [Google Scholar] [CrossRef]
- Provan, J.; Bennett, K.D. Phylogeographic insights into cryptic glacial refugia. Trends Ecol. Evol. 2008, 23, 564–571. [Google Scholar] [CrossRef]
- Taberlet, P.; Fumagalli, L.; Wust-Saucy, A.G.; Cosson, J.F. Comparative phylogeography and postglacial colonization routes in Europe. Mol. Ecol. 1998, 7, 453–464. [Google Scholar] [CrossRef]
- Hewitt, G.M. Post-glacial re-colonization of European biota. Biol. J. Linn. Soc. 1999, 68, 87–112. [Google Scholar] [CrossRef]
- Su, Y.; Wang, T.; Zheng, B.; Jiang, Y.; Chen, G.; Gu, H. Population genetic structure and phylogeographical pattern of a relict tree fern, Alsophila spinulosa (Cyatheaceae), inferred from cpDNA atpB-rbcL intergenic spacers. Theor. Appl. Genet. 2004, 109, 1459–1467. [Google Scholar] [CrossRef]
- Shepherd, L.D.; Perrie, L.R.; Brownsey, P.J. Fire and ice: Volcanic and glacial impacts on the phylogeography of the New Zealand forest fern Asplenium hookerianum. Mol. Ecol. 2007, 16, 4536–4549. [Google Scholar] [CrossRef]
- Rodríguez-Sánchez, F.; Guzmán, B.; Valido, A.; Vargas, P.; Arroyo, J. Late Neogene history of the laurel tree (Laurus L., Lauraceae) based on phylogeographical analyses of Mediterranean and Macaronesian populations. J. Biogeogr. 2009, 36, 1270–1281. [Google Scholar] [CrossRef]
- Wang, L.; Wu, Z.Q.; Bystriakova, N.; Ansell, S.W.; Xiang, Q.P.; Heinrichs, J.; Schneider, H.; Zhang, X.C. Phylogeography of the Sino-Himalayan fern Lepisorus clathratus on “the roof of the world.”. PLoS ONE 2011, 6, e25896. [Google Scholar] [CrossRef]
- García-Verdugo, C.; Calleja, J.A.; Vargas, P.; Silva, L.; Moreira, O.; Pulido, F. Polyploidy and microsatellite variation in the relict tree Prunus lusitanica L.: How effective are refugia in preserving genotypic diversity of clonal taxa? Mol. Ecol. 2013, 22, 1546–1557. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Qi, Z.-C.; Xu, X.-H.; Comes, H.P.; Koch, M.A.; Jin, X.-J.; Fu, C.-X.; Qiu, Y.-X. Understanding the formation of Mediterranean-African-Asian disjunctions: Evidence for Miocene climate-driven vicariance and recent long-distance dispersal in the Tertiary relict Smilax aspera (Smilacaceae). New Phytol. 2014, 204, 243–255. [Google Scholar] [CrossRef]
- Ohlsen, D.J.; Shepherd, L.D.; Perrie, L.R.; Brownsey, P.J.; Bayly, M.J. Genetic variation and phylogeography of the Australian and New Zealand fern Asplenium flabellifolium (Aspleniaceae). Aust. Syst. Bot. 2020, 33, 412–426. [Google Scholar] [CrossRef]
- Trewick, S.A.; Morgan-Richards, M.; Russell, S.J.; Henderson, S.; Rumsey, F.J.; Pintér, I.; Barrett, J.A.; Gibby, M.; Vogel, J.C. Polyploidy, phylogeography and Pleistocene refugia of the rockfern Asplenium ceterach: Evidence from chloroplast DNA. Mol. Ecol. 2002, 11, 2003–2012. [Google Scholar] [CrossRef] [PubMed]
- Hunt, H.V.; Ansell, S.W.; Russell, S.J.; Schneider, H.; Vogel, J.C. Genetic diversity and phylogeography in two diploid ferns, Asplenium fontanum subsp. fontanum and A. petrarchae subsp. bivalens, in the western Mediterranean. Mol. Ecol. 2009, 18, 4940–4954. [Google Scholar] [CrossRef] [PubMed]
- Jiménez, A.; Holderegger, R.; Csencsics, D.; Quintanilla, L.G. Microsatellites reveal substantial among-population genetic differentiation and strong inbreeding in the relict fern Dryopteris aemula. Ann. Bot. 2010, 106, 149–155. [Google Scholar] [CrossRef]
