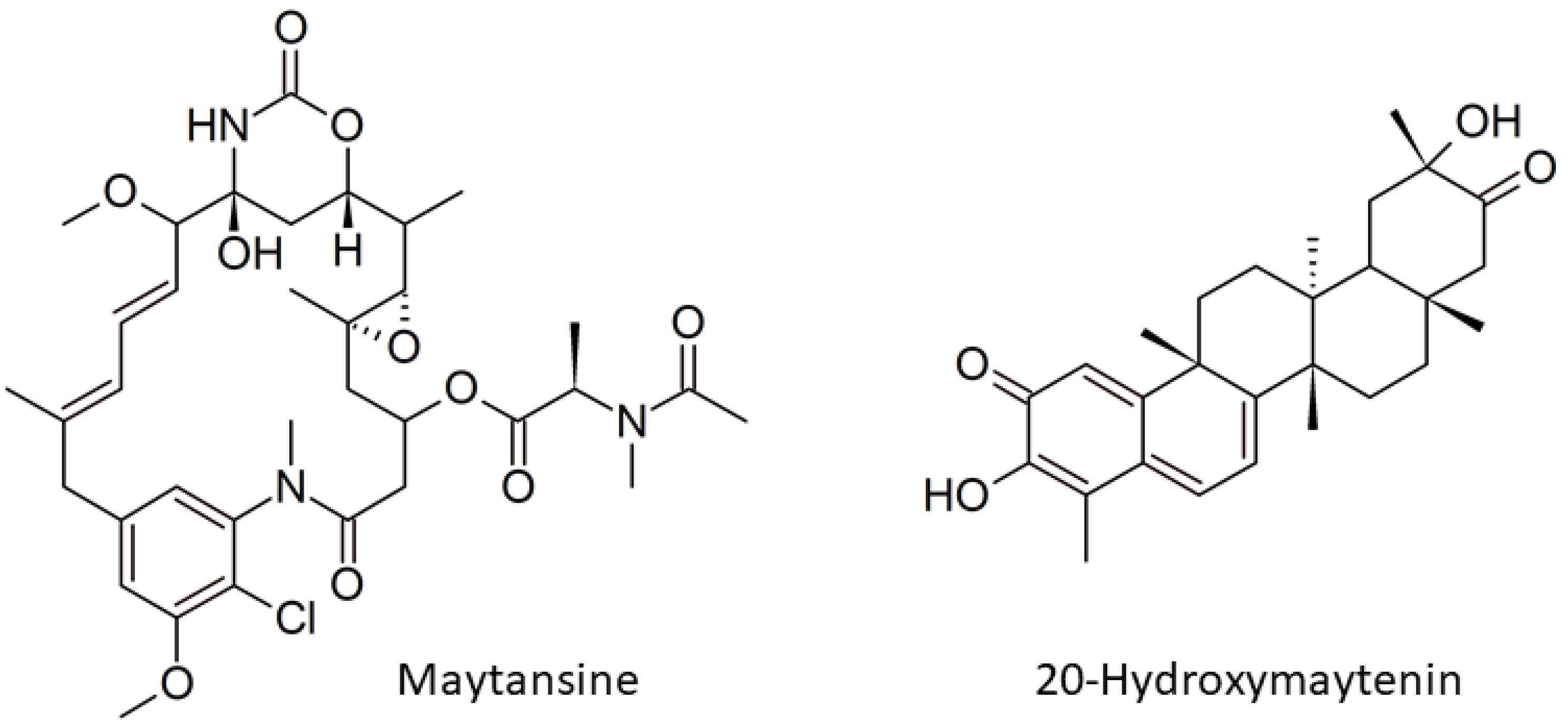
Genome Mining and Gene Expression Reveal Maytansine Biosynthetic Genes from Endophytic Communities Living inside Gymnosporia heterophylla (Eckl. and Zeyh.) Loes. and the Relationship with the Plant Biosynthetic Gene, Friedelin Synthase
Abstract
:1. Introduction
