Transcriptome Responses of Wild Arachis to UV-C Exposure Reveal Genes Involved in General Plant Defense and Priming
Abstract
:1. Introduction
2. Results
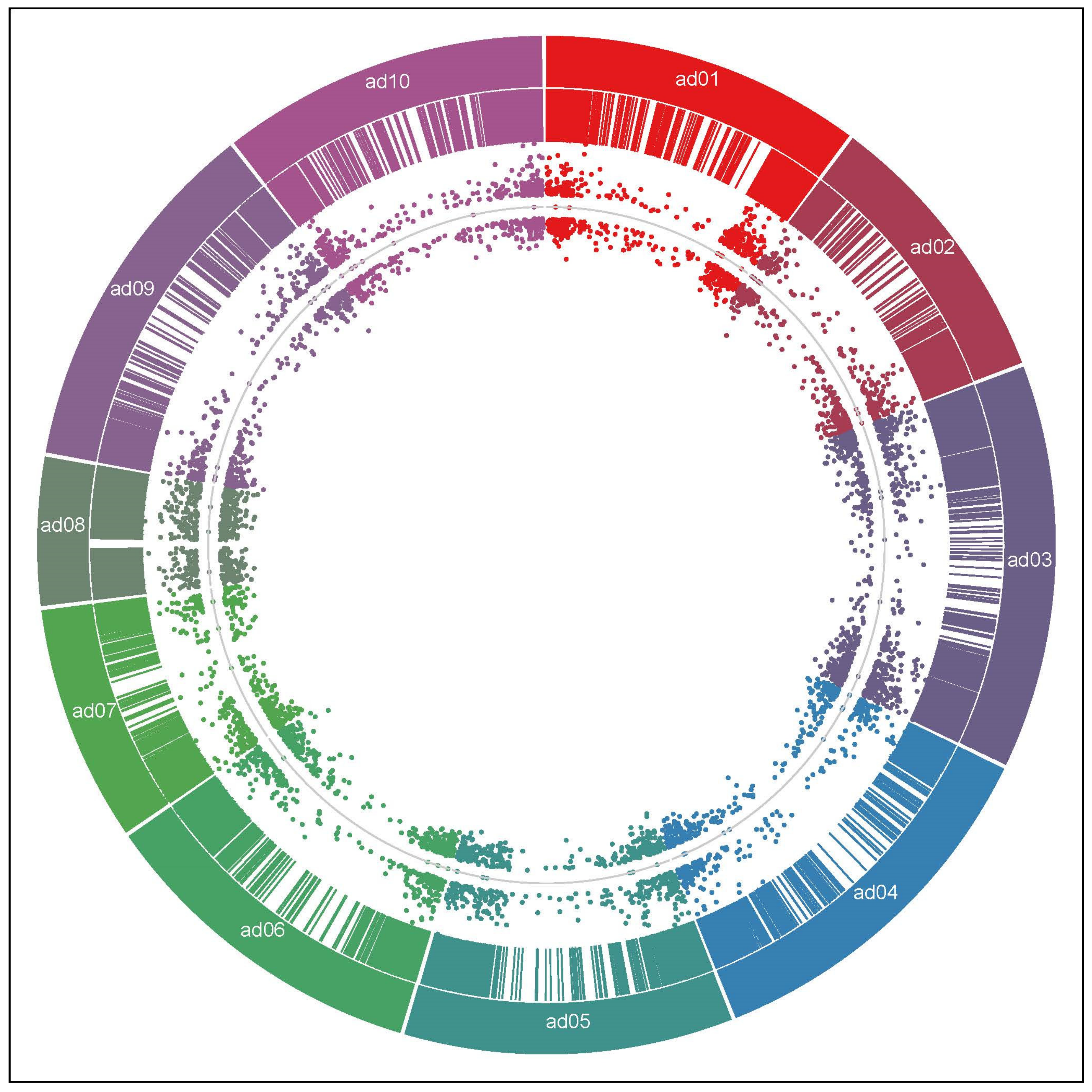
2.1. RNA-Seq Data Analysis
2.2. Functional Enrichment Analysis of DEGs
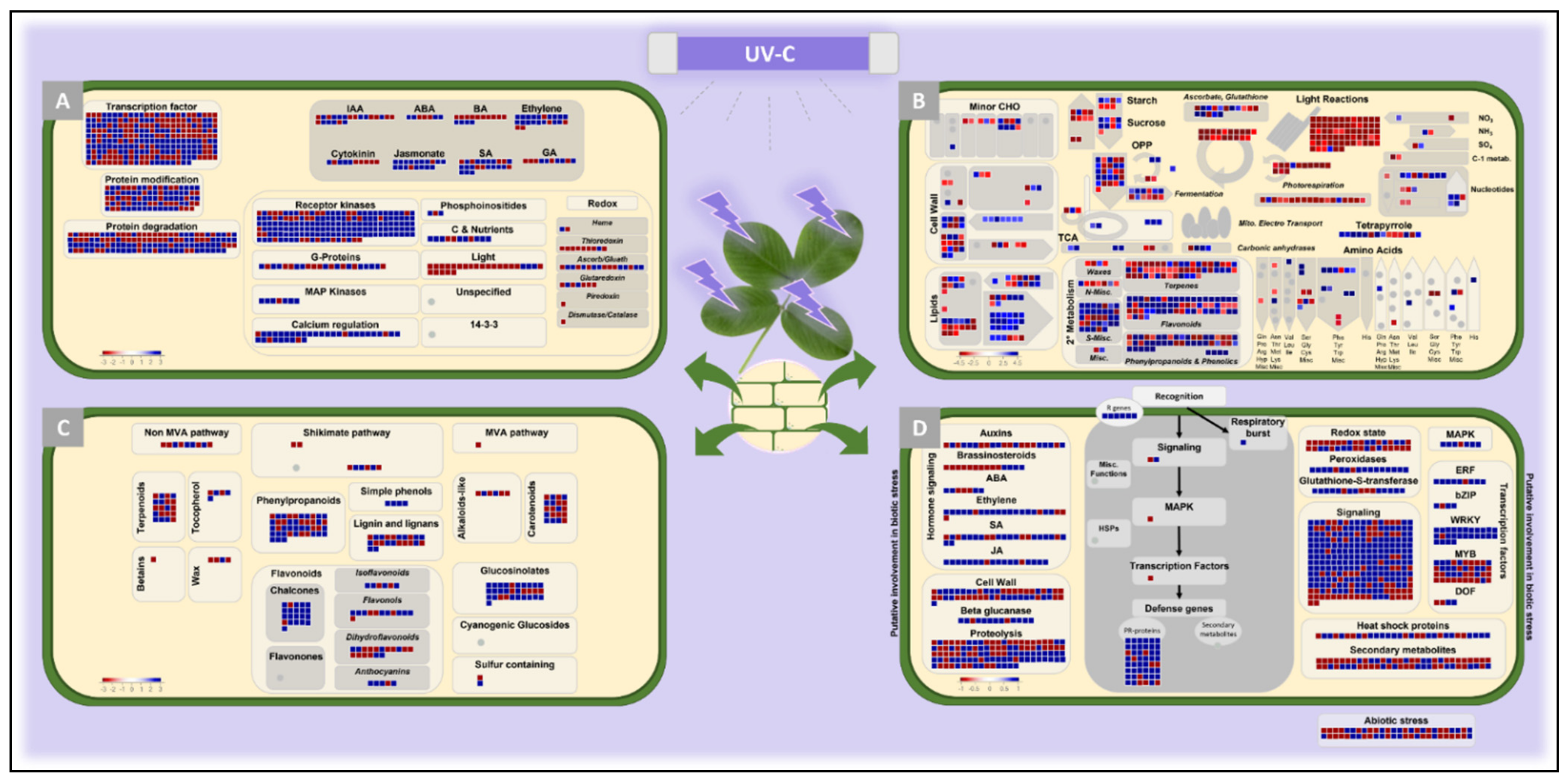
2.3. Functional Characterization by Mapman Analysis
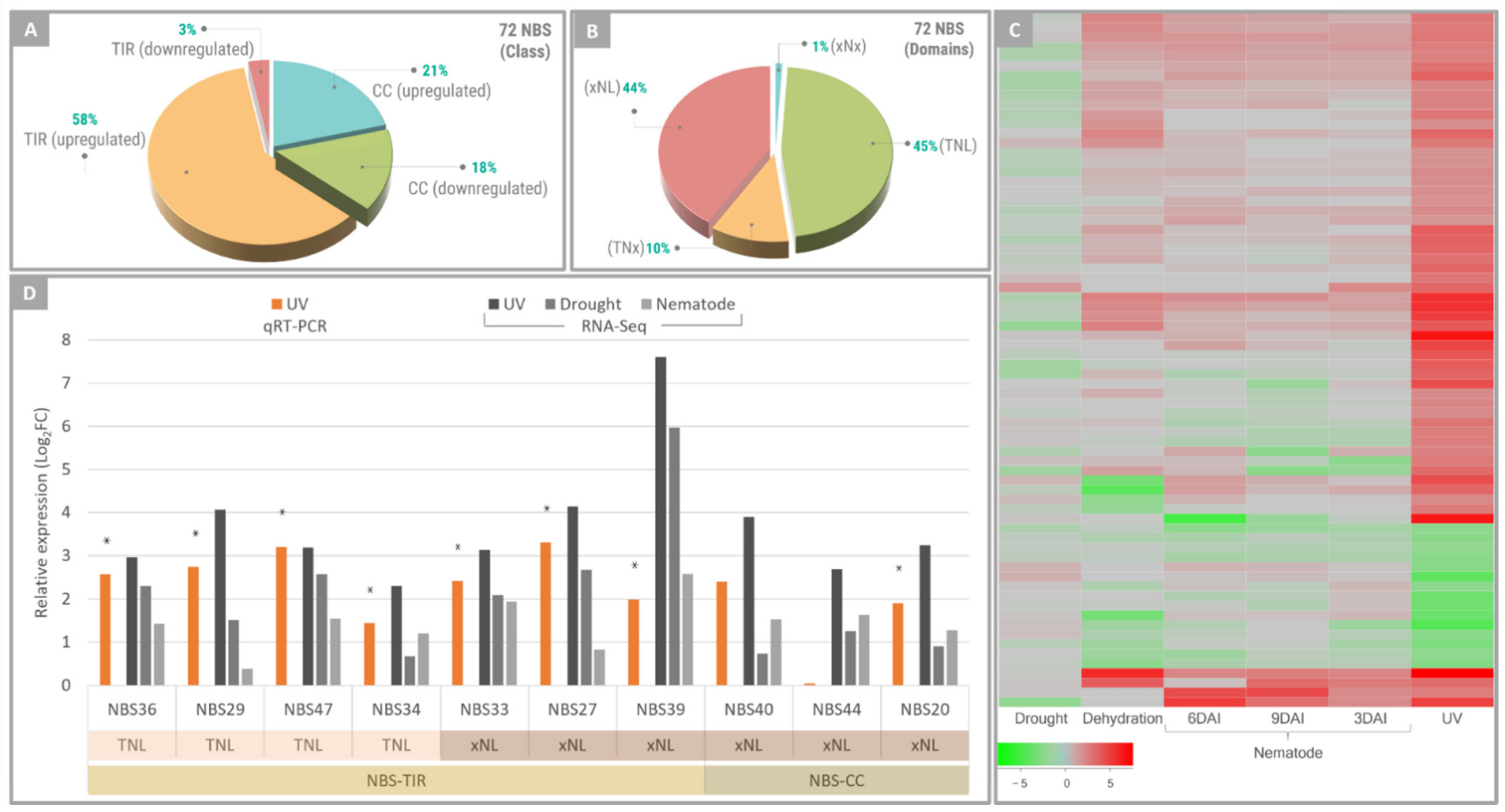
2.4. NBS-LRR Genes Responsive to UV Exposure
2.5. Comparative Transcriptome Meta-Analysis of A. stenosperma under Different Stresses
2.6. Identification of Gene Markers of Plant Priming
2.7. qRT-PCR Expression Analysis
3. Discussion
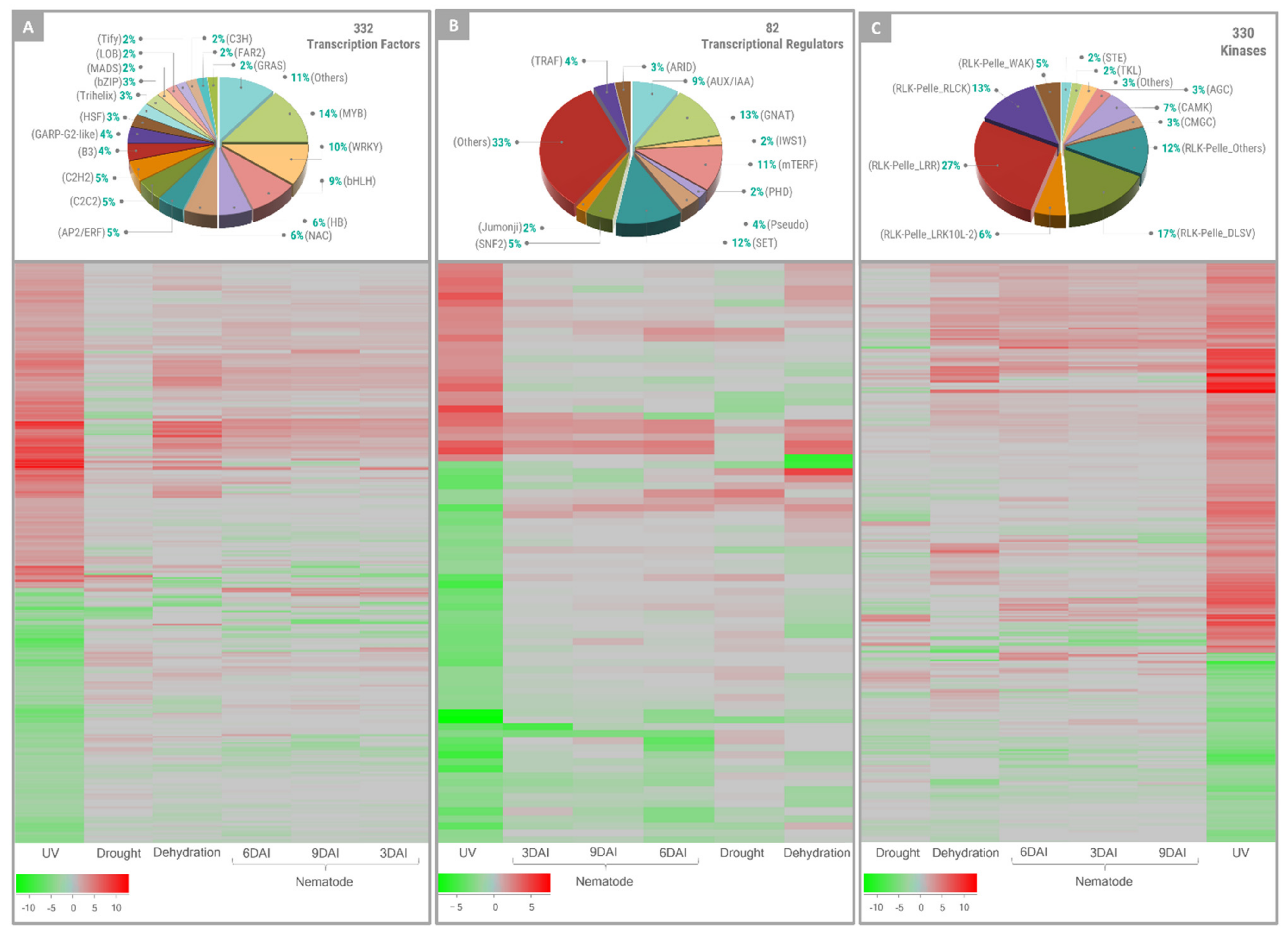
3.1. Exposure to UV-C Resulted in Altered Expression of Genes Encoding for TFs, Regulators, Kinases, and NBS-LRRs
3.2. A. stenosperma MetaDEGs Involved in Responses to Different Stress and Putatively Associated with Defense Priming
3.3. Secondary Metabolism Is Affected by Multiple Stresses in A. stenosperma
3.4. UV and Multiple Stresses Alter Cell Wall Dynamics in A. stenosperma
4. Materials and Methods
4.1. Plant Materials and Experimental Design
4.2. RNA Extraction and Library Sequencing
4.3. In silico Expression Profiling
4.4. Functional Analysis of A. stenosperma DEGs
4.5. Functional Analysis of A. stenosperma NBS-LRR Gene Family
4.6. Meta-Analysis of A. stenosperma RNA-Seq Data
4.7. Identification of Orthologs of Plant Priming Markers
4.8. Expression Analysis by qRT-PCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| DAI | Days after Inoculation |
| DEG | Differentially Expressed Gene |
| FC | Fold Change |
| FDR | False Discovery Rate |
| GO | Gene Ontology |
| HAI | Hours after Inoculation |
| J2 | Second-stage Juveniles |
| JA | Jasmonic Acid |
| PK | Protein Kinase |
| qRT-PCR | quantitative Reverse Transcription-olymerase Chain Reaction |
| RKN | Root-Knot Nematode |
| ROS | Reactive Oxygen Species |
| SA | Salicylic Acid |
| SAR | Systemic Acquired Resistance |
| TF | Transcription Factor |
| TR | Transcriptional Regulator |
| UV | Ultraviolet |
References
- Hossain, M.A.; Liu, F.; Burritt, D.; Fujita, M.; Huang, B. Priming-Mediated Stress and Cross-Stress Tolerance in Crop Plants; Academic Press: Cambridge, MA, USA, 2020; ISBN 0128178930. [Google Scholar]
- Hilker, M.; Schmülling, T. Stress priming, memory, and signalling in plants. Plant Cell Environ. 2019, 42, 753–761. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-K.; Chae, S.; Oh, N.-I.; Nguyen, N.H.; Cheong, J.-J. Recurrent drought conditions enhance the induction of drought stress memory genes in Glycine max L. Front. Genet. 2020, 11, 1248. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.T.Q.; Quynh, T.T.; Pham, H.B.V.; Le, T.V.; Phung, T.K.H.; Lee, S.H.; Cheong, J.J. Evaluation of proline, soluble sugar and ABA content in soybean Glycine max (L.) under drought stress memory. AIMS Bioeng. 2020, 7, 114–123. [Google Scholar] [CrossRef]
- Liu, H.; Able, A.J.; Able, J.A. Small RNAs and their targets are associated with the transgenerational effects of water-deficit stress in durum wheat. Sci. Rep. 2021, 11, 3613. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Sanchez, R.; Kundariya, H.; Maher, T.; Dopp, I.; Schwegel, R.; Virdi, K.; Axtell, M.J.; Mackenzie, S.A. Segregation of an MSH1 RNAi transgene produces heritable non-genetic memory in association with methylome reprogramming. Nat. Commun. 2020, 11, 2214. [Google Scholar] [CrossRef]
- Godwin, J.; Farrona, S. Plant epigenetic stress memory induced by drought: A physiological and molecular perspective. In Plant Epigenetics and Epigenomics; Springer: Berlin/Heidelberg, Germany, 2020; pp. 243–259. [Google Scholar]
- Baccelli, I.; Benny, J.; Caruso, T.; Martinelli, F. The priming fingerprint on the plant transcriptome investigated through meta-analysis of RNA-Seq data. Eur. J. Plant Pathol. 2020, 156, 779–797. [Google Scholar] [CrossRef]
- Kerchev, P.; van der Meer, T.; Sujeeth, N.; Verlee, A.; Stevens, C.V.; Van Breusegem, F.; Gechev, T. Molecular priming as an approach to induce tolerance against abiotic and oxidative stresses in crop plants. Biotechnol. Adv. 2020, 40, 107503. [Google Scholar] [CrossRef]