- Bystriakova, N.; Ansell, S.W.; Russell, S.J.; Grundmann, M.; Vogel, J.C.; Schneider, H. Present, past and future of the European rock fern Asplenium fontanum: Combining distribution modelling and population genetics to study the effect of climate change on geographic range and genetic diversity. Ann. Bot. 2014, 113, 453–465. [Google Scholar] [CrossRef]
- Maccagni, A.; Parisod, C.; Grant, J.R. Phylogeography of the moonwort fern Botrychium lunaria (Ophioglossaceae) based on chloroplast DNA in the Central-European Mountain System. Alp. Bot. 2017, 127, 185–196. [Google Scholar] [CrossRef]
- Ben-Menni Schuler, S.; García-López, M.d.C.; López-Flores, I.; Nieto-Lugilde, M.; Suárez-Santiago, V.N. Genetic diversity and population history of the Killarney fern, Vandenboschia speciosa (Hymenophyllaceae), at its southern distribution limit in continental Europe. Bot. J. Linn. Soc. 2017, 183, 94–105. [Google Scholar] [CrossRef][Green Version]
- Ben-Menni Schuler, S.; Picazo-Aragonés, J.; Rumsey, F.J.; Romero-García, A.T.; Suárez-Santiago, V.N. Macaronesia Acts as a Museum of Genetic Diversity of Relict Ferns: The Case of Diplazium caudatum (Athyriaceae). Plants 2021, 10, 2425. [Google Scholar] [CrossRef]
- Jermy, A.C. Origin and distribution of Pteridophytes in the Mediterranean area. Webbia 1984, 38, 397–416. [Google Scholar] [CrossRef]
- Vogel, J.C.; Rumsey, F.J.; Russell, S.J.; Cox, C.J.; Holmes, J.S.; Bujnoch, W.; Stark, C.; Barrett, J.A.; Gibby, M. Genetic structure, reproductive biology and ecology of isolated populations of Asplenium csikii (Aspleniaceae, Pteridophyta). Heredity 1999, 83, 604–612. [Google Scholar] [CrossRef]
- Quintanilla, L.G.; Pajarón, S.; Pangua, E.; Amigo, J. Allozyme variation in the sympatric ferns Culcita macrocarpa and Woodwardia radicans at the northern extreme of their ranges. Plant Syst. Evol. 2007, 263, 135–144. [Google Scholar] [CrossRef]
- Vanderpoorten, A.; Rumsey, F.J.; Carine, M.A. Does Macaronesia exist? Conflicting signal in the bryophyte and pteridophyte floras. Am. J. Bot. 2007, 94, 625–639. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Schneider, H. Evidence supporting Davallia canariensis as a late miocene relict endemic to macaronesia and atlantic Europe. Aust. Syst. Bot. 2013, 26, 378–385. [Google Scholar] [CrossRef]
- Rumsey, F.J.; Jermy, A.C.; Sheffield, E. The independent gametophytic stage of Trichomanes speciosum Willd. (Hymenophyllaceae), the Killarney Fern and its distribution in the British Isles. Watsonia 1998, 22, 1–19. [Google Scholar]
- Rumsey, F.J.; Vogel, J.C.; Russell, S.J.; Barrett, J.A.; Gibby, M. Climate, Colonisation and Celibacy: Population Structure in Central European Trichomanes speciosum (Pteridophyta). Bot. Acta 1998, 111, 481–489. [Google Scholar] [CrossRef]
- Makgomol, K.; Sheffield, E. Gametophyte morphology and ultrastructure of the extremely deep shade fern, Trichomanes speciosum. New Phytol. 2001, 151, 243–255. [Google Scholar] [CrossRef]
- Makgomol, K.; Sheffield, E. Development of gametophytes from gemmae of Killarney fern (Trichomanes speciosum Willd., Hymenophyllaceae, Pteridophyta). Fern Gaz. 2005, 17, 163–177. [Google Scholar]
- Rumsey, F.J.; Vogel, J.C.; Russell, S.J.; Barrett, J.A.; Gibby, M. Population structure the endangered fern and conservation biology of Trichomanes speciosum Willd. (Hymenophyllaceae) at its northern distributional limit. Biol. J. Linn. Soc. 1999, 66, 333–344. [Google Scholar] [CrossRef]
- Rumsey, F.J.; Barrett, J.A.; Gibby, M.; Russell, S.J.; Vogel, J.C. Reproductive strategies and population structure in the endangered pteridophyte Trichomanes speciosum (Hymenophyllaceae: Pteridophyta). Fern Gaz. 2005, 17, 205–215. [Google Scholar]
