Transcriptomic Reprogramming, Alternative Splicing and RNA Methylation in Potato (Solanum tuberosum L.) Plants in Response to Potato Virus Y Infection
Abstract
1. Introduction
2. Results and Discussion
2.1. Impact of Elevated Temperature on the Susceptibility of Potato Plants (cv Chicago) to PVY
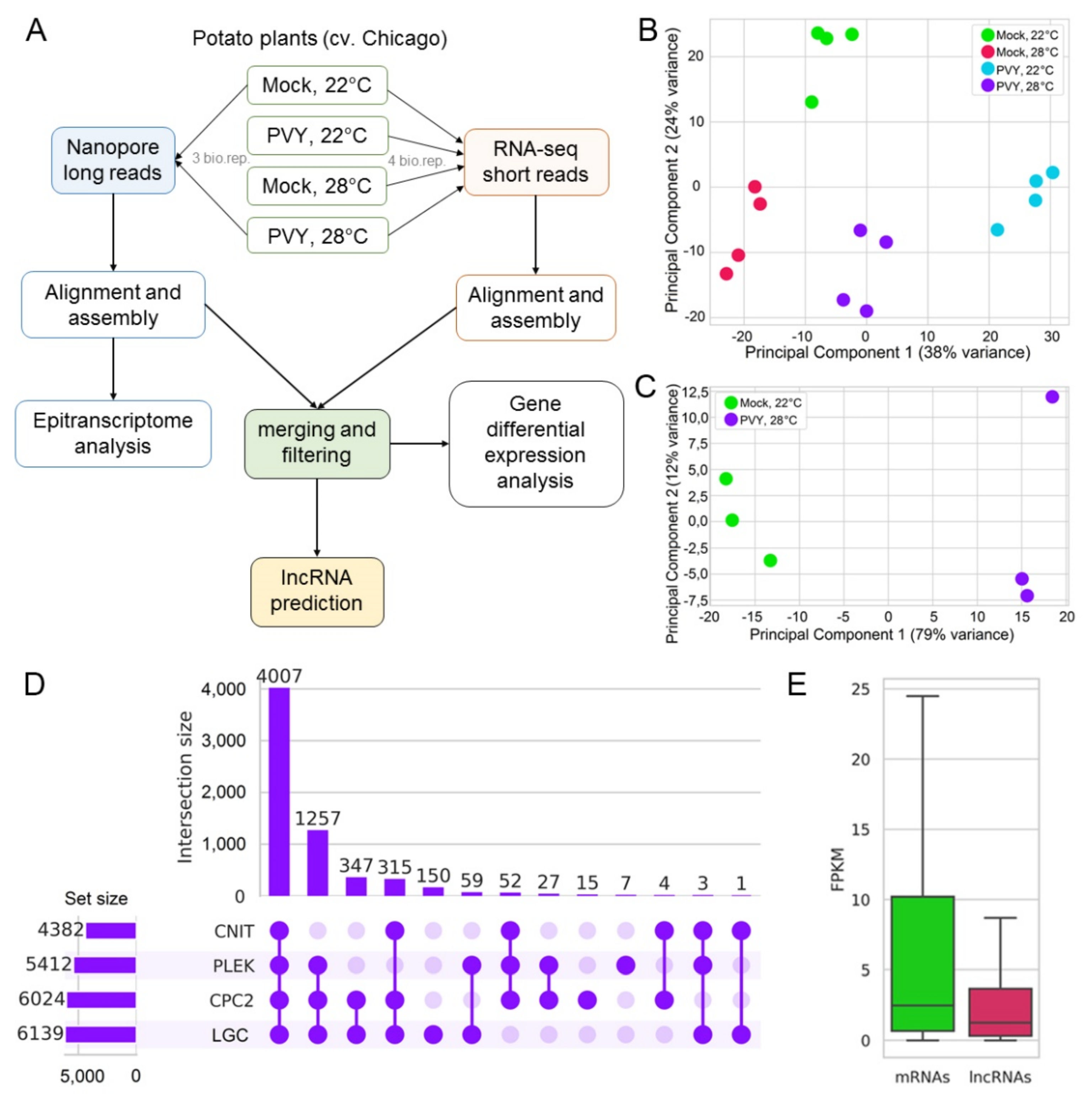
2.2. Transcriptome Analysis: Overview
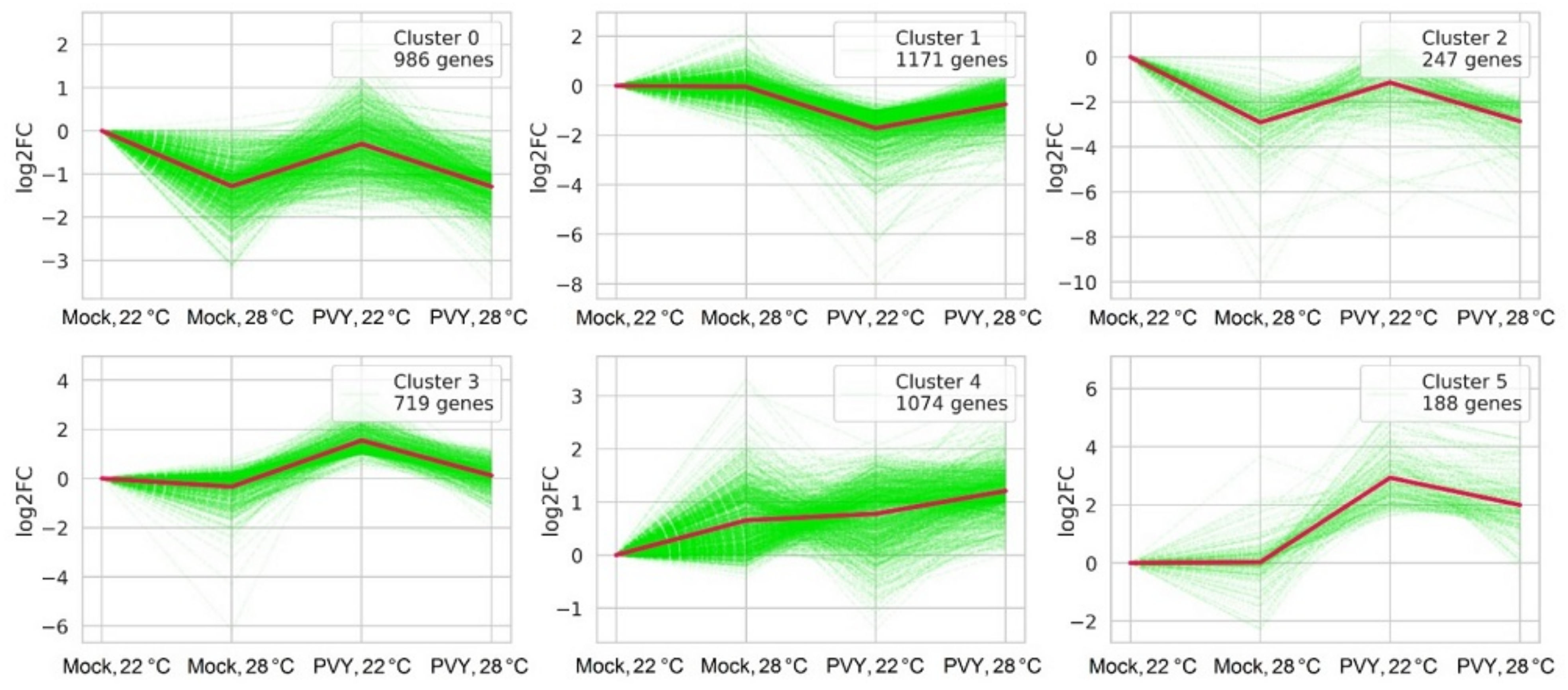
2.3. Differential Expression of Genes under Stress Conditions
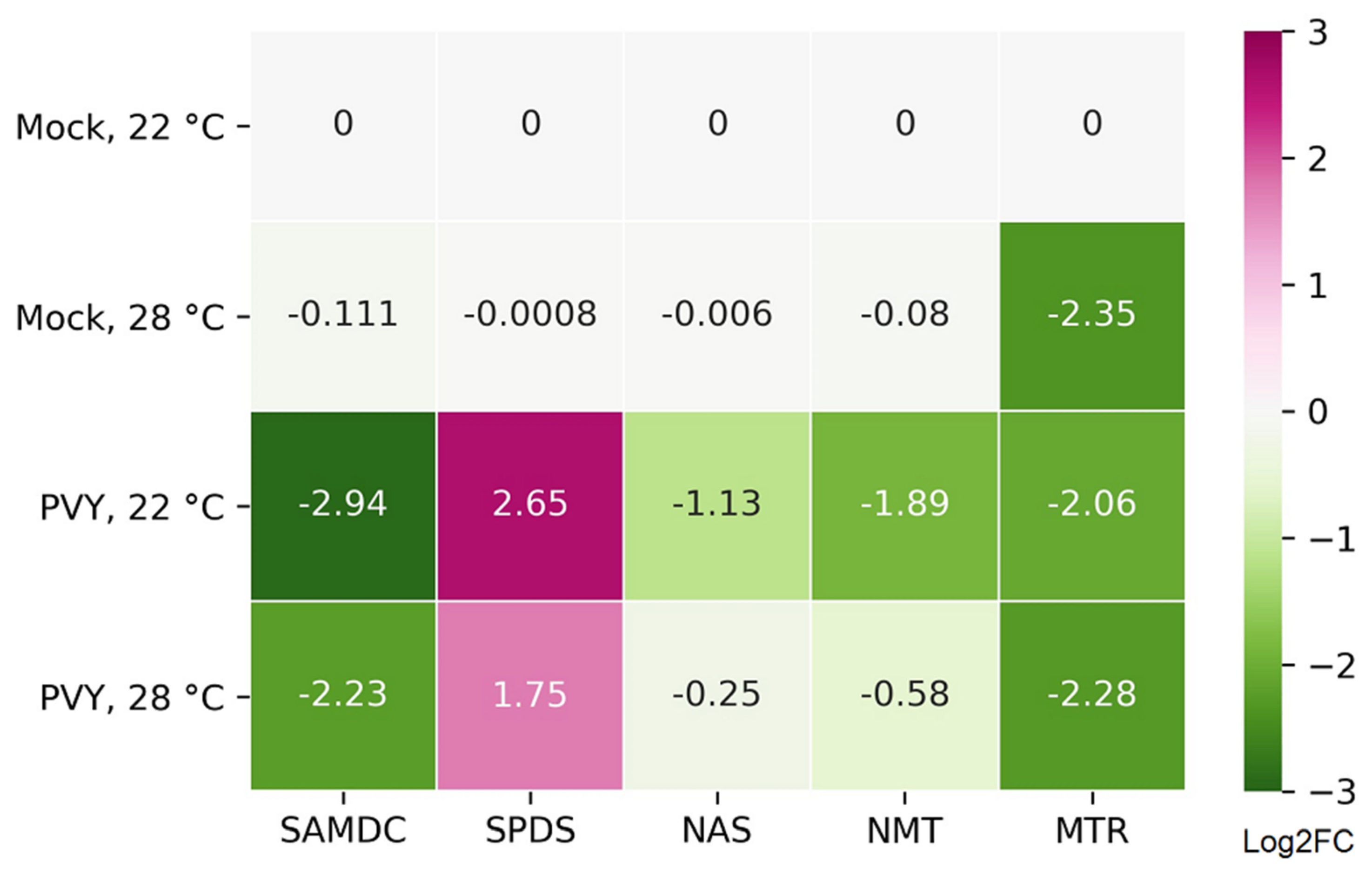
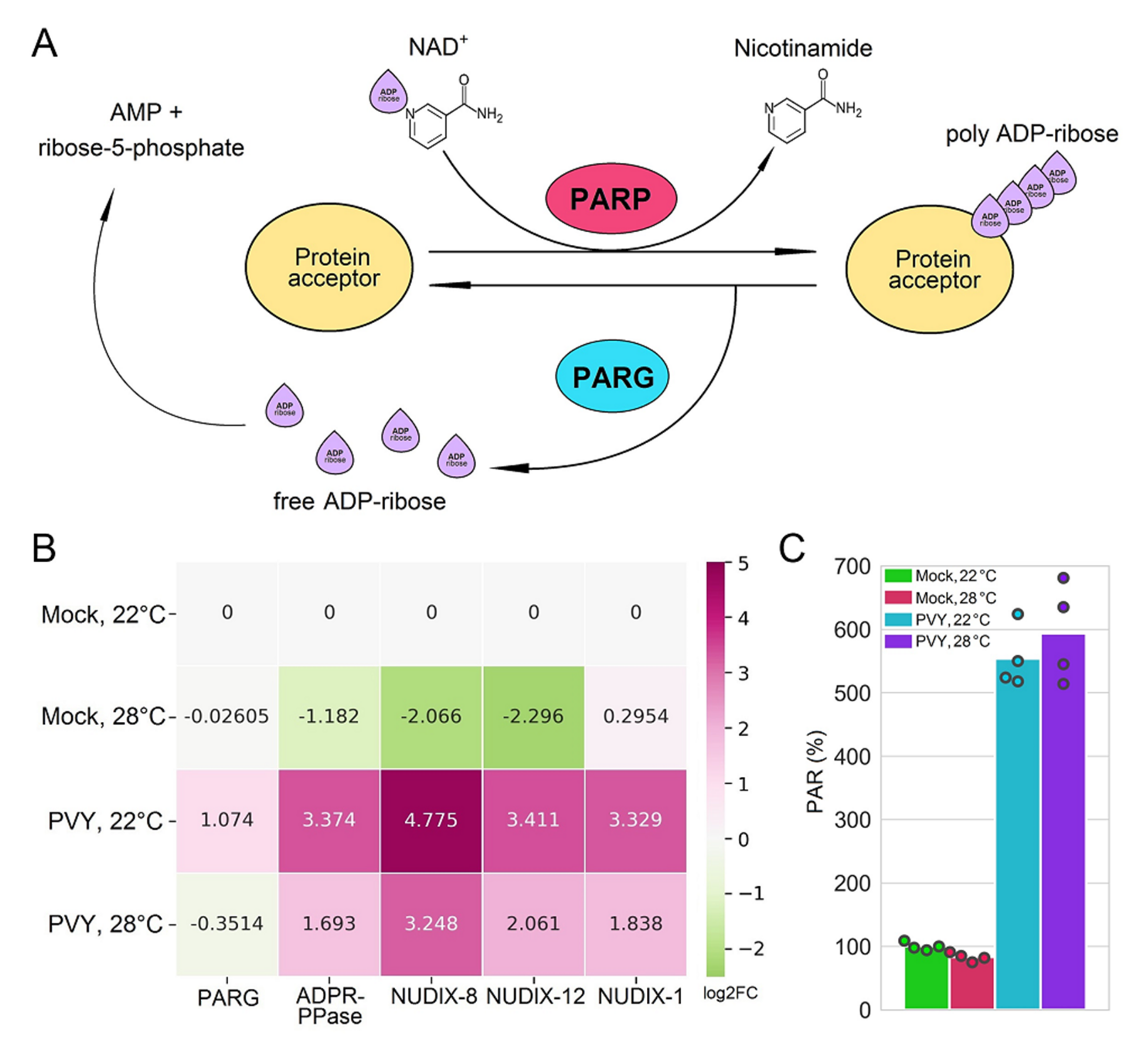
2.4. Functional Relevance of Differentially Expressed Protein-Coding RNAs
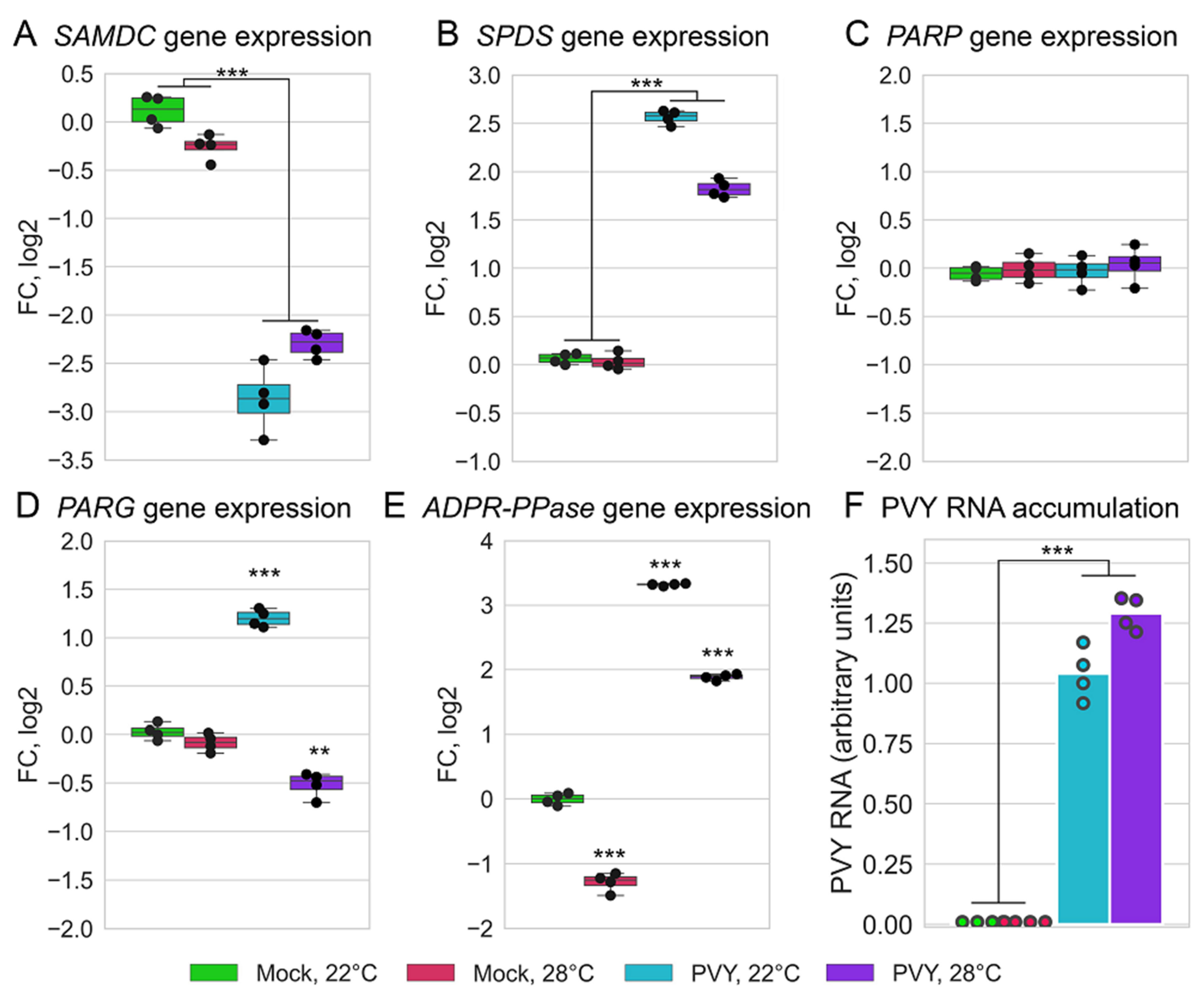
2.5. PVY Accumulation and Validation of Differentially Expressed Genes by qRT-PCR
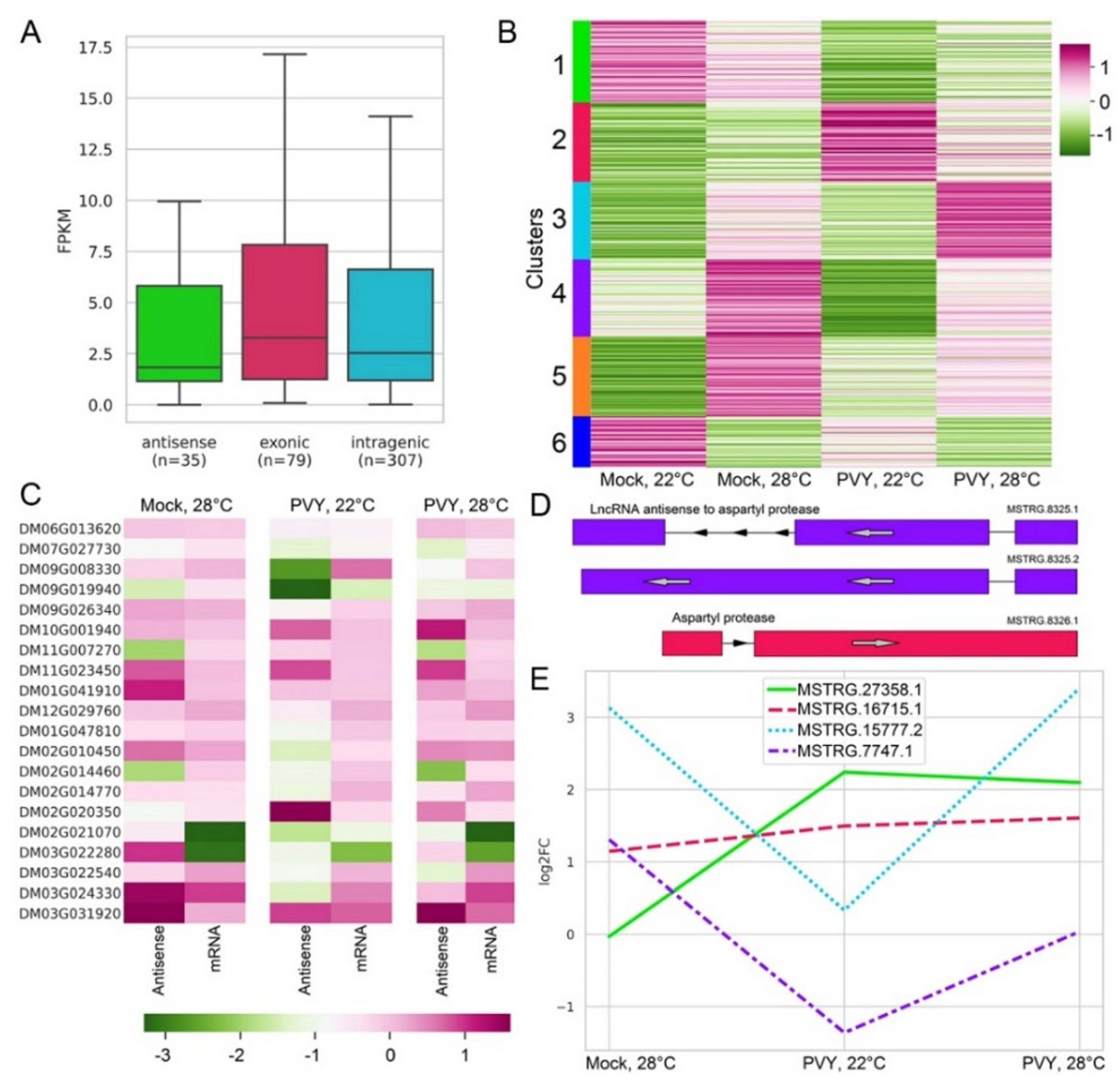
2.6. Differential Expression of LncRNAs under Stress Conditions
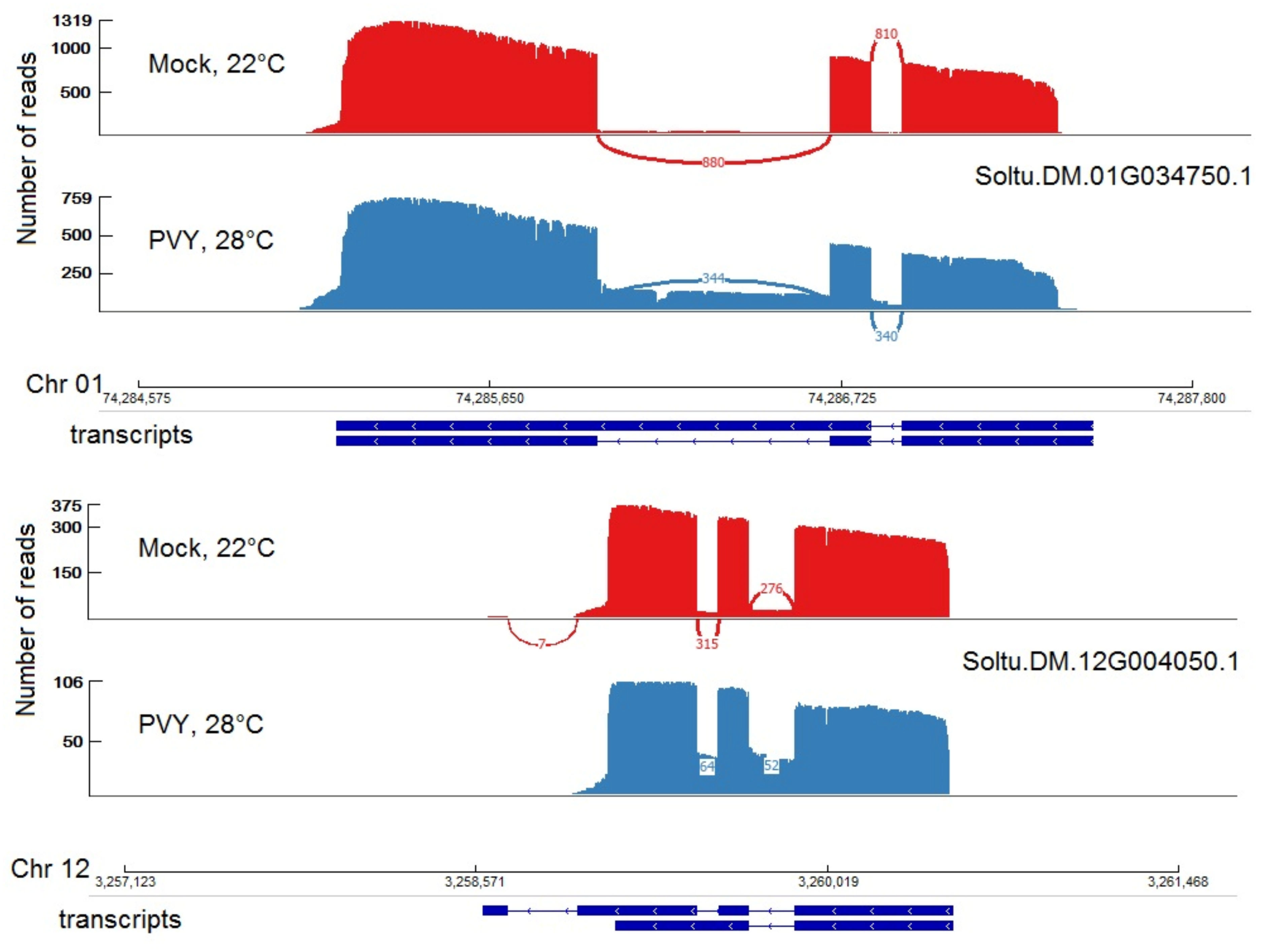
2.7. Differentially Expressed Isoforms: Alternative Splicing
2.8. Epitranscriptome Analysis: m6A RNA Methylation
3. Materials and Methods
3.1. Plant Material
3.2. RNA Isolation, Library Preparation, and Sequencing
3.3. Alignment to Genome and Transcriptome Assembly
3.4. Differential Expression Analysis
3.5. GO Analysis
3.6. Functional Annotation
3.7. Isoform Expression and Epitranscriptome Analysis
3.8. Immunological Detection of Poly ADP-Ribose (PAR)
3.9. RTqPCR
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Whitfield, A.E.; Falk, B.W.; Rotenberg, D. Insect Vector-Mediated Transmission of Plant Viruses. Virology 2015, 479–480, 278–289. [Google Scholar] [CrossRef] [PubMed]
- Torrance, L.; Talianksy, M.E. Potato Virus Y Emergence and Evolution from the Andes of South America to Become a Major Destructive Pathogen of Potato and Other Solanaceous Crops Worldwide. Viruses 2020, 12, 1430. [Google Scholar] [CrossRef]
- Kreuze, J.F.; Vaira, A.M.; Menzel, W.; Candresse, T.; Zavriev, S.K.; Hammond, J.; Hyun Ryu, K.; Report Consortium, I. ICTV Virus Taxonomy Profile: Alphaflexiviridae. J. Gen. Virol. 2020, 101, 699–700. [Google Scholar] [CrossRef]
- Niehl, A.; Wyrsch, I.; Boller, T.; Heinlein, M. Double-Stranded RNAs Induce a Pattern-Triggered Immune Signaling Pathway in Plants. New Phytol. 2016, 211, 1008–1019. [Google Scholar] [CrossRef]