- Conrath, U.; Beckers, G.J.M.; Langenbach, C.J.G.; Jaskiewicz, M.R. Priming for enhanced defense. Annu. Rev. Phytopathol. 2015, 53, 97–119. [Google Scholar] [CrossRef]
- Kollist, H.; Zandalinas, S.I.; Sengupta, S.; Nuhkat, M.; Kangasjärvi, J.; Mittler, R. Rapid responses to abiotic stress: Priming the landscape for the signal transduction network. Trends Plant Sci. 2019, 24, 25–37. [Google Scholar] [CrossRef] [Green Version]
- Vijayakumari, K.; Jisha, K.C.; Puthur, J.T. GABA/BABA priming: A means for enhancing abiotic stress tolerance potential of plants with less energy investments on defence cache. Acta Physiol. Plant. 2016, 38, 230. [Google Scholar] [CrossRef]
- Thomas, T.; Puthur, J. UV radiation priming: A means of amplifying the inherent potential for abiotic stress tolerance in crop plants. Environ. Exp. Bot. 2017, 138, 57–66. [Google Scholar] [CrossRef]
- Duarte-Sierra, A.; Tiznado-Hernández, M.E.; Jha, D.K.; Janmeja, N.; Arul, J. Abiotic stress hormesis: An approach to maintain quality, extend storability, and enhance phytochemicals on fresh produce during postharvest. Compr. Rev. Food Sci. Food Saf. 2020, 19, 3659–3682. [Google Scholar] [CrossRef] [PubMed]
- Urban, L.; Charles, F.; de Miranda, M.R.A.; Aarrouf, J. Understanding the physiological effects of UV-C light and exploiting its agronomic potential before and after harvest. Plant Physiol. Biochem. 2016, 105, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Pan, W.S.; Zheng, L.P.; Tian, H.; Li, W.Y.; Wang, J.W. Transcriptome responses involved in artemisinin production in Artemisia annua L. under UV-B radiation. J. Photochem. Photobiol. B Biol. 2014, 140, 292–300. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Charles, M.T.; Luo, Z.; Mimee, B.; Tong, Z.; Véronneau, P.; Roussel, D.; Rolland, D. Ultraviolet-C priming of strawberry leaves against subsequent Mycosphaerella fragariae infection involves the action of reactive oxygen species, plant hormones, and terpenes. Plant Cell Environ. 2019, 42, 815–831. [Google Scholar] [CrossRef] [PubMed]
- Agathokleous, E.; Kitao, M.; Calabrese, E.J. Hormesis: A compelling platform for sophisticated plant science. Trends Plant Sci. 2019, 24, 318–327. [Google Scholar] [CrossRef]
- Balmer, A.; Pastor, V.; Gamir, J.; Flors, V.; Mauch-Mani, B. The ‘prime-ome’: Towards a holistic approach to priming. Trends Plant Sci. 2015, 20, 443–452. [Google Scholar] [CrossRef]
- Westman, S.M.; Kloth, K.J.; Hanson, J.; Ohlsson, A.B.; Albrectsen, B.R. Defence priming in Arabidopsis—A meta-analysis. Sci. Rep. 2019, 9, 13309. [Google Scholar] [CrossRef]
- Carvalho, P.A.S.V.; de Carvalho Moretzsohn, M.; Brasileiro, A.C.M.; Guimarães, P.M.; da Silveira Agostini-Costa, T.; da Silva, J.P.; Gimenes, M.A. Presence of resveratrol in wild Arachis species adds new value to this overlooked genetic resource. Sci. Rep. 2020, 10, 12787. [Google Scholar] [CrossRef]
- Carvalho, P.V.; de Carvalho Moretzsohn, M.; Brasileiro, A.C.M.; Guimarães, P.M.; da Silveira Agostini Costa, T.; da Silva, J.P.; Gimenes, M.A. Evidences that polyploidization and hybridization affected resveratrol content in Arachis interspecific hybrids. J. Plant Breed. Crop Sci. 2019, 11, 265–270. [Google Scholar]
- Verhagen, B.W.M.; Trotel-Aziz, P.; Couderchet, M.; Höfte, M.; Aziz, A. Pseudomonas spp.-induced systemic resistance to Botrytis cinerea is associated with induction and priming of defence responses in grapevine. J. Exp. Bot. 2010, 61, 249–260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leal-Bertioli, S.C.M.; Farias, M.P.; Silva, P.I.; Guimarães, P.M.; Brasileiro, A.C.M.; Bertioli, D.J.; Araújo, A.C.G. Ultrastructure of the initial interaction of Puccinia arachidis and Cercosporidium personatum with leaves of Arachis hypogaea and Arachis stenosperma. J. Phytopathol. 2010, 158, 792–796. [Google Scholar] [CrossRef]
- Stalker, H.T. Utilizing wild species for peanut improvement. Crop Sci. 2017, 57, 1102–1120. [Google Scholar] [CrossRef]
- Proite, K.; Carneiro, R.; Falcão, R.; Gomes, A.; Leal-Bertioli, S.; Guimarães, P.; Bertioli, D. Post-infection development and histopathology of Meloidogyne arenaria race 1 on Arachis spp. Plant Pathol. 2008, 57, 974–980. [Google Scholar] [CrossRef]