2. Results
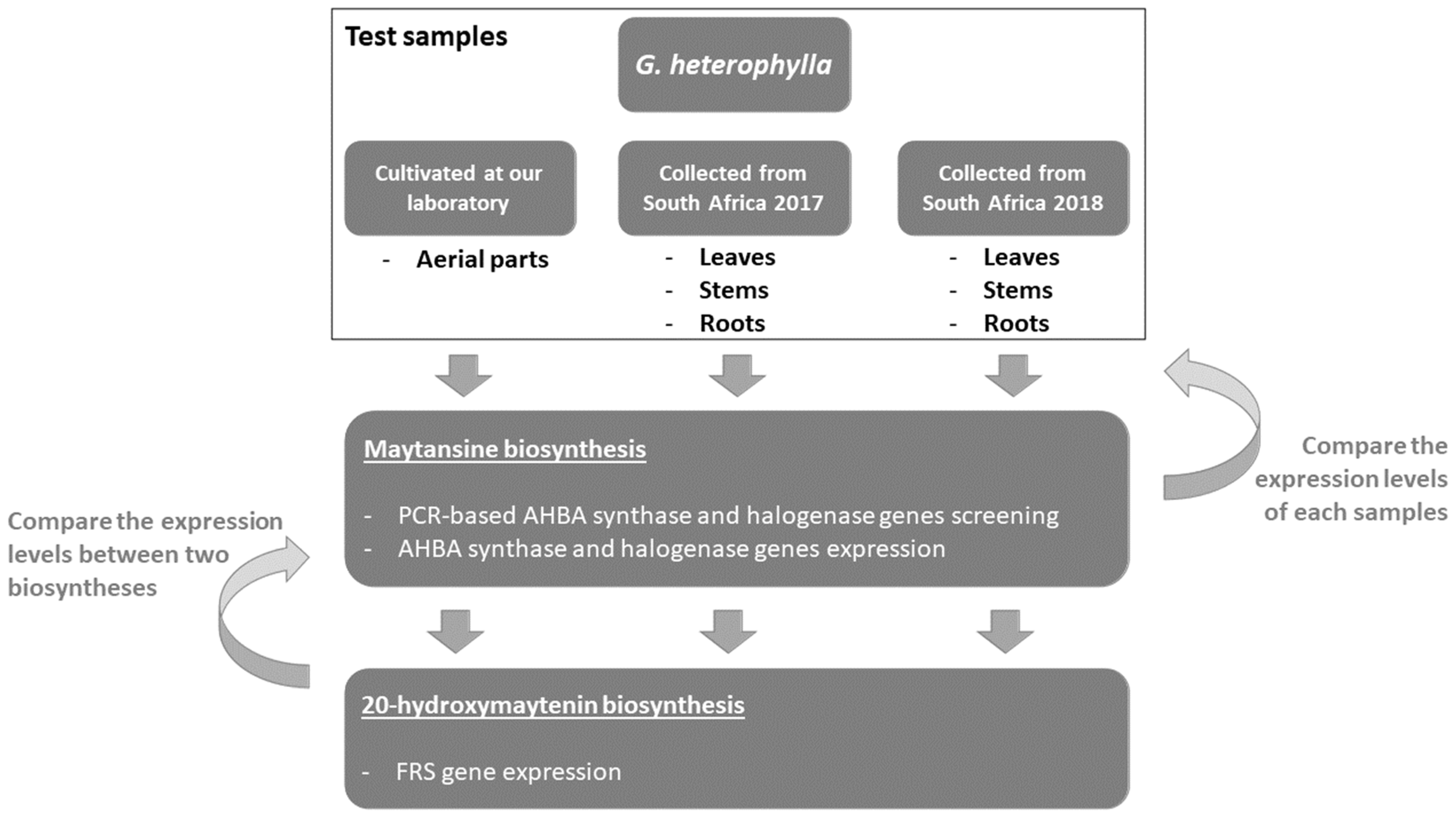
2.1. PCR-Based Genome Mining for Maytansine Biosynthetic Genes from G. heterophylla
2.1.1. Amino-Hydroxybenzoic Acid (AHBA) Synthase Gene from G. heterophylla
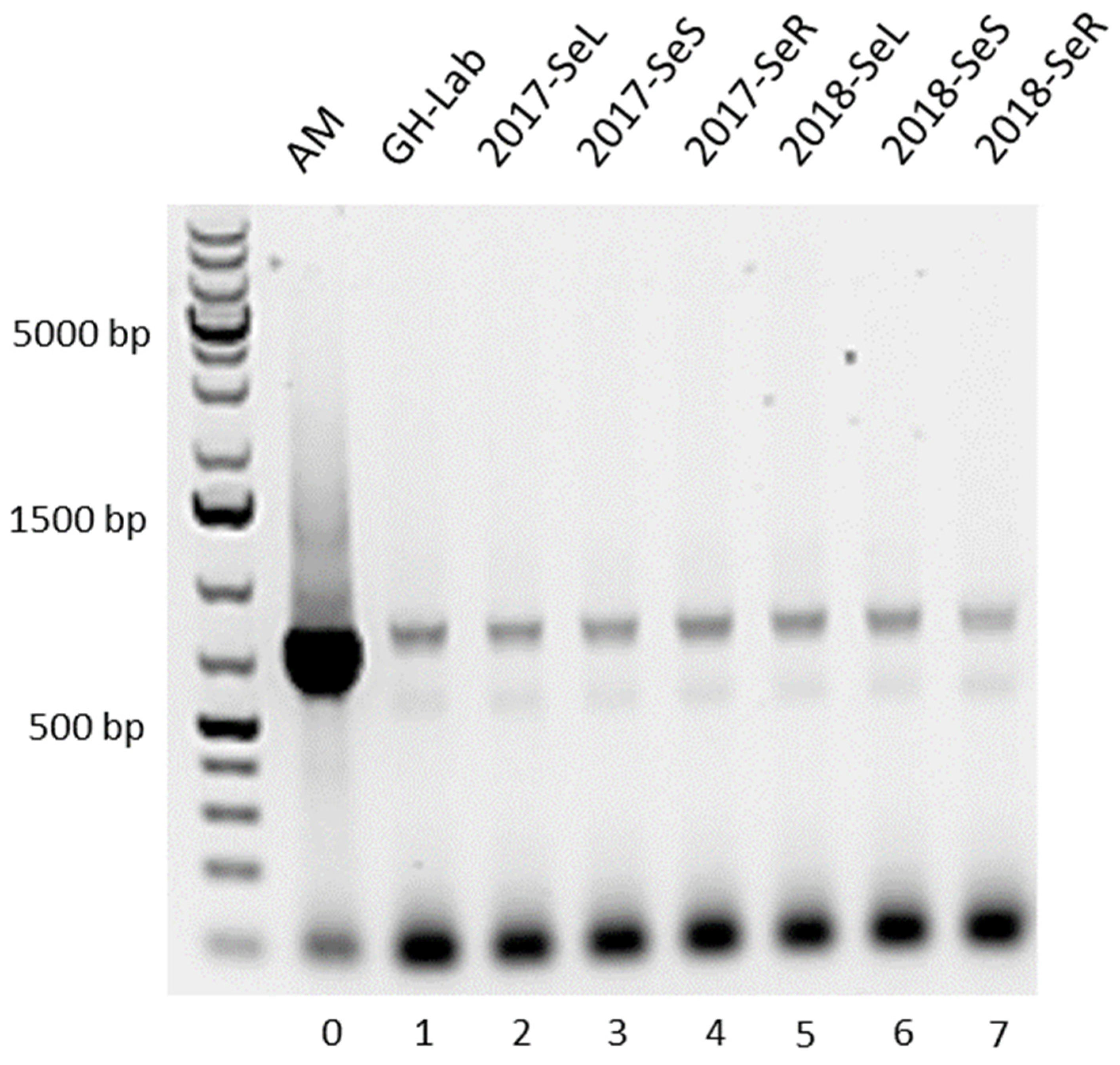
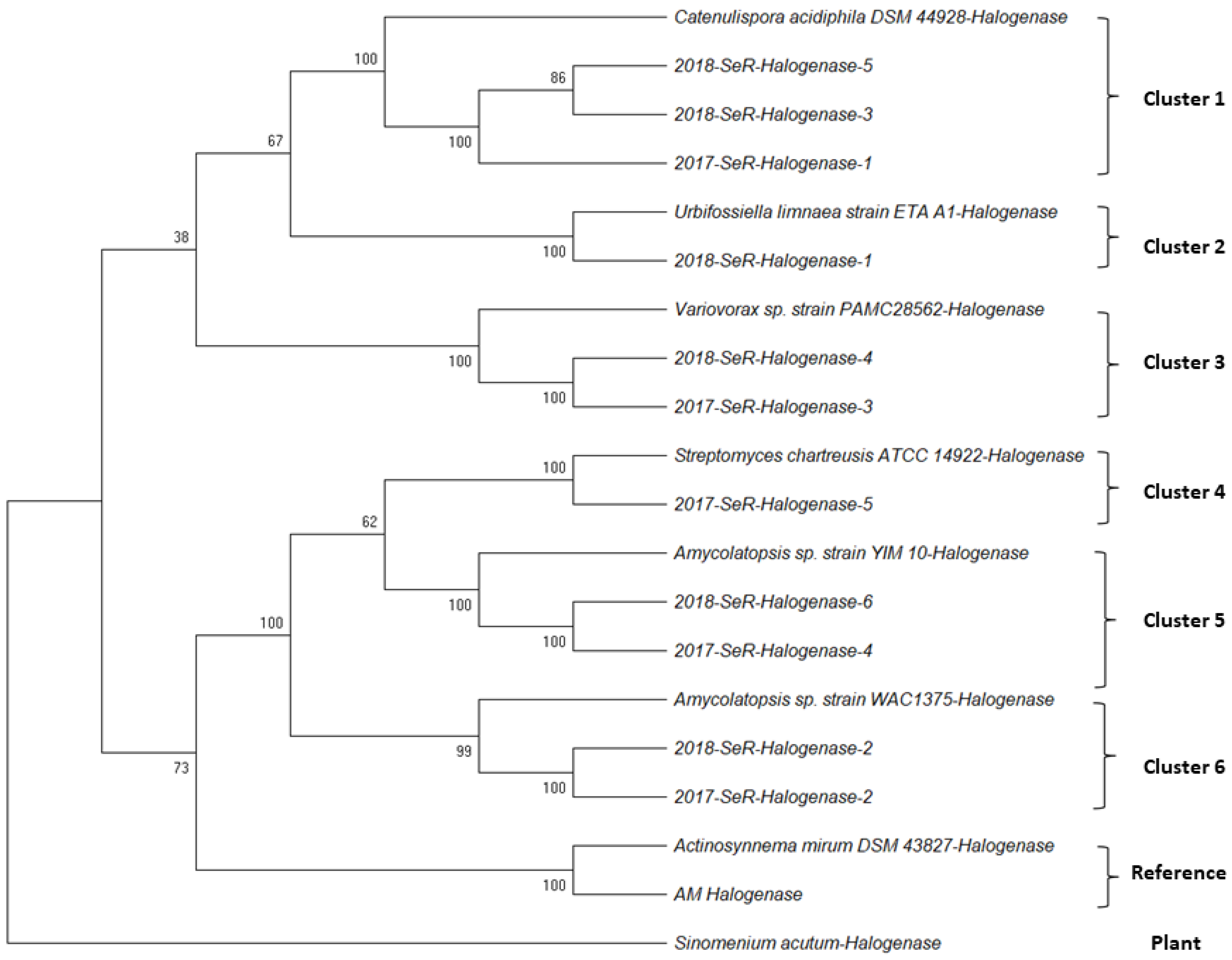