- Niehl, A.; Heinlein, M. Perception of Double-Stranded RNA in Plant Antiviral Immunity. Mol. Plant Pathol. 2019, 20, 1203–1210. [Google Scholar] [CrossRef]
- Kørner, C.J.; Klauser, D.; Niehl, A.; Domínguez-Ferreras, A.; Chinchilla, D.; Boller, T.; Heinlein, M.; Hann, D.R. The Immunity Regulator BAK1 Contributes to Resistance against Diverse RNA Viruses. Mol. Plant-Microbe Interact. 2013, 26, 1271–1280. [Google Scholar] [CrossRef]
- Nicaise, V.; Candresse, T. Plum Pox Virus Capsid Protein Suppresses Plant Pathogen-Associated Molecular Pattern (PAMP)-Triggered Immunity. Mol. Plant Pathol. 2017, 18, 878–886. [Google Scholar] [CrossRef] [PubMed]
- Zvereva, A.S.; Golyaev, V.; Turco, S.; Gubaeva, E.G.; Rajeswaran, R.; Schepetilnikov, M.V.; Srour, O.; Ryabova, L.A.; Boller, T.; Pooggin, M.M. Viral Protein Suppresses Oxidative Burst and Salicylic Acid-Dependent Autophagy and Facilitates Bacterial Growth on Virus-Infected Plants. New Phytol. 2016, 211, 1020–1034. [Google Scholar] [CrossRef]
- Baebler, Š.; Coll, A.; Gruden, K. Plant Molecular Responses to Potato Virus Y: A Continuum of Outcomes from Sensitivity and Tolerance to Resistance. Viruses 2020, 12, 217. [Google Scholar] [CrossRef]
- Vreugdenhil, D.; Bradshaw, J.; Gebhardt, C.; Govers, F.; Taylor, M.A.; MacKerron, D.K.L.; Ross, H.A. Potato Biology and Biotechnology: Advances and Perspectives; Elsevier: Amsterdam, The Netherlands, 2011; ISBN 978-0-08-052505-1. [Google Scholar]
- Gebhardt, C.; Valkonen, J.P. Organization of Genes Controlling Disease Resistance in the Potato Genome. Annu. Rev. Phytopathol. 2001, 39, 79–102. [Google Scholar] [CrossRef] [PubMed]
- Torrance, L.; Cowan, G.H.; McLean, K.; MacFarlane, S.; Al-Abedy, A.N.; Armstrong, M.; Lim, T.-Y.; Hein, I.; Bryan, G.J. Natural Resistance to Potato Virus Y in Solanum Tuberosum Group Phureja. TAG Theor. Appl. Genet. Theor. Angew. Genet. 2020, 133, 967–980. [Google Scholar] [CrossRef]
- Ding, S.-W. RNA-Based Antiviral Immunity. Nat. Rev. Immunol. 2010, 10, 632–644. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Li, Y. Dissection of RNAi-Based Antiviral Immunity in Plants. Curr. Opin. Virol. 2018, 32, 88–99. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Li, Y.; Ding, S.-W. Small RNA-Based Antimicrobial Immunity. Nat. Rev. Immunol. 2019, 19, 31–44. [Google Scholar] [CrossRef]