- Krapovickas, A.; Gregory, W.C.; Williams, D.E.; Simpson, C.E. Taxonomy of the genus Arachis (Leguminosae). Bonplandia 2007, 16, 7–205. [Google Scholar] [CrossRef] [Green Version]
- Vinson, C.C.; Mota, A.P.Z.; Oliveira, T.N.; Guimarães, L.A.; Leal-Bertioli, S.C.M.; Williams, T.C.R.; Nepomuceno, A.L.; Saraiva, M.A.P.; Araújo, A.C.G.; Guimarães, P.M.; et al. Early responses to dehydration in contrasting wild Arachis species. PLoS ONE 2018, 13, e0198191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leal-Bertioli, S.C.M.; Bertioli, D.J.; Guimarães, P.M.; Pereira, T.D.; Galhardo, I.; Silva, J.P.; Brasileiro, A.C.M.; Oliveira, R.S.; Silva, I.T.; Vadez, V.; et al. The effect of tetraploidization of wild Arachis on leaf morphology and other drought-related traits. Environ. Exp. Bot. 2012, 84, 17–24. [Google Scholar] [CrossRef]
- Carvalho, P.A.S.V.; Brasileiro, A.C.M.; Leal-Bertioli, S.C.M.; Bertioli, D.J.; Silva, J.P.; Agostini-Costa, T.S.; Gimenes, M.A. Coupled transcript and metabolite identification: Insights on induction and synthesis of resveratrol in peanut, wild relatives and synthetic allotetraploid. Genet. Mol. Res. 2017, 16, gmr16039802. [Google Scholar] [CrossRef]
- Lopes, R.M.; Silveira, D.; Gimenes, M.A.; Vasconcelos, P.A.S.; Alves, R.B.N.; Silva, J.P.; Agostini-Costa, T.S. Characterization of resveratrol content in ten wild species of section Arachis, genus Arachis. Genet. Resour. Crop Evol. 2013, 60, 2219–2226. [Google Scholar] [CrossRef] [Green Version]
- Luo, Y.Y.; Li, R.X.; Jiang, Q.S.; Bai, R.; Duan, D. Changes in the chlorophyll content of grape leaves could provide a physiological index for responses and adaptation to UV-C radiation. Nord. J. Bot. 2019, 37. [Google Scholar] [CrossRef]
- Xi, H.; Ma, L.; Liu, G.; Wang, N.; Wang, J.; Wang, L.; Dai, Z.; Li, S.; Wang, L. Transcriptomic analysis of grape (Vitis vinifera L.) leaves after exposure to ultraviolet C irradiation. PLoS ONE 2014, 9, e113772. [Google Scholar] [CrossRef] [PubMed]
- Karki, K.B.; Mishra, A.K.; Choi, S.-J.; Baek, K.-H. Effect of ultraviolet C irradiation on isoflavone concentrations in different cultivars of soybean (Glycine max). Plants 2020, 9, 1043. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Xu, J.; Wu, X.; Wang, Y.; Lin, Y.; Wu, D.; Zhang, H.; Qin, J. Molecular analysis of UV-C induced resveratrol accumulation in Polygonum cuspidatum leaves. Int. J. Mol. Sci. 2019, 20, 6185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ge, X.-M.; Hu, X.; Zhang, J.; Huang, Q.-M.; Gao, Y.; Li, Z.-Q.; Li, S.; He, J.-M. UV RESISTANCE LOCUS8 mediates ultraviolet-B-induced stomatal closure in an ethylene-dependent manner. Plant Sci. 2020, 301, 110679. [Google Scholar] [CrossRef]
- Barta, C.; Kálai, T.; Hideg, K.; Vass, I.; Hideg, É. Differences in the ROS-generating efficacy of various ultraviolet wavelengths in detached spinach leaves. Funct. Plant Biol. 2004, 31, 23–28. [Google Scholar] [CrossRef]
- Huang, L.K.; He, J.; Chow, W.S.; Whitecross, M.I.; Anderson, J.M. Responses of detached rice leaves (Oryza sativa L.) to moderate supplementary ultraviolet-B radiation allow early screening for relative sensitivity to ultraviolet-B irradiation. Funct. Plant Biol. 1993, 20, 285–297. [Google Scholar] [CrossRef]
- Biswal, B.; Joshi, P.N.; Kulandaivelu, G. Changes in leaf protein and pigment contents and photosynthetic activities during senescence of detached maize leaves: Influence of different ultraviolet radiations. Photosynthetica 1998, 34, 37–44. [Google Scholar] [CrossRef]
- Jiang, S.; Han, S.; He, D.; Cao, G.; Zhang, F.; Wan, X. Evaluating walnut (Juglans spp.) for resistance to walnut blight and comparisons between artificial inoculation assays and field studies. Australas. Plant Pathol. 2019, 48, 221–231. [Google Scholar] [CrossRef]
- Ma, B.; Uddin, W. Fitness and competitive ability of an azoxystrobin-resistant G143A mutant of Magnaporthe oryzae from perennial ryegrass. Plant Dis. 2009, 93, 1044–1049. [Google Scholar] [CrossRef] [Green Version]
- Miller-Butler, M.A.; Smith, B.J.; Babiker, E.M.; Kreiser, B.R.; Blythe, E.K. Comparison of whole plant and detached leaf screening techniques for identifying anthracnose resistance in strawberry plants. Plant Dis. 2018, 102, 2112–2119. [Google Scholar] [CrossRef] [Green Version]