2.1.2. Halogenase Gene from G. heterophylla
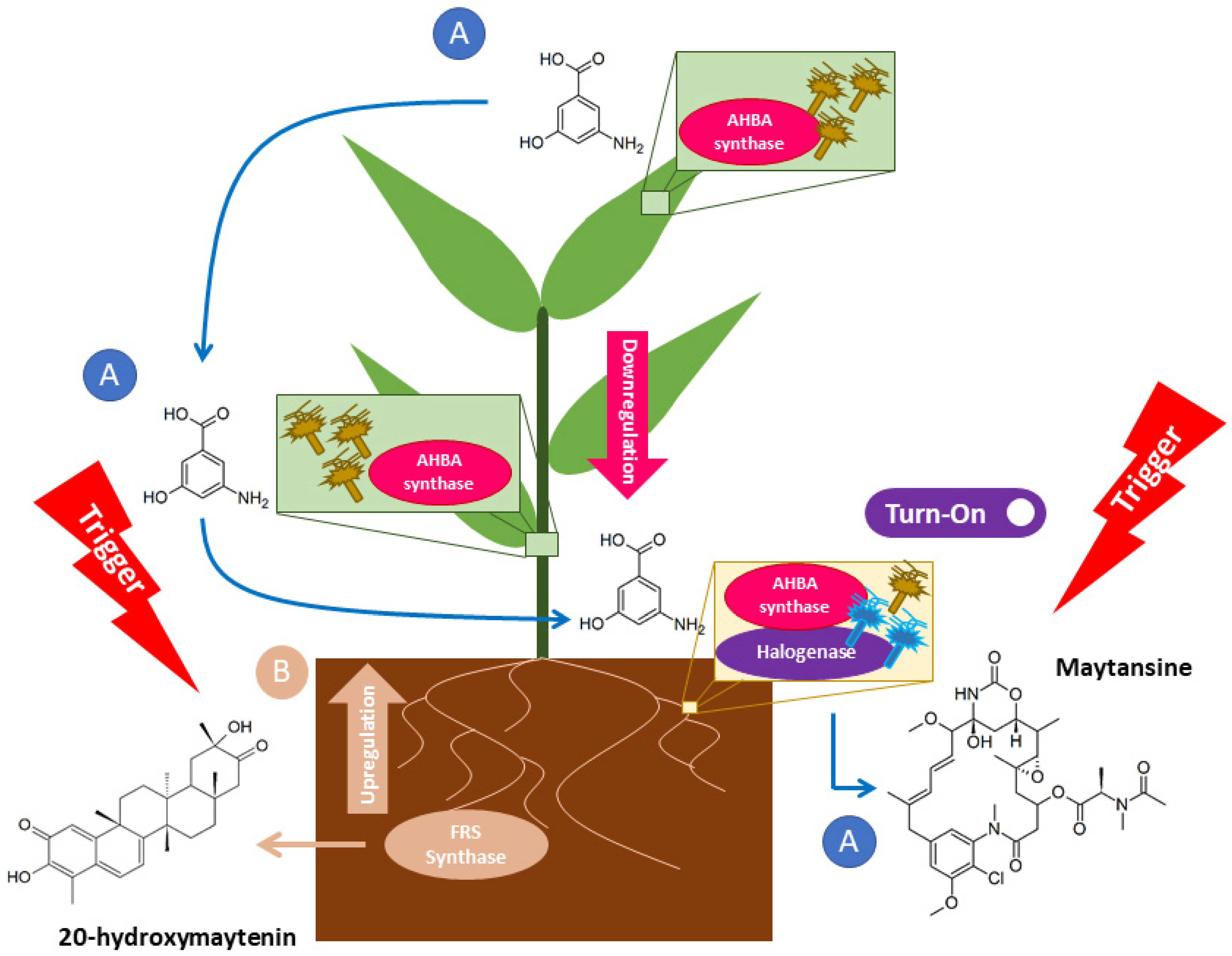
2.2. Maytansine and 20-Hydroxymaytenin Biosynthetic Genes Expressions from Endophytic Communities and G. heterophylla
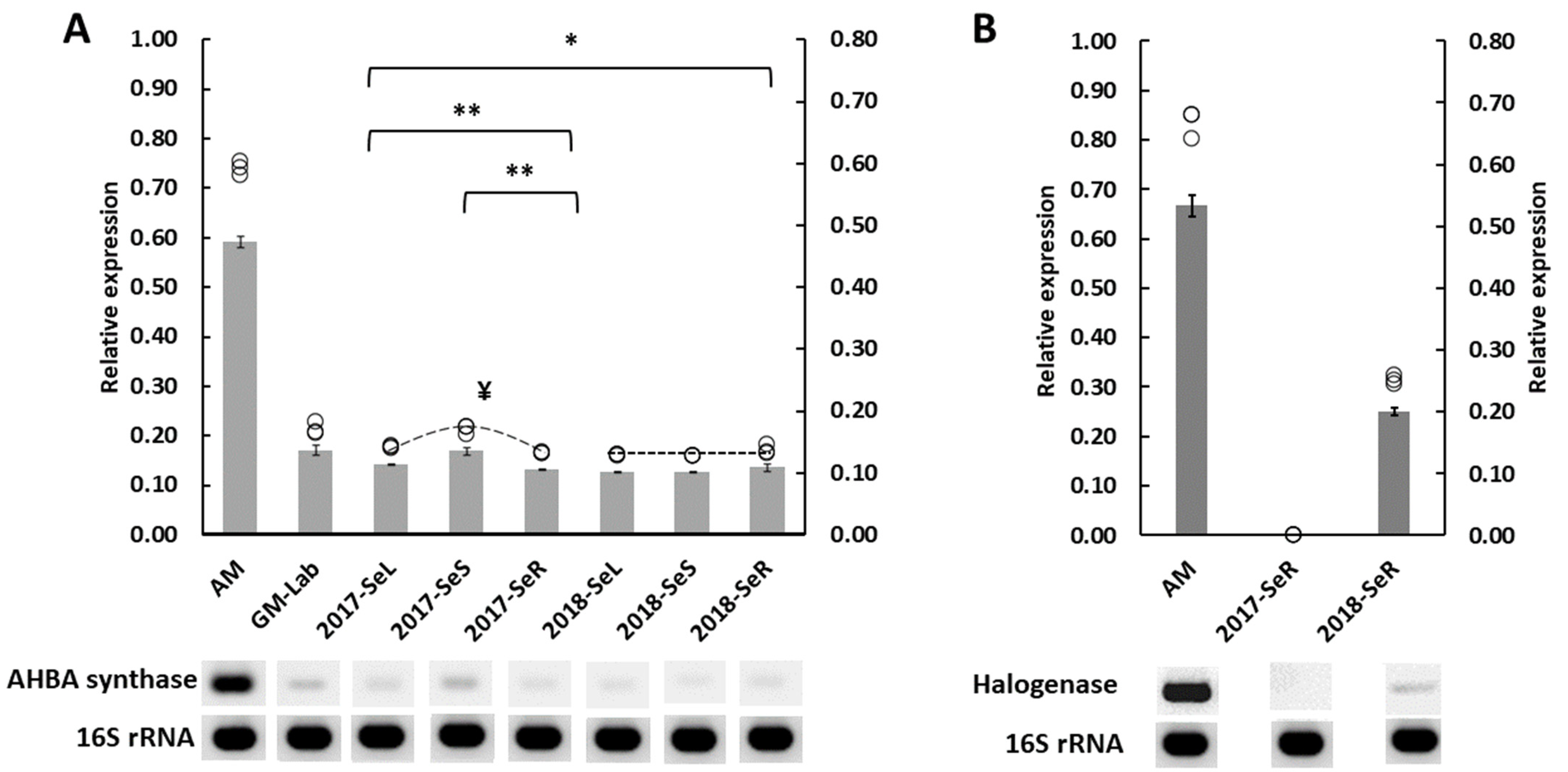
2.2.1. AHBA Synthase and Halogenase Genes