- Ghoshal, B.; Sanfaçon, H. Symptom Recovery in Virus-Infected Plants: Revisiting the Role of RNA Silencing Mechanisms. Virology 2015, 479–480, 167–179. [Google Scholar] [CrossRef] [PubMed]
- Baulcombe, D. RNA Silencing. Trends Biochem. Sci. 2005, 30, 290–293. [Google Scholar] [CrossRef] [PubMed]
- Mlotshwa, S.; Pruss, G.J.; Peragine, A.; Endres, M.W.; Li, J.; Chen, X.; Poethig, R.S.; Bowman, L.H.; Vance, V. DICER-LIKE2 Plays a Primary Role in Transitive Silencing of Transgenes in Arabidopsis. PLoS ONE 2008, 3, e1755. [Google Scholar] [CrossRef]
- Alvarado, V.; Scholthof, H.B. Plant Responses against Invasive Nucleic Acids: RNA Silencing and Its Suppression by Plant Viral Pathogens. Semin. Cell Dev. Biol. 2009, 20, 1032–1040. [Google Scholar] [CrossRef]
- Mäkinen, K.; De, S. The Significance of Methionine Cycle Enzymes in Plant Virus Infections. Curr. Opin. Plant Biol. 2019, 50, 67–75. [Google Scholar] [CrossRef]
- Fesenko, I.; Spechenkova, N.; Mamaeva, A.; Makhotenko, A.V.; Love, A.J.; Kalinina, N.O.; Taliansky, M. Role of the Methionine Cycle in the Temperature-Sensitive Responses of Potato Plants to Potato Virus Y. Mol. Plant Pathol. 2021, 22, 77–91. [Google Scholar] [CrossRef] [PubMed]
- Spechenkova, N.; Fesenko, I.A.; Mamaeva, A.; Suprunova, T.P.; Kalinina, N.O.; Love, A.J.; Taliansky, M. The Resistance Responses of Potato Plants to Potato Virus Y Are Associated with an Increased Cellular Methionine Content and an Altered SAM:SAH Methylation Index. Viruses 2021, 13, 955. [Google Scholar] [CrossRef] [PubMed]
- Shaw, J.; Love, A.J.; Makarova, S.S.; Kalinina, N.O.; Harrison, B.D.; Taliansky, M.E. Coilin, the Signature Protein of Cajal Bodies, Differentially Modulates the Interactions of Plants with Viruses in Widely Different Taxa. Nucl. Austin Tex 2014, 5, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Shaw, J.; Yu, C.; Makhotenko, A.V.; Makarova, S.S.; Love, A.J.; Kalinina, N.O.; MacFarlane, S.; Chen, J.; Taliansky, M.E. Interaction of a Plant Virus Protein with the Signature Cajal Body Protein Coilin Facilitates Salicylic Acid-Mediated Plant Defence Responses. New Phytol. 2019, 224, 439–453. [Google Scholar] [CrossRef]