- Twizeyimana, M.; Ojiambo, P.S.; Ikotun, T.; Paul, C.; Hartman, G.L.; Bandyopadhyay, R. Comparison of field, greenhouse, and detached-leaf evaluations of soybean germplasm for resistance to Phakopsora pachyrhizi. Plant Dis. 2007, 91, 1161–1169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bertioli, D.J.; Cannon, S.B.; Froenicke, L.; Huang, G.; Farmer, A.D.; Cannon, E.K.S.; Liu, X.; Gao, D.; Clevenger, J.; Dash, S.; et al. The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nat. Genet. 2016, 48, 438–446. [Google Scholar] [CrossRef] [PubMed]
- Guimarães, P.M.; Brasileiro, A.C.M.; Morgante, C.V.; Martins, A.C.Q.; Pappas, G.; Silva, O.B.; Togawa, R.; Leal-Bertioli, S.C.M.; Araújo, A.C.G.; Moretzsohn, M.C.; et al. Global transcriptome analysis of two wild relatives of peanut under drought and fungi infection. BMC Genom. 2012, 13, 387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guimarães, P.M.; Guimarães, L.A.; Morgante, C.V.; Silva, O.B.; Araújo, A.C.G.; Martins, A.C.Q.; Saraiva, M.A.P.; Oliveira, T.N.; Togawa, R.C.; Leal-Bertioli, S.C.M.; et al. Root transcriptome analysis of wild peanut reveals candidate genes for nematode resistance. PLoS ONE 2015, 10, e0140937. [Google Scholar] [CrossRef] [PubMed]
- Shumayla; Tyagi, S.; Upadhyay, S.K. Receptor-like kinases and environmental stress in plants. In Molecular Approaches in Plant Biology and Environmental Challenges; Springer: Berlin/Heidelberg, Germany, 2019; pp. 79–102. [Google Scholar]
- Mota, A.P.Z.; Vidigal, B.; Danchin, E.G.J.; Togawa, R.C.; Leal-Bertioli, S.C.M.; Bertioli, D.J.; Araújo, A.C.G.; Brasileiro, A.C.M.; Guimarães, P.M. Comparative root transcriptome of wild Arachis reveals NBS-LRR genes related to nematode resistance. BMC Plant Biol. 2018, 18, 159. [Google Scholar] [CrossRef]
- Meyers, B.C.; Dickerman, A.W.; Michelmore, R.W.; Sivaramakrishnan, S.; Sobral, B.W.; Young, N.D. Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. Plant J. 1999, 20, 317–332. [Google Scholar] [CrossRef]
- Yin, X.; Singer, S.D.; Qiao, H.; Liu, Y.; Jiao, C.; Wang, H.; Li, Z.; Fei, Z.; Wang, Y.; Fan, C. Insights into the mechanisms underlying ultraviolet-C induced resveratrol metabolism in grapevine (V. amurensis Rupr.) cv.“Tonghua-3.”. Front. Plant Sci. 2016, 7, 503. [Google Scholar]
- Yoon, M.Y.; Kim, M.Y.; Shim, S.; Kim, K.D.; Ha, J.; Shin, J.H.; Kang, S.; Lee, S.-H. Transcriptomic profiling of soybean in response to high-intensity UV-B irradiation reveals stress defense signaling. Front. Plant Sci. 2016, 7, 1917. [Google Scholar] [CrossRef] [Green Version]
- Morgante, C.V.; Brasileiro, A.C.M.; Roberts, P.A.; Guimarães, L.A.; Araújo, A.C.G.; Fonseca, L.N.; Leal-Bertioli, S.C.M.; Bertioli, D.J.; Guimarães, P.M. A survey of genes involved in Arachis stenosperma resistance to Meloidogyne arenaria race 1. Funct. Plant Biol. 2013, 40, 1298–1309. [Google Scholar] [CrossRef] [Green Version]
- Mota, A.P.Z.; Brasileiro, A.C.M.; Vidigal, B.; Oliveira, T.N.; Martins, A.d.C.Q.; Passos Saraiva, M.A.; Araújo, A.C.G.; Togawa, R.C.; Grossi-de-Sá, M.F.; Guimarães, P.M. Defining the combined stress response in wild Arachis. Sci. Rep. 2021, 11, 11097. [Google Scholar] [CrossRef]
- Mota, A.P.Z.; Fernandez, D.; Arraes, F.B.M.; Petitot, A.-S.; Melo, B.P.; Sa, M.E.L.; Grynberg, P.; Saraiva, M.A.P.; Guimarães, P.M.; Brasileiro, A.C.M.; et al. Evolutionarily conserved plant genes responsive to root-knot nematodes identified by comparative genomics. Mol. Genet. Genom. 2020, 295, 1063–1078. [Google Scholar] [CrossRef] [PubMed]
- Zarattini, M.; Farjad, M.; Launay, A.; Cannella, D.; Soulié, M.-C.; Bernacchia, G.; Fagard, M. Every cloud has a silver lining: How abiotic stresses affect gene expression in plant-pathogen interactions. J. Exp. Bot. 2021, 72, 1020–1033. [Google Scholar] [CrossRef] [PubMed]
- Frost, C.J.; Mescher, M.C.; Dervinis, C.; Davis, J.M.; Carlson, J.E.; De Moraes, C.M. Priming defense genes and metabolites in hybrid poplar by the green leaf volatile cis-3-hexenyl acetate. New Phytol. 2008, 180, 722–734. [Google Scholar] [CrossRef]