Expression from Endophytic Bacteria in G. heterophylla
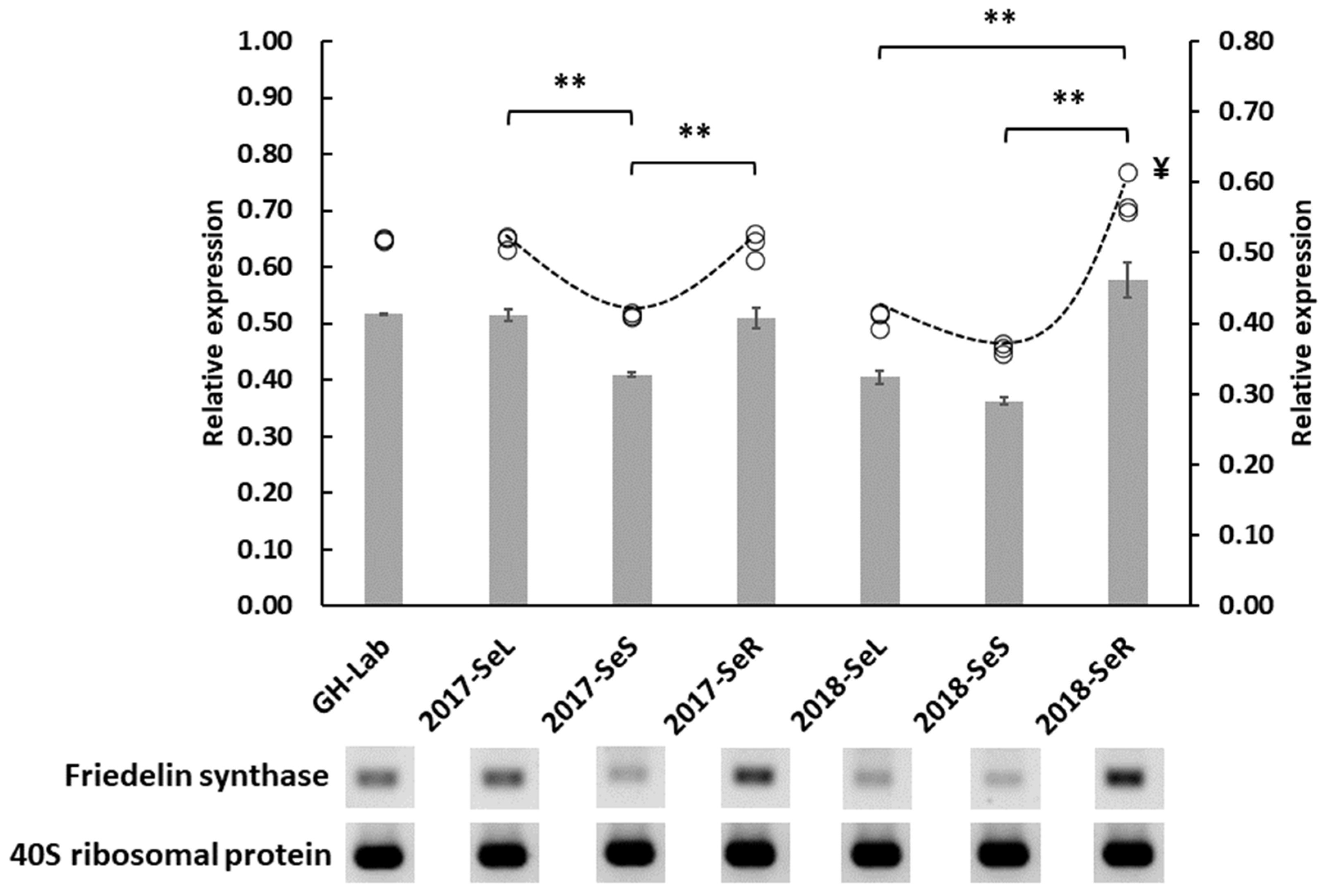
2.2.2. Friedelin Synthase or FRS Gene Expression from G. heterophylla
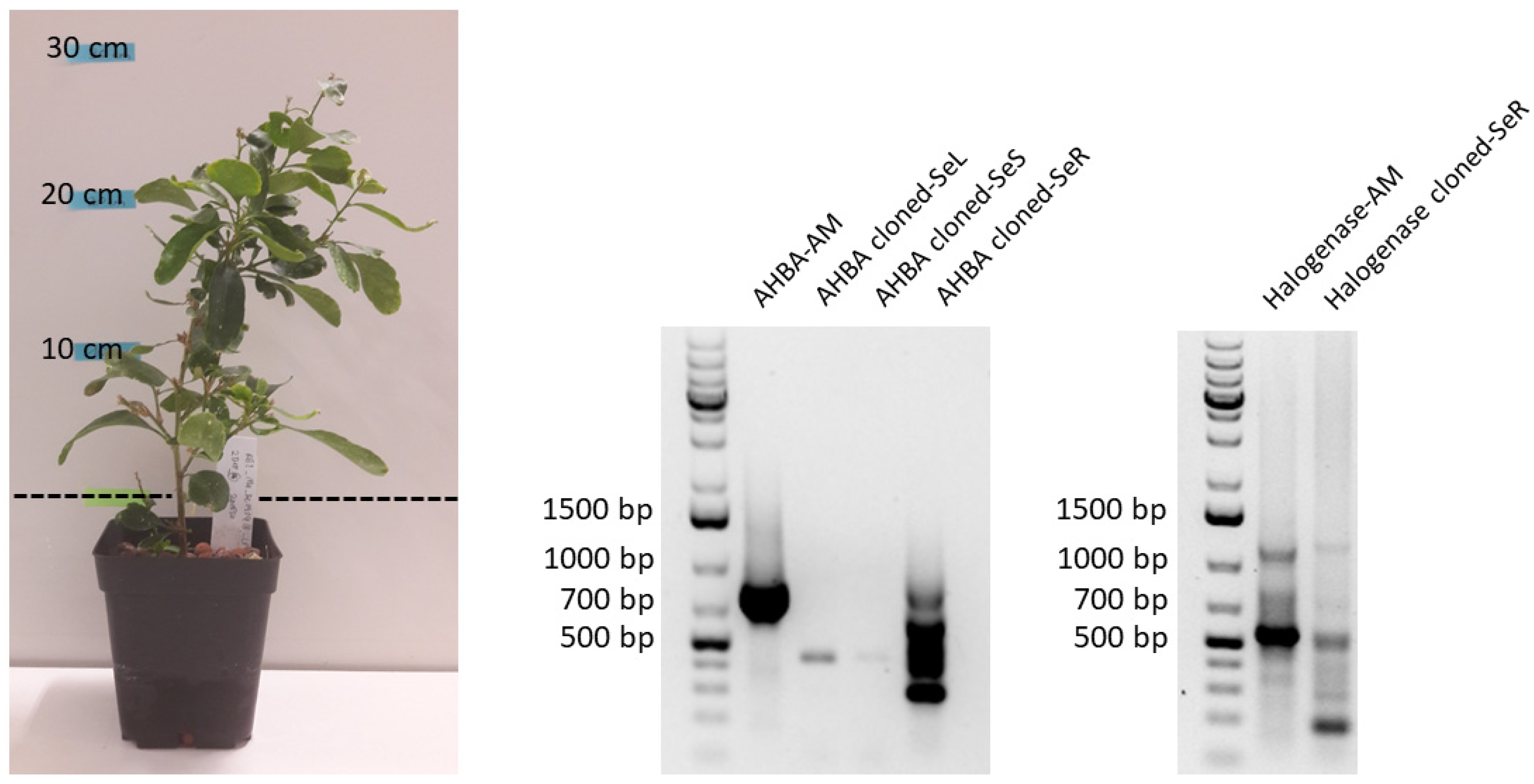
2.3. Transmission of Maytansine Producible Endophytes to a Vegetatively Cloned G. heterophylla Plant