- Song, L.; Wang, J.; Jia, H.; Kamran, A.; Qin, Y.; Liu, Y.; Hao, K.; Han, F.; Zhang, C.; Li, B.; et al. Identification and Functional Characterization of NbMLP28, a Novel MLP-like Protein 28 Enhancing Potato Virus Y Resistance in Nicotiana Benthamiana. BMC Microbiol. 2020, 20, 55. [Google Scholar] [CrossRef]
- Van Dam, J.; Kooman, P.L.; Struik, P.C. Effects of Temperature and Photoperiod on Early Growth and Final Number of Tubers in Potato (Solanum tuberosum L.). Potato Res. 1996, 39, 51–62. [Google Scholar] [CrossRef]
- Bita, C.E.; Gerats, T. Plant Tolerance to High Temperature in a Changing Environment: Scientific Fundamentals and Production of Heat Stress-Tolerant Crops. Front. Plant Sci. 2013, 4, 273. [Google Scholar] [CrossRef]
- Prasch, C.M.; Sonnewald, U. Simultaneous Application of Heat, Drought, and Virus to Arabidopsis Plants Reveals Significant Shifts in Signaling Networks. Plant Physiol. 2013, 162, 1849–1866. [Google Scholar] [CrossRef]
- Bradshaw, J.E.; Ramsay, G. Utilisation of the Commonwealth Potato Collection in Potato Breeding. Euphytica 2005, 146, 9–19. [Google Scholar] [CrossRef]
- Solomon-Blackburn, R.M.; Bradshaw, J.E. Resistance to Potato Virus Y in a Multitrait Potato Breeding Scheme without Direct Selection in Each Generation. Potato Res. 2007, 50, 87–95. [Google Scholar] [CrossRef]
- Anfoka, G.; Moshe, A.; Fridman, L.; Amrani, L.; Rotem, O.; Kolot, M.; Zeidan, M.; Czosnek, H.; Gorovits, R. Tomato Yellow Leaf Curl Virus Infection Mitigates the Heat Stress Response of Plants Grown at High Temperatures. Sci. Rep. 2016, 6, 19715. [Google Scholar] [CrossRef]
- Makarova, S.; Makhotenko, A.; Spechenkova, N.; Love, A.J.; Kalinina, N.O.; Taliansky, M. Interactive Responses of Potato (Solanum tuberosum L.) Plants to Heat Stress and Infection With Potato Virus Y. Front. Microbiol. 2018, 9, 2582. [Google Scholar] [CrossRef] [PubMed]
- Szittya, G. Low Temperature Inhibits RNA Silencing-Mediated Defence by the Control of SiRNA Generation. EMBO J. 2003, 22, 633–640. [Google Scholar] [CrossRef] [PubMed]