- Rumsey, F.J.; Sheffield, E. Inter-generational ecological niche separation and the “independent gametophyte” phenomenom. In Pteridology in Perspective; Camus, J.M., Gibby, M., Johns, R.J., Eds.; Royal Botanic Gardens, Kew: London, UK, 1996; pp. 563–570. [Google Scholar]
- Sang, T.; Crawford, D.J.; Stuessy, T.F. Chloroplast DNA Phylogeny, Reticulate Evolution, and Biogeography of Paeonia (Paeoniaceae). Syst. Bot. 1997, 84, 1120–1136. [Google Scholar] [CrossRef]
- Ruiz-Ruano, F.J.; Navarro-Domínguez, B.; Camacho, J.P.M.; Garrido-Ramos, M.A. Full plastome sequence of the fern Vandenboschia speciosa (Hymenophyllales): Structural singularities and evolutionary insights. J. Plant Res. 2019, 132, 3–17. [Google Scholar] [CrossRef]
- Ebihara, A.; Ishikawa, H.; Matsumoto, S.; Lin, S.-J.; Iwatsuki, K.; Takamiya, M.; Watano, Y.; Ito, M. Nuclear DNA, chloroplast DNA, and ploidy analysis clarified biological complexity of the Vandenboschia radicans complex (Hymenophyllaceae) in Japan and adjacent areas. Am. J. Bot. 2005, 92, 1535–1547. [Google Scholar] [CrossRef]
- Ebihara, A.; Matsumoto, S.; Ito, M. Hybridization involving independent gametophytes in the Vandenboschia radicans complex (Hymenophyllaceae): A new perspective on the distribution of fern hybrids. Mol. Ecol. 2009, 18, 4904–4911. [Google Scholar] [CrossRef]
- Nitta, J.H.; Ebihara, A.; Ito, M. Reticulate evolution in the Crepidomanes minutum species complex (Hymenophyllaceae). Am. J. Bot. 2011, 98, 1782–1800. [Google Scholar] [CrossRef]
- Manton, I. Problems of Cytology and Evolution in the Pteridophyta; Cambridge University Press: Cambridge, UK, 1950. [Google Scholar]
- Ní Dhúill, E. Conservation Biology of the Threatened Killarney Fern (Trichomanes speciosum Willd.) in Ireland. Ph.D. Thesis, Trinity College, University of Dublin, Dublin, Ireland, 2014. [Google Scholar]
- Hall, T. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Schneider, H.; Navarro-Gomez, A.; Russell, S.J.; Ansell, S.; Grundmann, M.; Vogel, J. Exploring the utility of three nuclear regions to reconstruct reticulate evolution in the fern genus Asplenium. J. Syst. Evol. 2013, 51, 142–153. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Lefort, V.; Longueville, J.E.; Gascuel, O. SMS: Smart Model Selection in PhyML. Mol. Biol. Evol. 2017, 34, 2422–2424. [Google Scholar] [CrossRef]
- Gascuel, O. BIONJ: An improved version of the NJ algorithm based on a simple model of sequence data. Mol. Biol. Evol. 1997, 14, 685–695. [Google Scholar] [CrossRef]
- Anisimova, M.; Gascuel, O. Approximate Likelihood-Ratio Test for branches: A fast, accurate, and powerful alternative. Syst. Biol. 2006, 55, 539–552. [Google Scholar] [CrossRef] [PubMed]
- Excoffier, L.; Smouse, P.E.; Quattro, J.M. Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics 1992, 131, 479–491. [Google Scholar] [CrossRef] [PubMed]
- Clement, M.; Posada, D.; Crandall, K.A. TCS: A computer program to estimate gene genealogies. Mol. Ecol. 2000, 9, 1657–1659. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef]
- Nylander, J.A.A. MrModeltest v2; Program Distributed by the Author; Evolutionary Biology Centre, Uppsala University: Uppsala, Sweden, 2004. [Google Scholar]
- Akaike, H. Information theory and an extension of the maximum likelihood principle. In Proceedings of the Second International Symposium on Information Theory; Petrov, B.N., Caski, F., Eds.; Akademiai Kiado: Budapest, Hungary, 1973; pp. 267–281. [Google Scholar]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef]