- Bernsdorff, F.; Döring, A.-C.; Gruner, K.; Schuck, S.; Bräutigam, A.; Zeier, J. Pipecolic acid orchestrates plant systemic acquired resistance and defense priming via salicylic acid-dependent and-independent pathways. Plant Cell 2016, 28, 102–129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Li, C.; Yi, J.; Yang, Y.; Lei, C.; Gong, M. Transcriptome response to drought, rehydration and re-dehydration in potato. Int. J. Mol. Sci. 2020, 21, 159. [Google Scholar] [CrossRef] [Green Version]
- Valletta, A.; Iozia, L.M.; Leonelli, F. Impact of environmental factors on stilbene biosynthesis. Plants 2021, 10, 90. [Google Scholar] [CrossRef] [PubMed]
- Yildiz, I.; Mantz, M.; Hartmann, M.; Zeier, T.; Kessel, J.; Thurow, C.; Gatz, C.; Petzsch, P.; Köhrer, K.; Zeier, J. The mobile SAR signal N-hydroxypipecolic acid induces NPR1-dependent transcriptional reprogramming and immune priming. Plant Physiol. 2021, 186, 1679–1705. [Google Scholar] [CrossRef]
- Guimarães, L.A.; Mota, A.P.Z.; Araújo, A.C.G.; Alencar Figueiredo, L.F.; Pereira, B.M.; Passos Saraiva, M.A.; Silva, R.B.; Danchin, E.G.J.J.; Guimarães, P.M.; Brasileiro, A.C.M. Genome-wide analysis of expansin superfamily in wild Arachis discloses a stress-responsive expansin-like B gene. Plant Mol. Biol. 2017, 94, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Martins, A.C.Q.; Mehta, A.; Murad, A.M.; Mota, A.P.Z.; Saraiva, M.A.P.; Araújo, A.C.G.; Miller, R.N.G.; Brasileiro, A.C.M.; Guimarães, P.M. Proteomics unravels new candidate genes for Meloidogyne resistance in wild Arachis. J. Proteom. 2020, 217, 103690. [Google Scholar] [CrossRef]
- Bertioli, D.J.; Seijo, G.; Freitas, F.O.; Valls, J.F.M.; Leal-Bertioli, S.C.M.; Moretzsohn, M.C. An overview of peanut and its wild relatives. Plant Genet. Resour. 2011, 9, 134–149. [Google Scholar] [CrossRef] [Green Version]
- Zhuang, W.; Chen, H.; Yang, M.; Wang, J.; Pandey, M.K.; Zhang, C.; Chang, W.-C.; Zhang, L.; Zhang, X.; Tang, R. The genome of cultivated peanut provides insight into legume karyotypes, polyploid evolution and crop domestication. Nat. Genet. 2019, 51, 865–876. [Google Scholar] [CrossRef] [PubMed]
- Guimarães, P.M.; Brasileiro, A.C.M.; Mehta, A.; Araújo, A.C.G. Functional genomics in peanut wild relatives. In The Peanut Genome—Compendium of Plant Genomes; Varshney, R.K., Pandey, M.K., Puppala, N., Eds.; Springer International Publishing: Berlin/Heidelberg, Germany, 2017; pp. 149–164. [Google Scholar]
- Duarte-Sierra, A.; Charles, M.T.; Arul, J. UV-C hormesis: A means of controlling diseases and delaying senescence in fresh fruits and vegetables during storage. In Postharvest Pathology of Fresh Horticultural Produce; CRC Press: Boca Raton, FL, USA, 2019; pp. 539–594. ISBN 1315209187. [Google Scholar]
- Pecinka, A.; Rosa, M.; Schikora, A.; Berlinger, M.; Hirt, H.; Luschnig, C.; Scheid, O.M. Transgenerational stress memory is not a general response in Arabidopsis. PLoS ONE 2009, 4, e5202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Onik, J.C.; Xie, Y.; Duan, Y.; Hu, X.; Wang, Z.; Lin, Q. UV-C treatment promotes quality of early ripening apple fruit by regulating malate metabolizing genes during postharvest storage. PLoS ONE 2019, 14, e0215472. [Google Scholar] [CrossRef]
- Zhang, K.; Li, W.; Ju, Y.; Wang, X.; Sun, X.; Fang, Y.; Chen, K. Transcriptomic and metabolomic basis of short-and long-term post-harvest UV-C application in regulating grape berry quality development. Foods 2021, 10, 625. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Wang, Q.; Li, L.; Grierson, D.; Yuan, S.; Zheng, S.; Wang, Y.; Wang, B.; Bai, C.; Fu, A. UV-C irradiation delays the physiological changes of bell pepper fruit during storage. Postharvest Biol. Technol. 2021, 180, 111506. [Google Scholar] [CrossRef]
- Kan, J.; Hui, Y.; Lin, X.; Liu, Y.; Jin, C. Postharvest UV-C treatment of peach fruit: Changes in transcriptome profile focusing on genes involved in softening and senescence. J. Food Process. Preserv. 2021, 45, e15813. [Google Scholar] [CrossRef]
- Aboul Fotouh, M.M.; Moawad, F.G.; El-Naggar, H.A.; El-Din, M.A.T.; Eldeen, H.A.S. Influence of seed treatment with UV-C on saline stress tolerance in green beans (Phaseolus vulgaris L.). Environ. Sci 2014, 9, 391–414. [Google Scholar]