- Chellappan, P.; Vanitharani, R.; Ogbe, F.; Fauquet, C.M. Effect of Temperature on Geminivirus-Induced RNA Silencing in Plants. Plant Physiol. 2005, 138, 1828–1841. [Google Scholar] [CrossRef]
- Tuttle, J.R.; Idris, A.M.; Brown, J.K.; Haigler, C.H.; Robertson, D. Geminivirus-Mediated Gene Silencing from Cotton Leaf Crumple Virus Is Enhanced by Low Temperature in Cotton. Plant Physiol. 2008, 148, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Lewsey, M.G.; Murphy, A.M.; MacLean, D.; Dalchau, N.; Westwood, J.H.; Macaulay, K.; Bennett, M.H.; Moulin, M.; Hanke, D.E.; Powell, G.; et al. Disruption of Two Defensive Signaling Pathways by a Viral RNA Silencing Suppressor. Mol. Plant-Microbe Interact. 2010, 23, 835–845. [Google Scholar] [CrossRef]
- Love, A.J.; Geri, C.; Laird, J.; Carr, C.; Yun, B.-W.; Loake, G.J.; Tada, Y.; Sadanandom, A.; Milner, J.J. Cauliflower Mosaic Virus Protein P6 Inhibits Signaling Responses to Salicylic Acid and Regulates Innate Immunity. PLoS ONE 2012, 7, e47535. [Google Scholar] [CrossRef]
- Laird, J.; McInally, C.; Carr, C.; Doddiah, S.; Yates, G.; Chrysanthou, E.; Khattab, A.; Love, A.J.; Geri, C.; Sadanandom, A.; et al. Identification of the Domains of Cauliflower Mosaic Virus Protein P6 Responsible for Suppression of RNA Silencing and Salicylic Acid Signalling. J. Gen. Virol. 2013, 94, 2777–2789. [Google Scholar] [CrossRef]
- Csorba, T.; Kontra, L.; Burgyán, J. Viral Silencing Suppressors: Tools Forged to Fine-Tune Host-Pathogen Coexistence. Virology 2015, 479–480, 85–103. [Google Scholar] [CrossRef]
- Shams-Bakhsh, M.; Canto, T.; Palukaitis, P. Enhanced Resistance and Neutralization of Defense Responses by Suppressors of RNA Silencing. Virus Res. 2007, 130, 103–109. [Google Scholar] [CrossRef][Green Version]
- Rizhsky, L.; Liang, H.; Shuman, J.; Shulaev, V.; Davletova, S.; Mittler, R. When Defense Pathways Collide. The Response of Arabidopsis to a Combination of Drought and Heat Stress. Plant Physiol. 2004, 134, 1683–1696. [Google Scholar] [CrossRef]
- Statello, L.; Guo, C.-J.; Chen, L.-L.; Huarte, M. Gene Regulation by Long Non-Coding RNAs and Its Biological Functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef] [PubMed]
- G:Profiler. Available online: http://biit.cs.ut.ee/gprofiler/gost (accessed on 13 December 2021).