- Bouckaert, R.; Heled, J.; Kühnert, D.; Vaughan, T.; Wu, C.H.; Xie, D.; Suchard, M.A.; Rambaut, A.; Drummond, A.J. BEAST 2: A Software Platform for Bayesian Evolutionary Analysis. PLoS Comput. Biol. 2014, 10, e1003537. [Google Scholar] [CrossRef]
- Bozukov, V.S. Genus Trichomanes in the European and Bulgarian Cenozoic macroflora. Phytol. Balc. 2008, 14, 3–9. [Google Scholar]
- Levin, R.A.; Wagner, W.L.; Hoch, P.C.; Nepokroeff, M.; Pires, J.C.; Zimmer, E.A.; Sytsma, K.J. Family-level relationships of Onagraceae based on chloroplast rbcL and ndhF data. Am. J. Bot. 2003, 90, 107–115. [Google Scholar] [CrossRef]
- Schuettpelz, E.; Pryer, K.M. Evidence for a Cenozoic radiation of ferns in an angiosperm-dominated canopy. Proc. Natl. Acad. Sci. USA 2009, 106, 11200–11205. [Google Scholar] [CrossRef]
- Testo, W.; Sundue, M. A 4000-species dataset provides new insight into the evolution of ferns. Mol. Phylogenet. Evol. 2016, 105, 200–211. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. JModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef]
- Drummond, A.J.; Ho, S.Y.W.; Phillips, M.J.; Rambaut, A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006, 4, e88. [Google Scholar] [CrossRef]
- Yang, Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef]
- Bouckaert, R.; Vaughan, T.G.; Barido-Sottani, J.; Duchêne, S.; Fourment, M.; Gavryushkina, A.; Heled, J.; Jones, G.; Kühnert, D.; De Maio, N.; et al. BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2019, 15, e1006650. [Google Scholar] [CrossRef]
- Fu, Y.-X. Statistical Tests of Neutrality of Mutations Against Population Growth, Hitchhiking and Background Selection. Genetics 1997, 147, 915–925. [Google Scholar] [CrossRef]
- Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [CrossRef]
- Drummond, A.J.; Rambaut, A.; Shapiro, B.; Pybus, O.G. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol. Biol. Evol. 2005, 22, 1185–1192. [Google Scholar] [CrossRef]
- Müller, N.F.; Rasmussen, D.; Stadler, T. MASCOT: Parameter and state inference under the marginal structured coalescent approximation. Bioinformatics 2018, 34, 3843–3848. [Google Scholar] [CrossRef]
- Barido-Sottani, J.; Bošková, V.; Plessis, L.D.; Kühnert, D.; Magnus, C.; Mitov, V.; Müller, N.F.; Pečerska, J.; Rasmussen, D.A.; Zhang, C.; et al. Taming the BEAST—A Community Teaching Material Resource for BEAST 2. Syst. Biol. 2018, 67, 170–174. [Google Scholar] [CrossRef]
- Hijmans, R.J.; Cameron, S.E.; Parra, J.L.; Jones, P.G.; Jarvis, A. Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. 2005, 25, 1965–1978. [Google Scholar] [CrossRef]
- van Vuuren, D.P.; Edmonds, J.; Kainuma, M.; Riahi, K.; Thomson, A.; Hibbard, K.; Hurtt, G.C.; Kram, T.; Krey, V.; Lamarque, J.-F.; et al. The representative concentration pathways: An overview. Clim. Chang. 2011, 109, 5. [Google Scholar] [CrossRef]
- Louis-Arsène, F. Trichomanes speciosum Willd. en Bretagne. Bull. la Société Bot. Fr. 1953, 100, 6. [Google Scholar] [CrossRef]
- Marchetti, D. Notule pteridologiche italiche. i (1-31). Ann. Del Mus. Civ. Di Rovereto Sez. Archeol. Stor. Sci. Nat. 2002, 16, 371–392. [Google Scholar]
- Krukowski, M.; Świerkosz, K. Discovery of the gametophytes of Trichomanes speciosum (Hymenophyllaceae: Pteridophyta) in Poland and its biogeographical importance. Fern Gaz. 2004, 17, 79–85. [Google Scholar] [CrossRef]