- Ouhibi, C.; Attia, H.; Rebah, F.; Msilini, N.; Chebbi, M.; Aarrouf, J.; Urban, L.; Lachaal, M. Salt stress mitigation by seed priming with UV-C in lettuce plants: Growth, antioxidant activity and phenolic compounds. Plant Physiol. Biochem. 2014, 83, 126–133. [Google Scholar] [CrossRef]
- Ma, T.; Gao, H.; Zhang, D.; Shi, Y.; Zhang, T.; Shen, X.; Wu, L.; Xiang, L.; Chen, S. Transcriptome analyses revealed the ultraviolet B irradiation and phytohormone gibberellins coordinately promoted the accumulation of artemisinin in Artemisia annua L. Chin. Med. 2020, 15, 67. [Google Scholar] [CrossRef]
- Kovalchuk, I.; Molinier, J.; Yao, Y.; Arkhipov, A.; Kovalchuk, O. Transcriptome analysis reveals fundamental differences in plant response to acute and chronic exposure to ionizing radiation. Mutat. Res. Mol. Mech. Mutagen. 2007, 624, 101–113. [Google Scholar] [CrossRef]
- Vandenbussche, F.; Yu, N.; Li, W.; Vanhaelewyn, L.; Hamshou, M.; Van Der Straeten, D.; Smagghe, G. An ultraviolet B condition that affects growth and defense in Arabidopsis. Plant Sci. 2018, 268, 54–63. [Google Scholar] [CrossRef] [PubMed]
- Cheong, Y.H.; Chang, H.-S.S.; Gupta, R.; Wang, X.; Zhu, T.; Luan, S. Transcriptional profiling reveals novel interactions between wounding, pathogen, abiotic stress, and hormonal responses in Arabidopsis. Plant Physiol. 2002, 129, 661–677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pontin, M.A.; Piccoli, P.N.; Francisco, R.; Bottini, R.; Martinez-Zapater, J.M.; Lijavetzky, D. Transcriptome changes in grapevine (Vitis vinifera L.) cv. Malbec leaves induced by ultraviolet-B radiation. BMC Plant Biol. 2010, 10, 224. [Google Scholar] [CrossRef] [Green Version]
- Surjadinata, B.B.; Jacobo-Velázquez, D.A.; Cisneros-Zevallos, L. UVA, UVB and UVC light enhances the biosynthesis of phenolic antioxidants in fresh-cut carrot through a synergistic effect with wounding. Molecules 2017, 22, 668. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ortega-Hernández, E.; Welti-Chanes, J.; Jacobo-Velázquez, D.A. Effects of UVB light, wounding stress, and storage time on the accumulation of betalains, phenolic compounds, and ascorbic acid in red prickly pear (Opuntia ficus-indica cv. Rojo Vigor). Food Bioprocess Technol. 2018, 11, 2265–2274. [Google Scholar] [CrossRef]
- Ortega-Hernández, E.; Nair, V.; Welti-Chanes, J.; Cisneros-Zevallos, L.; Jacobo-Velázquez, D.A. Wounding and UVB light synergistically induce the biosynthesis of phenolic compounds and ascorbic acid in red prickly pears (Opuntia ficus-indica cv. Rojo Vigor). Int. J. Mol. Sci. 2019, 20, 5327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashrafi-Dehkordi, E.; Alemzadeh, A.; Tanaka, N.; Razi, H. Meta-analysis of transcriptomic responses to biotic and abiotic stress in tomato. PeerJ 2018, 6, e4631. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Wu, H.; Hu, M.; Wang, Z.; Jiang, X.; Guan, L.; Bai, W.; Lei, K. Global analysis of gene expression profiles in glutinous rice 89-1 (Oryza sativa L.) seedlings exposed to chilling stress. Plant Mol. Biol. Rep. 2021, 39, 626–639. [Google Scholar] [CrossRef]
- Robles, P.; Quesada, V. Research progress in the molecular functions of plant mTERF proteins. Cells 2021, 10, 205. [Google Scholar] [CrossRef]
- Hake, K.; Romeis, T. Protein kinase-mediated signalling in priming: Immune signal initiation, propagation, and establishment of long-term pathogen resistance in plants. Plant Cell Environ. 2019, 42, 904–917. [Google Scholar] [CrossRef]
- Tian, D.; Traw, M.B.; Chen, J.Q.; Kreitman, M.; Bergelson, J. Fitness costs of R-gene-mediated resistance in Arabidopsis thaliana. Nature 2003, 423, 74–77. [Google Scholar] [CrossRef] [PubMed]
- Ramkumar, G.; Madhav, M.S.; Biswal, A.K.; Rama Devi, S.J.S.; Sakthivel, K.; Madhan Mohan, K.; Umakanth, B.; Mangrauthi, S.K.; Sundaram, R.M.; Viraktamath, B.C. Genome-wide identification and characterization of transcription factor binding motifs of NBS-LRR genes in rice and Arabidopsis. J. Genomes Exomes 2014, 3, 7–15. [Google Scholar]