- Li, G.; Chen, T.; Zhang, Z.; Li, B.; Tian, S. Roles of Aquaporins in Plant-Pathogen Interaction. Plants 2020, 9, 1134. [Google Scholar] [CrossRef]
- Fu, Y.; Tang, J.; Yao, G.-F.; Huang, Z.-Q.; Li, Y.-H.; Han, Z.; Chen, X.-Y.; Hu, L.-Y.; Hu, K.-D.; Zhang, H. Central Role of Adenosine 5′-Phosphosulfate Reductase in the Control of Plant Hydrogen Sulfide Metabolism. Front. Plant Sci. 2018, 9, 1404. [Google Scholar] [CrossRef]
- Lee, J.S.; Brown, W.E.; Graham, J.S.; Pearce, G.; Fox, E.A.; Dreher, T.W.; Ahern, K.G.; Pearson, G.D.; Ryan, C.A. Molecular Characterization and Phylogenetic Studies of a Wound-Inducible Proteinase Inhibitor I Gene in Lycopersicon Species. Proc. Natl. Acad. Sci. USA 1986, 83, 7277–7281. [Google Scholar] [CrossRef] [PubMed]
- Baebler, Š.; Krečič-Stres, H.; Rotter, A.; Kogovšek, P.; Cankar, K.; Kok, E.J.; Gruden, K.; Kovač, M.; Žel, J.; Pompe-Novak, M.; et al. PVY NTN Elicits a Diverse Gene Expression Response in Different Potato Genotypes in the First 12 h after Inoculation. Mol. Plant Pathol. 2009, 10, 263–275. [Google Scholar] [CrossRef] [PubMed]
- Baebler, Š.; Stare, K.; Kovač, M.; Blejec, A.; Prezelj, N.; Stare, T.; Kogovšek, P.; Pompe-Novak, M.; Rosahl, S.; Ravnikar, M.; et al. Dynamics of Responses in Compatible Potato—Potato Virus Y Interaction Are Modulated by Salicylic Acid. PLoS ONE 2011, 6, e29009. [Google Scholar] [CrossRef]
- Stare, T.; Stare, K.; Weckwerth, W.; Wienkoop, S.; Gruden, K. Comparison between Proteome and Transcriptome Response in Potato (Solanum tuberosum L.) Leaves Following Potato Virus Y (PVY) Infection. Proteomes 2017, 5, 14. [Google Scholar] [CrossRef] [PubMed]
- Babu, M.; Gagarinova, A.G.; Brandle, J.E.; Wang, A. Association of the Transcriptional Response of Soybean Plants with Soybean Mosaic Virus Systemic Infection. J. Gen. Virol. 2008, 89, 1069–1080. [Google Scholar] [CrossRef] [PubMed]
- Pompe-Novak, M.; Gruden, K.; Baebler, Š.; Krečič-Stres, H.; Kovač, M.; Jongsma, M.; Ravnikar, M. Potato Virus Y Induced Changes in the Gene Expression of Potato (Solanum tuberosum L.). Physiol. Mol. Plant Pathol. 2005, 67, 237–247. [Google Scholar] [CrossRef]
- García-Marcos, A.; Pacheco, R.; Martiáñez, J.; González-Jara, P.; Díaz-Ruíz, J.R.; Tenllado, F. Transcriptional Changes and Oxidative Stress Associated with the Synergistic Interaction between Potato Virus X and Potato Virus Y and Their Relationship with Symptom Expression. Mol. Plant-Microbe Interact. 2009, 22, 1431–1444. [Google Scholar] [CrossRef]
- Hanssen, I.M.; van Esse, H.P.; Ballester, A.-R.; Hogewoning, S.W.; Parra, N.O.; Paeleman, A.; Lievens, B.; Bovy, A.G.; Thomma, B.P.H.J. Differential Tomato Transcriptomic Responses Induced by Pepino Mosaic Virus Isolates with Differential Aggressiveness. Plant Physiol. 2011, 156, 301–318. [Google Scholar] [CrossRef]
- Whitham, S.A.; Yang, C.; Goodin, M.M. Global Impact: Elucidating Plant Responses to Viral Infection. Mol. Plant-Microbe Interact. 2006, 19, 1207–1215. [Google Scholar] [CrossRef]
- Chen, D.; Shao, Q.; Yin, L.; Younis, A.; Zheng, B. Polyamine Function in Plants: Metabolism, Regulation on Development, and Roles in Abiotic Stress Responses. Front. Plant Sci. 2019, 9, 1945. [Google Scholar] [CrossRef]
- Firpo, M.R.; Mounce, B.C. Diverse Functions of Polyamines in Virus Infection. Biomolecules 2020, 10, 628. [Google Scholar] [CrossRef]
- Briggs, A.G.; Bent, A.F. Poly(ADP-Ribosyl)Ation in Plants. Trends Plant Sci. 2011, 16, 372–380. [Google Scholar] [CrossRef]
- Love, A.J.; Yu, C.; Petukhova, N.V.; Kalinina, N.O.; Chen, J.; Taliansky, M.E. Cajal Bodies and Their Role in Plant Stress and Disease Responses. RNA Biol. 2017, 14, 779–790. [Google Scholar] [CrossRef] [PubMed]
- Vainonen, J.P.; Shapiguzov, A.; Vaattovaara, A.; Kangasjärvi, J. Plant PARPs, PARGs and PARP-like Proteins. Curr. Protein Pept. Sci. 2016, 17, 713–723. [Google Scholar] [CrossRef] [PubMed]
- Kotova, E.; Jarnik, M.; Tulin, A.V. Poly (ADP-Ribose) Polymerase 1 Is Required for Protein Localization to Cajal Body. PLoS Genet. 2009, 5, e1000387. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Kraus, W.L. On PAR with PARP: Cellular Stress Signaling through Poly(ADP-Ribose) and PARP-1. Genes Dev. 2012, 26, 417–432. [Google Scholar] [CrossRef]
- Ji, Y.; Tulin, A.V. Post-Transcriptional Regulation by Poly(ADP-Ribosyl)Ation of the RNA-Binding Proteins. Int. J. Mol. Sci. 2013, 14, 16168–16183. [Google Scholar] [CrossRef] [PubMed]
- Zaynab, M.; Fatima, M.; Abbas, S.; Umair, M.; Sharif, Y.; Raza, M.A. Long Non-Coding RNAs as Molecular Players in Plant Defense against Pathogens. Microb. Pathog. 2018, 121, 277–282. [Google Scholar] [CrossRef]
- Mindrebo, J.T.; Nartey, C.M.; Seto, Y.; Burkart, M.D.; Noel, J.P. Unveiling the Functional Diversity of the Alpha/Beta Hydrolase Superfamily in the Plant Kingdom. Curr. Opin. Struct. Biol. 2016, 41, 233–246. [Google Scholar] [CrossRef]
- Fang, Y.; Fullwood, M.J. Roles, Functions, and Mechanisms of Long Non-Coding RNAs in Cancer. Genom. Proteom. Bioinform. 2016, 14, 42–54. [Google Scholar] [CrossRef]
- Lakhotia, N.; Joshi, G.; Bhardwaj, A.R.; Katiyar-Agarwal, S.; Agarwal, M.; Jagannath, A.; Goel, S.; Kumar, A. Identification and Characterization of MiRNAome in Root, Stem, Leaf and Tuber Developmental Stages of Potato (Solanum tuberosum L.) by High-Throughput Sequencing. BMC Plant Biol. 2014, 14, 6. [Google Scholar] [CrossRef]