- Loriot, S.; Geslin, J. Trichomanes speciosum Willd. (Hymenophylacceae, Pteridophyta) dans le Massif Armoricain. E.R.I.C.A Bull. Bot. Armor. 2005, 19, 3–22. [Google Scholar]
- Eichler, M.; Kempf, M. Nachuntersuchungen zur Verbreitung des Prächtigen Dünnfarns (Trichomanes speciosum) in Hessen (Art des Anhangs II der FFH-Richtlinie) im Jahr (2011)—Endbericht (2012); Servicezentrum Forsteinrichtung und Naturschutz (FENA): Hessen, Germany, 2012. [Google Scholar]
- Sánchez Velázquez, T. Trichomanes speciosum (Pteridophyta Hymenophyllaceae) en la isla de Gran Canaria, Islas Canarias. Botánica Macaronésica 2013, 28, 83–92. [Google Scholar]
- Sanz-Azkue, I.; Díez-López, J.; Olariaga-Ibarguren, I. Inventory and mapping of red-listed vascular flora in Hernani municipality (Gipuzkoa, Basque Country). Munibe. Ciencias Nat. 2013, 61, 7–32. [Google Scholar]
- Phillips, S.J.; Anderson, R.P.; Schapire, R.E. Maximum entropy modeling of species geographic distributions. Ecol. Modell. 2006, 190, 231–259. [Google Scholar] [CrossRef]
- Martínez, B.; Viejo, R.M.; Carreño, F.; Aranda, S.C. Habitat distribution models for intertidal seaweeds: Responses to climatic and non-climatic drivers. J. Biogeogr. 2012, 39, 1877–1890. [Google Scholar] [CrossRef]
- Pinson, J.B.; Chambers, S.M.; Nitta, J.H.; Kuo, L.Y.; Sessa, E.B. The separation of generations: Biology and biogeography of Long-Lived sporophyteless fern gametophytes. Int. J. Plant Sci. 2017, 178, 1–18. [Google Scholar] [CrossRef]
- Ebihara, A.; Dubuisson, J.-Y.; Iwatsuki, K.; Ito, M. Systematics of Trichomanes (Hymenophyllaceae: Pteridophyta), progress and future interests. Fern Gaz. 2007, 18, 53–58. [Google Scholar]
- Soltis, P.S.; Soltis, D.E. The role of hybridization in plant speciation. Annu. Rev. Plant Biol. 2009, 60, 561–588. [Google Scholar] [CrossRef]
- Mosbrugger, V.; Utescher, T.; Dilcher, D.L. Cenozoic continental climatic evolution of Central Europe. Proc. Natl. Acad. Sci. USA 2005, 102, 14964–14969. [Google Scholar] [CrossRef]
- Zachos, J.; Pagani, H.; Sloan, L.; Thomas, E.; Billups, K. Trends, rhythms, and aberrations in global climate 65 Ma to present. Science 2001, 292, 686–693. [Google Scholar] [CrossRef]
- Palamarev, E.; Petkova, A. The Paleogene macroflora of the Rhodopes region. I. Polypodiophyta–Polypodiopsida. Fitologiya 1990, 38, 3–22. [Google Scholar]
- Lafontaine, J.D.; Wood, D.M. A zoogeographic analysis of the Noctuidae (Lepidoptera) of Beringia, and some inferences about past Beringian habitats. Mem. Entomol. Soc. Canada 1988, 120, 109–123. [Google Scholar] [CrossRef]
- Kryshtofovich, A. A Final Link between the Tertiary Floras of Asia and Europe (Contribution to the Age of the Arcto-Tertiary Floras of the Northern Holarctic). New Phytol. 1935, 34, 339–344. [Google Scholar] [CrossRef]
- Krijgsman, W.; Hilgen, F.J.; Raffi, I.; Sierro, F.J.; Wilson, D.S. Chronology, causes and progression of the Messinian salinity crisis. Nature 1999, 400, 652–655. [Google Scholar] [CrossRef]
- Kovar-Eder, J.; Kvaček, Z.; Martinetto, E.; Roiron, P. Late Miocene to Early Pliocene vegetation of southern Europe (7-4 Ma) as reflected in the megafossil plant record. Palaeogeogr. Palaeoclimatol. Palaeoecol. 2006, 238, 321–339. [Google Scholar] [CrossRef]
- Utescher, T.; Erdei, B.; François, L.; Mosbrugger, V. Tree diversity in the Miocene forests of Western Eurasia. Palaeogeogr. Palaeoclimatol. Palaeoecol. 2007, 253, 226–250. [Google Scholar] [CrossRef]
- Suc, J.P. Origin and evolution of the mediterranean vegetation and climate in Europe. Nature 1984, 307, 429–432. [Google Scholar] [CrossRef]