- Zhu, Q.-H.; Fan, L.; Liu, Y.; Xu, H.; Llewellyn, D.; Wilson, I. miR482 regulation of NBS-LRR defense genes during fungal pathogen infection in cotton. PLoS ONE 2013, 8, e84390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vinson, C.C.; Mota, A.P.Z.; Porto, B.N.; Oliveira, T.N.; Sampaio, I.; Lacerda, A.L.; Danchin, E.G.J.; Guimarães, P.M.; Williams, T.C.R.; Brasileiro, A.C.M. Characterization of raffinose metabolism genes uncovers a wild Arachis galactinol synthase conferring tolerance to abiotic stresses. Sci. Rep. 2020, 10, 1–19. [Google Scholar]
- Cohen, S.P.; Leach, J.E. Abiotic and biotic stresses induce a core transcriptome response in rice. Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef]
- Tugizimana, F.; Mhlongo, M.I.; Piater, L.A.; Dubery, I.A. Metabolomics in plant priming research: The way forward? Int. J. Mol. Sci. 2018, 19, 1759. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Jiang, W. UV treatment improved the quality of postharvest fruits and vegetables by inducing resistance. Trends Food Sci. Technol. 2019, 92, 71–80. [Google Scholar] [CrossRef]
- An, J.; Kim, S.H.; Bahk, S.; Vuong, U.T.; Nguyen, N.T.; Do, H.L.; Kim, S.H.; Chung, W.S. Naringenin induces pathogen resistance against Pseudomonas syringae through the activation of NPR1 in Arabidopsis. Front. Plant Sci. 2021, 12, 672552. [Google Scholar] [CrossRef]
- Shokat, S.; Novák, O.; Široká, J.; Singh, S.; Gill, K.S.; Roitsch, T.; Großkinsky, D.K.; Liu, F. Elevated CO2 modulates the effect of heat stress responses in Triticum aestivum by differential expression of an isoflavone reductase-like gene. J. Exp. Bot. 2021, 72, 7594–7609. [Google Scholar] [CrossRef]
- Holmes, E.C.; Chen, Y.-C.; Mudgett, M.B.; Sattely, E.S. Arabidopsis UGT76B1 glycosylates N-hydroxy-pipecolic acid and inactivates systemic acquired resistance in tomato. Plant Cell 2021, 33, 750–765. [Google Scholar] [CrossRef]
- Benedetti, M.; Verrascina, I.; Pontiggia, D.; Locci, F.; Mattei, B.; De Lorenzo, G.; Cervone, F. Four Arabidopsis berberine bridge enzyme-like proteins are specific oxidases that inactivate the elicitor-active oligogalacturonides. Plant J. 2018, 94, 260–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rui, Y.; Dinneny, J.R. A wall with integrity: Surveillance and maintenance of the plant cell wall under stress. New Phytol. 2020, 225, 1428–1439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bacete, L.; Mélida, H.; Miedes, E.; Molina, A. Plant cell wall-mediated immunity: Cell wall changes trigger disease resistance responses. Plant J. 2018, 93, 614–636. [Google Scholar] [CrossRef]
- De la Rubia, A.G.; Mélida, H.; Centeno, M.L.; Encina, A.; García-Angulo, P. Immune priming triggers cell wall remodeling and increased resistance to halo blight disease in common bean. Plants 2021, 10, 1514. [Google Scholar] [CrossRef] [PubMed]
- Brasileiro, A.C.M.; Lacorte, C.; Pereira, B.M.; Oliveira, T.N.; Ferreira, D.S.; Mota, A.P.Z.; Saraiva, M.A.P.; Araújo, A.C.G.; Paulino Silva, L.; Guimarães, P.M. Ectopic expression of an expansin-like B gene from wild Arachis enhances tolerance to both abiotic and biotic stresses. Plant J. 2021, 107, 1681–1696. [Google Scholar] [CrossRef]
- Gujjar, R.S.; Pathak, A.D.; Karkute, S.G.; Supaibulwattana, K. Multifunctional proline rich proteins and their role in regulating cellular proline content in plants under stress. Biol. Plant 2019, 63, 448–454. [Google Scholar] [CrossRef]
- Arcuri, M.L.C.; Fialho, L.C.; Nunes-Laitz, A.V.; Fuchs-Ferraz, M.C.P.; Wolf, I.R.; Valente, G.T.; Marino, C.L.; Maia, I.G. Genome-wide identification of multifunctional laccase gene family in Eucalyptus grandis: Potential targets for lignin engineering and stress tolerance. Trees 2020, 34, 745–758. [Google Scholar] [CrossRef]
- Ge, X.; Li, G.-J.; Wang, S.-B.; Zhu, H.; Zhu, T.; Wang, X.; Xia, Y. AtNUDT7, a negative regulator of basal immunity in Arabidopsis, modulates two distinct defense response pathways and is involved in maintaining redox homeostasis. Plant Physiol. 2007, 145, 204–215. [Google Scholar] [CrossRef] [Green Version]
- Yoosomboon, P.; Sojikul, P.; Viboonjun, U.; Narangajavana, J. Salicylic acid-induced syntaxin gene expression coexists with enhanced resistance against Colletotrichum gloeosporioides infection in cassava. Trop. Plant Biol. 2021, 14, 50–62. [Google Scholar] [CrossRef]