- Öztürk Gökçe, Z.N.; Aksoy, E.; Bakhsh, A.; Demirel, U.; Çalışkan, S.; Çalışkan, M.E. Combined Drought and Heat Stresses Trigger Different Sets of MiRNAs in Contrasting Potato Cultivars. Funct. Integr. Genom. 2021, 21, 489–502. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, L.-F.; Bhutto, S.H.; He, X.-R.; Yang, X.-M.; Zhou, X.-H.; Lin, X.-Y.; Rajput, A.A.; Li, G.-B.; Zhao, J.-H.; et al. Blocking MiR530 Improves Rice Resistance, Yield, and Maturity. Front. Plant Sci. 2021, 12, 729560. [Google Scholar] [CrossRef]
- Xia, R.; Xu, J.; Meyers, B.C. The Emergence, Evolution, and Diversification of the MiR390-TAS3—ARF Pathway in Land Plants. Plant Cell 2017, 29, 1232–1247. [Google Scholar] [CrossRef]
- Casadevall, R.; Rodriguez, R.E.; Debernardi, J.M.; Palatnik, J.F.; Casati, P. Repression of Growth Regulating Factors by the MicroRNA396 Inhibits Cell Proliferation by UV-B Radiation in Arabidopsis Leaves. Plant Cell 2013, 25, 3570–3583. [Google Scholar] [CrossRef] [PubMed]
- Szczygieł-Sommer; Gaj The MiR396–GRF Regulatory Module Controls the Embryogenic Response in Arabidopsis via an Auxin-Related Pathway. Int. J. Mol. Sci. 2019, 20, 5221. [CrossRef] [PubMed]
- Zhang, C.; Wang, D.; Yang, C.; Kong, N.; Shi, Z.; Zhao, P.; Nan, Y.; Nie, T.; Wang, R.; Ma, H.; et al. Genome-Wide Identification of the Potato WRKY Transcription Factor Family. PLoS ONE 2017, 12, e0181573. [Google Scholar] [CrossRef]
- Parker, M.T.; Knop, K.; Sherwood, A.V.; Schurch, N.J.; Mackinnon, K.; Gould, P.D.; Hall, A.J.; Barton, G.J.; Simpson, G.G. Nanopore Direct RNA Sequencing Maps the Complexity of Arabidopsis MRNA Processing and M6A Modification. eLife 2020, 9, e49658. [Google Scholar] [CrossRef] [PubMed]
- Litholdo, C.G.; Bousquet-Antonelli, C. Chemical RNA Modifications: The Plant Epitranscriptome. In Epigenetics in Plants of Agronomic Importance: Fundamentals and Applications; Alvarez-Venegas, R., De-la-Peña, C., Casas-Mollano, J.A., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 291–310. ISBN 978-3-030-14759-4. [Google Scholar]
- Covelo-Molares, H.; Bartosovic, M.; Vanacova, S. RNA Methylation in Nuclear Pre-mRNA Processing. Wiley Interdiscip. RNA 2018, 9, e1489. [Google Scholar] [CrossRef] [PubMed]
- Howard, J.M.; Sanford, J.R. The RNAissance Family: SR Proteins as Multifaceted Regulators of Gene Expression: The RNAissance Family. Wiley Interdiscip. Rev. RNA 2015, 6, 93–110. [Google Scholar] [CrossRef]
- Martínez-Pérez, M.; Aparicio, F.; López-Gresa, M.P.; Bellés, J.M.; Sánchez-Navarro, J.A.; Pallás, V. Arabidopsis M6A Demethylase Activity Modulates Viral Infection of a Plant Virus and the M6A Abundance in Its Genomic RNAs. Proc. Natl. Acad. Sci. USA 2017, 114, 10755–10760. [Google Scholar] [CrossRef]
- Brocard, M.; Ruggieri, A.; Locker, N. M6A RNA Methylation, a New Hallmark in Virus-Host Interactions. J. Gen. Virol. 2017, 98, 2207–2214. [Google Scholar] [CrossRef]
- Gibson, R.W.; Jones, M.G.; Fish, N. Resistance to Potato Leaf Roll Virus and Potato Virus Y in Somatic Hybrids between Dihaploid Solanum tuberosum and S. brevidens. TAG Theor. Appl. Genet. Theor. Angew. Genet. 1988, 76, 113–117. [Google Scholar] [CrossRef]
- Lanfear, R.; Schalamun, M.; Kainer, D.; Wang, W.; Schwessinger, B. MinIONQC: Fast and Simple Quality Control for MinION Sequencing Data. Bioinformatics 2019, 35, 523–525. [Google Scholar] [CrossRef]
- Pham, G.M.; Hamilton, J.P.; Wood, J.C.; Burke, J.T.; Zhao, H.; Vaillancourt, B.; Ou, S.; Jiang, J.; Buell, C.R. Construction of a Chromosome-Scale Long-Read Reference Genome Assembly for Potato. GigaScience 2020, 9, giaa100. [Google Scholar] [CrossRef]
- Li, H. Minimap2: Pairwise Alignment for Nucleotide Sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map Format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Quinlan, A.R.; Hall, I.M. BEDTools: A Flexible Suite of Utilities for Comparing Genomic Features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Park, C.; Bennett, C.; Thornton, M.; Kim, D. Rapid and Accurate Alignment of Nucleotide Conversion Sequencing Reads with HISAT-3N. Genome Res. 2021, 31, 1290–1295. [Google Scholar] [CrossRef] [PubMed]
- Kovaka, S.; Zimin, A.V.; Pertea, G.M.; Razaghi, R.; Salzberg, S.L.; Pertea, M. Transcriptome Assembly from Long-Read RNA-Seq Alignments with StringTie2. Genome Biol. 2019, 20, 278. [Google Scholar] [CrossRef]
- Pertea, G.; Pertea, M. GFF Utilities: GffRead and GffCompare. F1000Research 2020, 9, 304. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. FeatureCounts: An Efficient General Purpose Program for Assigning Sequence Reads to Genomic Features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- NanoCount 0.2.4.Post1. Available online: https://zenodo.org/record/4486652#.YbnQm70zaHs (accessed on 13 December 2021).
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-Optimal Probabilistic RNA-Seq Quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef]
- Picard. Available online: http://broadinstitute.github.io/picard/ (accessed on 13 December 2021).
- CountToFPKM. Available online: https://cran.r-project.org/web/packages/countToFPKM/index.html (accessed on 13 December 2021).
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Zhu, A.; Ibrahim, J.G.; Love, M.I. Heavy-Tailed Prior Distributions for Sequence Count Data: Removing the Noise and Preserving Large Differences. Bioinformatics 2019, 35, 2084–2092. [Google Scholar] [CrossRef] [PubMed]
- Scikit-Learn: Machine Learning in Python. Available online: https://jmlr.csail.mit.edu/papers/v12/pedregosa11a.html (accessed on 13 December 2021).
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. G:Profiler: A Web Server for Functional Enrichment Analysis and Conversions of Gene Lists (2019 Update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef]
- CRAN. Available online: https://cran.r-project.org/web/packages/tagcloud/index.html (accessed on 13 December 2021).