- Benito Garzón, M.; Sainz Ollero, H. Potencialidad del Elemento Lauroide en la Península Ibérica. Master’s Thesis, Universidad Autónoma de Madrid, Madrid, Spain, 2002. [Google Scholar]
- Beatty, G.E.; Provan, J. Post-glacial dispersal, rather than in situ glacial survival, best explains the disjunct distribution of the Lusitanian plant species Daboecia cantabrica (Ericaceae). J. Biogeogr. 2013, 40, 335–344. [Google Scholar] [CrossRef]
- Beatty, G.E.; Provan, J. Phylogeographical analysis of two cold-tolerant plants with disjunct Lusitanian distributions does not support in situ survival during the last glaciation. J. Biogeogr. 2014, 41, 2185–2193. [Google Scholar] [CrossRef]
- Durães, C.J. Genetic Variation and Evolutionary History of the Macaronesian fern Asplenium hemionitis L. Ph.D. Thesis, Edinburgh University, Edinburgh, UK, 2005. [Google Scholar]
- Perrie, L.R.; Ohlsen, D.J.; Shepherd, L.D.; Garrett, M.; Brownsey, P.J.; Bayly, M.J. Tasmanian and Victorian populations of the fern Asplenium hookerianum result from independent dispersals from New Zealand. Aust. Syst. Bot. 2010, 23, 387–392. [Google Scholar] [CrossRef]
- Ranker, T.A.; Floyd, S.K.; Trapp, P.G. Multiple Colonizations of Asplenium adiantum-nigrum Onto the Hawaiian Archipelago. Evolution 1994, 48, 1364–1370. [Google Scholar] [CrossRef]
- Kondraskov, P.; Schütz, N.; Schüßler, C.; De Sequeira, M.M.; Guerra, A.S.; Caujapé-Castells, J.; Jaén-Molina, R.; Marrero-Rodríguez, Á.; Koch, M.A.; Linder, P.; et al. Biogeography of mediterranean hotspot biodiversity: Re-evaluating the tertiary relict’ hypothesis of macaronesian laurel forests. PLoS ONE 2015, 10, e0132091. [Google Scholar] [CrossRef]
- Góis-Marques, C.A.; Madeira, J.; Menezes de Sequeira, M. Inventory and review of the Mio–Pleistocene São Jorge flora (Madeira Island, Portugal): Palaeoecological and biogeographical implications. J. Syst. Palaeontol. 2018, 16, 159–177. [Google Scholar] [CrossRef]
- García-Talavera, F. La Macaronesia. Consideraciones geológicas, biogeográficas y paleoecológicas. In Ecología y Cultura en Canarias; Fernández-Palacios, J.M., Bacallado, J.J., Belmonte, J.A., Eds.; Museo de la Ciencia, Cabildo Insular de Tenerife: Santa Cruz de Tenerife, Spain, 1999; pp. 37–63. ISBN 84-88594-20-8. [Google Scholar]
- Fernández-Palacios, J.M.; De Nascimento, L.; Otto, R.; Delgado, J.D.; García-Del-Rey, E.; Arévalo, J.R.; Whittaker, R.J. A reconstruction of Palaeo-Macaronesia, with particular reference to the long-term biogeography of the Atlantic island laurel forests. J. Biogeogr. 2011, 38, 226–246. [Google Scholar] [CrossRef]
- Góis-Marques, C.A.; de Nascimento, L.; Menezes de Sequeira, M.; Fernández-Palacios, J.M.; Madeira, J. The Quaternary plant fossil record from the volcanic Azores Archipelago (Portugal, North Atlantic Ocean): A review. Hist. Biol. 2018, 31, 1267–1283. [Google Scholar] [CrossRef]
- Robinson, R.C. Invasive and problem ferns: A European perspective. Int. Urban Ecol. 2009, 4, 83–90. [Google Scholar]
| psbA-trnH | gapC-572 | gapC-575 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Code | ha | Priv | Hd | π | ha | Priv | Hd | π | ha | Priv | Hd | π |
| Andalusia | 2 | 0.425 | 0.0019 | 4 | 1 | 0.193 | 0.0087 | 8 | 4 | 0.340 | 0.0045 | |
| ALM | 1 | 0 | 0 | 2 | 0.285 | 0.0124 | 2 | 1 | 0.182 | 0.0019 | ||
| COQ | 1 | 0 | 0 | 2 | 1 | 0.222 | 0.0096 | 3 | 1 | 0.416 | 0.0048 | |
| CRM | 1 | 0 | 0 | |||||||||