- Shen, M.-H.; Singh, R.K. Effect of rotating peanuts on aflatoxin detoxification by ultraviolet C light and irradiation uniformity evaluated by AgCl-based dosimeter. Food Control 2021, 120, 107533. [Google Scholar] [CrossRef]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prüfer, K.; Muetzel, B.; Do, H.-H.; Weiss, G.; Khaitovich, P.; Rahm, E.; Pääbo, S.; Lachmann, M.; Enard, W. FUNC: A package for detecting significant associations between gene sets and ontological annotations. BMC Bioinform. 2007, 8, 41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, Y.; Jiao, C.; Sun, H.; Rosli, H.; Pombo, M.A.; Zhang, P.; Banf, M.; Dai, X.; Martin, G.B.; Giovannoni, J.J. iTAK: A program for genome-wide prediction and classification of plant transcription factors, transcriptional regulators, and protein kinases. Mol. Plant 2016, 9, 1667–1670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Warnes, G.R.; Bolker, B.; Bonebakker, L.; Gentleman, R.; Liaw, W.H.A.; Lumley, T.; Venables, B. gplots: Various R Programming Tools for Plotting Data, R package version 2.17. 0. 2015., computer software. Available online: https://rdrr.io/cran/gplots/ (accessed on 1 December 2021).
- Emms, D.M.; Kelly, S. OrthoFinder: Solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 2015, 16, 157. [Google Scholar] [CrossRef] [Green Version]
- Zhao, S.; Fernald, R.D. Comprehensive algorithm for quantitative real-time polymerase chain reaction. J. Comput. Biol. 2005, 12, 1047–1064. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W.; Horgan, G.W.; Dempfle, L. Relative expression software tool (REST©) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002, 30, 36. [Google Scholar] [CrossRef]
- Morgante, C.V.; Guimarães, P.M.; Martins, A.; Araújo, A.C.G.; Leal-Bertioli, S.C.M.; Bertioli, D.J.; Brasileiro, A.C.M. Reference genes for quantitative reverse transcription-polymerase chain reaction expression studies in wild and cultivated peanut. BMC Res. Notes 2011, 4, 339. [Google Scholar] [CrossRef] [Green Version]



| Protein Family (Orthogroup) a | Universal Gene Markers (Arabidopsis thaliana) | metaDEGs (Arachis duranensis Gene Models) | BLAST |
|---|---|---|---|
| OG0000001 | AT4G21390 | Aradu.9LI83; Aradu.BTP7B; Aradu.NI7KT | Receptor-like serine/threonine kinase |
| OG0000057 | AT1G07650 | Aradu.4W18S; Aradu.KB9NK | Leucine-rich repeat (LRR) receptor-like serine/threonine-protein kinase |
| OG0000067 | AT4G38620 | Aradu.CT448; Aradu.18EWZ | MYB-like transcription factor |
| OG0000286 | AT4G02340 | Aradu.JJ61J | Epoxide hydrolase |
| OG0001961 | AT5G48380 | Aradu.8IX7E | BAK1-interacting receptor-like kinase 1 |
| OG0001992 | AT3G62870 | Aradu.32KIU; Aradu.9L616 | 60S ribosomal protein L7A (RPL7aB) |
| OG0006245 | AT2G44500 | Aradu.YX17V | O-fucosyltransferase family protein |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martins, A.C.Q.; Mota, A.P.Z.; Carvalho, P.A.S.V.; Passos, M.A.S.; Gimenes, M.A.; Guimaraes, P.M.; Brasileiro, A.C.M. Transcriptome Responses of Wild Arachis to UV-C Exposure Reveal Genes Involved in General Plant Defense and Priming. Plants 2022, 11, 408. https://doi.org/10.3390/plants11030408
Martins ACQ, Mota APZ, Carvalho PASV, Passos MAS, Gimenes MA, Guimaraes PM, Brasileiro ACM. Transcriptome Responses of Wild Arachis to UV-C Exposure Reveal Genes Involved in General Plant Defense and Priming. Plants. 2022; 11(3):408. https://doi.org/10.3390/plants11030408
Chicago/Turabian StyleMartins, Andressa Cunha Quintana, Ana Paula Zotta Mota, Paula Andrea Sampaio Vasconcelos Carvalho, Mario Alfredo Saraiva Passos, Marcos Aparecido Gimenes, Patricia Messenberg Guimaraes, and Ana Cristina Miranda Brasileiro. 2022. "Transcriptome Responses of Wild Arachis to UV-C Exposure Reveal Genes Involved in General Plant Defense and Priming" Plants 11, no. 3: 408. https://doi.org/10.3390/plants11030408
APA StyleMartins, A. C. Q., Mota, A. P. Z., Carvalho, P. A. S. V., Passos, M. A. S., Gimenes, M. A., Guimaraes, P. M., & Brasileiro, A. C. M. (2022). Transcriptome Responses of Wild Arachis to UV-C Exposure Reveal Genes Involved in General Plant Defense and Priming. Plants, 11(3), 408. https://doi.org/10.3390/plants11030408