- Jones, P.; Binns, D.; Chang, H.-Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-Scale Protein Function Classification. Bioinform. Oxf. Engl. 2014, 30, 1236–1240. [Google Scholar] [CrossRef]
- Kalvari, I.; Nawrocki, E.P.; Ontiveros-Palacios, N.; Argasinska, J.; Lamkiewicz, K.; Marz, M.; Griffiths-Jones, S.; Toffano-Nioche, C.; Gautheret, D.; Weinberg, Z.; et al. Rfam 14: Expanded Coverage of Metagenomic, Viral and MicroRNA Families. Nucleic Acids Res. 2021, 49, D192–D200. [Google Scholar] [CrossRef]
- Nawrocki, E.P. Annotating Functional RNAs in Genomes Using Infernal. In RNA Sequence, Structure, and Function: Computational and Bioinformatic Methods; Gorodkin, J., Ruzzo, W.L., Eds.; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2014; Volume 1097, pp. 163–197. ISBN 978-1-62703-708-2. [Google Scholar]
- Guo, J.-C.; Fang, S.-S.; Wu, Y.; Zhang, J.-H.; Chen, Y.; Liu, J.; Wu, B.; Wu, J.-R.; Li, E.-M.; Xu, L.-Y.; et al. CNIT: A Fast and Accurate Web Tool for Identifying Protein-Coding and Long Non-Coding Transcripts Based on Intrinsic Sequence Composition. Nucleic Acids Res. 2019, 47, W516–W522. [Google Scholar] [CrossRef]
- Li, A.; Zhang, J.; Zhou, Z. PLEK: A Tool for Predicting Long Non-Coding RNAs and Messenger RNAs Based on an Improved k-Mer Scheme. BMC Bioinform. 2014, 15, 311. [Google Scholar] [CrossRef]
- Xu, X.; Liu, S.; Yang, Z.; Zhao, X.; Deng, Y.; Zhang, G.; Pang, J.; Zhao, C.; Zhang, W. A Systematic Review of Computational Methods for Predicting Long Noncoding RNAs. Brief. Funct. Genom. 2021, 20, 162–173. [Google Scholar] [CrossRef]
- Lex, A.; Gehlenborg, N.; Strobelt, H.; Vuillemot, R.; Pfister, H. UpSet: Visualization of Intersecting Sets. IEEE Trans. Vis. Comput. Graph. 2014, 20, 1983–1992. [Google Scholar] [CrossRef] [PubMed]
- Bonnet, E.; He, Y.; Billiau, K.; Van de Peer, Y. TAPIR, a Web Server for the Prediction of Plant MicroRNA Targets, Including Target Mimics. Bioinformatics 2010, 26, 1566–1568. [Google Scholar] [CrossRef] [PubMed]
- Hendra, C.; Pratanwanich, P.N.; Wan, Y.K.; Goh, W.S.S.; Thiery, A.; Göke, J. Detection of M6A from Direct RNA Sequencing Using a Multiple Instance Learning Framework. Bioinformatics 2021. Preprint . [Google Scholar] [CrossRef]
- Pratanwanich, P.N.; Yao, F.; Chen, Y.; Koh, C.W.Q.; Wan, Y.K.; Hendra, C.; Poon, P.; Goh, Y.T.; Yap, P.M.L.; Chooi, J.Y.; et al. Identification of Differential RNA Modifications from Nanopore Direct RNA Sequencing with XPore. Nat. Biotechnol. 2021, 39, 1394–1402. [Google Scholar] [CrossRef]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative Genomics Viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef]
- De Block, M.; Verduyn, C.; De Brouwer, D.; Cornelissen, M. Poly(ADP-Ribose) Polymerase in Plants Affects Energy Homeostasis, Cell Death and Stress Tolerance. Plant J. Cell Mol. Biol. 2005, 41, 95–106. [Google Scholar] [CrossRef]
- Affar, E.B.; Duriez, P.J.; Shah, R.G.; Winstall, E.; Germain, M.; Boucher, C.; Bourassa, S.; Kirkland, J.B.; Poirier, G.G. Immunological Determination and Size Characterization of Poly(ADP-Ribose) Synthesized in Vitro and in Vivo. Biochim. Biophys. Acta 1999, 1428, 137–146. [Google Scholar] [CrossRef]
- Nicot, N.; Hausman, J.-F.; Hoffmann, L.; Evers, D. Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J. Exp. Bot. 2005, 56, 2907–2914. [Google Scholar] [CrossRef] [PubMed]

| Identification | NCBI Accession | Number of Reads | Number of Read Pairs | Q20 (%) | Aligned to the Genome (%) |
|---|---|---|---|---|---|
| Mock, 22 °C_1 | SRR17129393 | 72,472,982 | 108,709,473 | 97.86 | 93.20 |
| Mock, 22 °C_2 | SRR17129392 | 72,465,906 | 108,698,859 | 97.66 | 93.45 |
| Mock, 22 °C_3 | SRR17129381 | 75,141,894 | 112,712,841 | 97.57 | 92.96 |
| Mock, 22 °C_4 | SRR17129378 | 95,142,250 | 142,713,375 | 97.84 | 92.24 |
| Mock, 28 °C_1 | SRR17129377 | 75,125,564 | 112,688,346 | 97.78 | 93.08 |
| Mock, 28 °C_2 | SRR17129376 | 100,155,380 | 150,233,070 | 97.71 | 91.76 |
| Mock, 28 °C_3 | SRR17129375 | 89,289,122 | 133,933,683 | 98.09 | 92.25 |
| Mock, 28 °C_4 | SRR17129374 | 86,033,238 | 129,049,857 | 97.70 | 91.88 |
| PVY, 22 °C_1 | SRR17129373 | 72,632,520 | 108,948,780 | 97.79 | 93.21 |
| PVY, 22 °C_2 | SRR17129372 | 72,454,368 | 108,681,552 | 97.96 | 93.33 |
| PVY, 22 °C_3 | SRR17129391 | 74,958,594 | 112,437,891 | 97.77 | 92.88 |
| PVY, 22 °C_4 | SRR17129390 | 75,086,442 | 112,629,663 | 97.71 | 93.02 |
| PVY, 28 °C_1 | SRR17129389 | 83,177,956 | 124,766,934 | 97.83 | 91.46 |
| PVY, 28 °C_2 | SRR17129388 | 75,185,626 | 112,778,439 | 97.67 | 92.66 |
| PVY, 28 °C_3 | SRR17129387 | 72,652,314 | 108,978,471 | 97.90 | 93.21 |
| PVY, 28 °C_4 | SRR17129386 | 75,062,806 | 112,594,209 | 97.60 | 92.51 |
| Identification | NCBI Accession | Total Reads | Reads N50 | Median Q Score | Median Length (bp) | Aligned to the Genome (%) |
|---|---|---|---|---|---|---|
| PVY 28 °C_1 | SRR17129382 | 2,912,102 | 1038 | 10.5 | 796 | 99.07 |
| PVY 28 °C_2 | SRR17129380 | 2,231,219 | 1089 | 9.9 | 838 | 99.2 |
| PVY 28 °C_3 | SRR17129379 | 2,297,949 | 1011 | 9.9 | 785 | 98.8 |
| Mock 22 °C_1 | SRR17129385 | 2,408,614 | 1099 | 10.4 | 858 | 99.25 |
| Mock 22 °C_2 | SRR17129384 | 2,462,883 | 983 | 10.6 | 779 | 99.09 |
| Mock 22 °C_3 | SRR17129383 | 748,042 | 983 | 10.6 | 753 | 98.96 |
| Number of Reads | Genes | Transcripts | Max Isoform Number per Gene | Isoforms per Gene | Mean Exon Number | Annotated Genes | |
|---|---|---|---|---|---|---|---|
| Long reads | 13,060,809 | 25,252 | 46,488 | 36 | 1.84 | 5.9 | 21,498 |
| Short reads | 1,267,036,962 | 25,646 | 47,174 | 53 | 1.84 | 6.4 | 21,398 |
| Combined transcriptome | 1,280,097,771 | 26,975 | 49,089 | 48 | 1.82 | 5.9 | 22,305 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Glushkevich, A.; Spechenkova, N.; Fesenko, I.; Knyazev, A.; Samarskaya, V.; Kalinina, N.O.; Taliansky, M.; Love, A.J. Transcriptomic Reprogramming, Alternative Splicing and RNA Methylation in Potato (Solanum tuberosum L.) Plants in Response to Potato Virus Y Infection. Plants 2022, 11, 635. https://doi.org/10.3390/plants11050635
Glushkevich A, Spechenkova N, Fesenko I, Knyazev A, Samarskaya V, Kalinina NO, Taliansky M, Love AJ. Transcriptomic Reprogramming, Alternative Splicing and RNA Methylation in Potato (Solanum tuberosum L.) Plants in Response to Potato Virus Y Infection. Plants. 2022; 11(5):635. https://doi.org/10.3390/plants11050635
Chicago/Turabian StyleGlushkevich, Anna, Nadezhda Spechenkova, Igor Fesenko, Andrey Knyazev, Viktoriya Samarskaya, Natalia O. Kalinina, Michael Taliansky, and Andrew J. Love. 2022. "Transcriptomic Reprogramming, Alternative Splicing and RNA Methylation in Potato (Solanum tuberosum L.) Plants in Response to Potato Virus Y Infection" Plants 11, no. 5: 635. https://doi.org/10.3390/plants11050635
APA StyleGlushkevich, A., Spechenkova, N., Fesenko, I., Knyazev, A., Samarskaya, V., Kalinina, N. O., Taliansky, M., & Love, A. J. (2022). Transcriptomic Reprogramming, Alternative Splicing and RNA Methylation in Potato (Solanum tuberosum L.) Plants in Response to Potato Virus Y Infection. Plants, 11(5), 635. https://doi.org/10.3390/plants11050635