| MCH | 2 | 0.355 | 0.0016 | 1 | 0 | 0 | 4 | 1 | 0.371 | 0.0058 | ||
| SCD | 1 | 0 | 0 | |||||||||
| SDN | 1 | 0 | 0 | |||||||||
| VIF | 1 | 0 | 0 | 2 | 0.250 | 0.0108 | 3 | 1 | 0.464 | 0.0054 | ||
| Azores | 4 | 2 | 0.233 | 0.0008 | 7 | 2 | 0.326 | 0.0153 | 8 | 3 | 0.371 | 0.0048 |
| CAR | 3 | 2 | 0.377 | 0.0009 | 2 | 0.200 | 0.0087 | 3 | 1 | 0.345 | 0.0055 | |
| CID | 2 | 0.200 | 0.0009 | 3 | 0.524 | 0.0248 | 3 | 1 | 0.345 | 0.0039 | ||
| CON | 2 | 0.200 | 0.0009 | 3 | 1 | 0.464 | 0.0217 | 3 | 1 | 0.318 | 0.0036 | |
| NAT | 2 | 0.200 | 0.0009 | 2 | 1 | 0.222 | 0.0097 | 3 | 0.472 | 0.0055 | ||
| Basque Country | 5 | 1 | 0.673 | 0.0030 | 6 | 1 | 0.331 | 0.0155 | 4 | 3 | 0.221 | 0.0025 |
| AZK | 3 | 0.644 | 0.0029 | 4 | 1 | 0.643 | 0.0326 | 2 | 1 | 0.286 | 0.0031 | |
| ERR | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | |||
| ITU | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | |||
| USO | 5 | 1 | 0.767 | 0.0036 | 3 | 0.378 | 0.0174 | 3 | 2 | 0.464 | 0.0054 | |
| Canary Isl. | 3 | 1 | 0.405 | 0.0013 | 5 | 3 | 0.192 | 0.0087 | 8 | 4 | 0.348 | 0.0041 |
| ANC | 1 | 0 | 0 | 1 | 0 | 0 | 2 | 0.333 | 0.0036 | |||
| CED | 1 | 0 | 0 | 2 | 1 | 0.286 | 0.0124 | 3 | 1 | 0.524 | 0.0062 | |
| IJU | 3 | 0.711 | 0.0032 | 3 | 1 | 0.417 | 0.0193 | 2 | 0.222 | 0.0024 | ||
| PIJ | 2 | 0.467 | 0.0010 | 2 | 1 | 0.182 | 0.0079 | 3 | 1 | 0.345 | 0.0040 | |
| ZAR | 2 | 0.467 | 0.0010 | 1 | 0 | 0 | 4 | 2 | 0.396 | 0.0047 | ||
| Czech Republic | 1 | 0 | 0 | 3 | 1 | 0.257 | 0.0116 | 6 | 5 | 0.447 | 0.0076 | |
| HAR | 1 | 0 | 0 | 2 | 1 | 0.400 | 0.0174 | 1 | 0 | 0 | ||
| MUZ | 1 | 0 | 0 | 2 | 0.667 | 0.0290 | 5 | 4 | 0.786 | 0.0163 | ||
| SKA | 1 | 0 | 0 | 1 | 0 | 0 | 2 | 1 | 0.333 | 0.0036 | ||
| Galicia | 3 | 0.567 | 0.0025 | 4 | 1 | 0.249 | 0.0113 | 6 | 3 | 0.490 | 0.0060 | |
| EUM | 2 | 0.533 | 0.0024 | 2 | 0.133 | 0.0058 | 6 | 3 | 0.778 | 0.0109 | ||
| SEI | 2 | 0.356 | 0.0016 | 3 | 1 | 0.464 | 0.0218 | 1 | 0 | 0 | ||
| Ire-Wal-Bri * | 4 | 1 | 0.611 | 0.0026 | 5 | 3 | 0.197 | 0.0089 | 10 | 7 | 0.379 | 0.0051 |
| COR | 2 | 1 | 0.200 | 0.0004 | 3 | 1 | 0.295 | 0.0134 | 2 | 1 | 0.222 | 0.0024 |
| DEV | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | |||
| LIM | 1 | 0 | 0 | 1 | 0 | 0 | 5 | 2 | 0.576 | 0.0072 | ||
| TAU | 2 | 0.533 | 0.0024 | 2 | 1 | 0.667 | 0.0290 | 1 | 0 | 0 | ||
| WAT | 2 | 0.467 | 0.0021 | 2 | 1 | 0.222 | 0.0097 | 5 | 4 | 0.667 | 0.0109 | |
| Italy | 1 | 0 | 0 | 7 | 3 | 0.521 | 0.0261 | 6 | 4 | 0.515 | 0.0077 | |
| SER | 1 | 0 | 0 | 5 | 2 | 0.667 | 0.0348 | 3 | 2 | 0.417 | 0.0072 | |
| STA | 1 | 0 | 0 | 3 | 1 | 0.378 | 0.0174 | 4 | 2 | 0.643 | 0.0081 | |
| Luxembourg | 1 | 0 | 0 | 4 | 2 | 0.331 | 0.0153 | 2 | 1 | 0.100 | 0.0011 | |
| ARD | 1 | 0 | 0 | 3 | 1 | 0.524 | 0.0248 | 1 | 0 | 0 | ||
| BEA | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | |||
| ROL | 1 | 0 | 0 | 2 | 1 | 0.333 | 0.0145 | 2 | 1 | 0.333 | 0.0036 | |
| Madeira | 2 | 0.351 | 0.0015 | 2 | 0.133 | 0.0058 | 1 | 0 | 0 | |||
| FRI | 2 | 0.467 | 0.0021 | 2 | 0.200 | 0.0087 | 1 | 0 | 0 | |||
| POR | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | |||
| URZ | 2 | 0.400 | 0.0018 | |||||||||
| Vosges du Nord | 2 | 0.356 | 0.0016 | 4 | 1 | 0.533 | 0.0261 | 4 | 0.350 | 0.0041 | ||
| BIT | 2 | 0.600 | 0.0027 | 2 | 0.400 | 0.0174 | 1 | 0 | 0 | |||
| PIE | 1 | 0 | 0 | 3 | 1 | 0.700 | 0.0348 | 4 | 0.714 | 0.0093 | ||
| Source of Variation | d.f | Sum of Squares | Variance Components | Percentage of Variation | Fixation Indices | p-Value * |
|---|---|---|---|---|---|---|
| trnH-psbA | ||||||
| Among generations | 1 | 4.042 | 0.00773 Va | 1.084 | FCT = 0.011 | 0.21495 |
| Among populations within generations | 62 | 160.115 | 0.49222 Vb | 68.954 | FSC = 0.7 | <0.001 |
| Within populations | 245 | 52.400 | 0.21388 Vc | 29.962 | FST = 0.7 | <0.001 |
| Total | 308 | 216.557 | 0.71383 | |||
| Among geographical regions | 10 | 48.120 | 0.14279 Va | 38.94 | FCT = 0.39 | <0.001 |
| Among populations within regions | 29 | 25.525 | 0.09971 Vb | 27.19 | FSC = 0.45 | <0.001 |
| Within populations | 269 | 33.417 | 0.12423 Vc | 33.87 | FST = 0.66 | <0.001 |
| Total | 308 | 107.061 | 0.36672 | |||
| Among evolutionary units (3 units) | 2 | 34.062 | 0.16218 Va | 39.47 | FCT = 0.39 | <0.001 |
| Among populations within units | 37 | 39.583 | 0.12447 Vb | 30.29 | FSC = 0.5 | <0.001 |
| Within populations | 269 | 33.417 | 0.12423 Vc | 30.23 | FST = 0.7 | <0.001 |
| Total | 308 | 107.061 | 0.41087 | |||
| Among evolutionary units (2 units) | 1 | 31.022 | 0.23645 Va | 50.77 | FCT = 0.51 | <0.001 |
| Among populations within units | 32 | 33.948 | 0.13004 Vb | 27.92 | FSC = 0.57 | <0.001 |
| Within populations | 220 | 21.833 | 0.09924 Vc | 21.31 | FST = 0.78 | <0.001 |
| Total | 253 | 86.803 | 0.46572 | |||
| gapCp-572 copy | ||||||
| Among geographical regions | 10 | 1.298 | 0.00037 Va | 0.27 | FCT = 0.003 | 0.34851 |
| Among populations within groups | 25 | 3.066 | −0.00218 Vb | 0 | FSC = −0.016 | 0.72851 |
| Within populations | 235 | 32.544 | 0.13848 Vc | 99.73 | FST = −0.013 | 0.76386 |
| Total | 270 | 36.908 | 0.13667 | |||
| gapCp-575 copy | ||||||
| Among regions (11 groups) | 10 | 1.651 | −0.00084 Va | −0.50 | FCT = −0.005 | 0.69554 |
| Among populations within groups | 24 | 4.518 | 0.00230 Vb | 1.36 | FSC = 0.013 | 0.14871 |
| Within populations | 275 | 46.183 | 0.16794 Vc | 99.14 | FST = 0.008 | 0.25485 |
| Total | 309 | 52.352 | 0.1694 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ben-Menni Schuler, S.; Hamza, H.; Blanca, G.; Romero-García, A.T.; Suárez-Santiago, V.N. Phylogeographical Analyses of a Relict Fern of Palaeotropical Flora (Vandenboschia speciosa): Distribution and Diversity Model in Relation to the Geological and Climate Events of the Late Miocene and Early Pliocene. Plants 2022, 11, 839. https://doi.org/10.3390/plants11070839
Ben-Menni Schuler S, Hamza H, Blanca G, Romero-García AT, Suárez-Santiago VN. Phylogeographical Analyses of a Relict Fern of Palaeotropical Flora (Vandenboschia speciosa): Distribution and Diversity Model in Relation to the Geological and Climate Events of the Late Miocene and Early Pliocene. Plants. 2022; 11(7):839. https://doi.org/10.3390/plants11070839
Chicago/Turabian StyleBen-Menni Schuler, Samira, Hammadi Hamza, Gabriel Blanca, Ana Teresa Romero-García, and Víctor N. Suárez-Santiago. 2022. "Phylogeographical Analyses of a Relict Fern of Palaeotropical Flora (Vandenboschia speciosa): Distribution and Diversity Model in Relation to the Geological and Climate Events of the Late Miocene and Early Pliocene" Plants 11, no. 7: 839. https://doi.org/10.3390/plants11070839
APA StyleBen-Menni Schuler, S., Hamza, H., Blanca, G., Romero-García, A. T., & Suárez-Santiago, V. N. (2022). Phylogeographical Analyses of a Relict Fern of Palaeotropical Flora (Vandenboschia speciosa): Distribution and Diversity Model in Relation to the Geological and Climate Events of the Late Miocene and Early Pliocene. Plants, 11(7), 839. https://doi.org/10.3390/plants11070839