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. PCR-Based Genome Mining Experiment Amplifying Maytansine Biosynthetic Genes from G. heterophylla Originating from South Africa
4.2.1. Preparation of G. heterophylla Plant Materials
4.2.2. Plant Genomic DNA Extraction and PCR Amplification for AHBA Synthase Gene from G. heterophylla
4.2.3. PCR Amplification for Halogenase Gene from G. heterophylla and the Vector-Cloned Library Construction
4.3. Maytasine and 20-Hydroxymaytenin Biosynthetic Genes Expression
4.3.1. Maytansine Biosynthetic (AHBA Synthase and Halogenase) Genes Expression
4.3.2. 20-Hydroxymaytenin Biosynthetic (FRS) Gene Expression
4.3.3. Bioinformatic and Statistical Analysis
4.4. Data Availability
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Salvatore, M.M.; Andolfi, A.; Nicoletti, R. The Thin Line between Pathogenicity and Endophytism: The Case of Lasiodiplodia theobromae. Agriculture 2020, 10, 488. [Google Scholar] [CrossRef]
- Khan, A.L.; Gilani, S.A.; Waqas, M.; Al-Hosni, K.; Al-Khiziri, S.; Kim, Y.-H.; Ali, L.; Kang, S.-M.; Asaf, S.; Shahzad, R.; et al. Endophytes from Medicinal Plants and Their Potential for Producing Indole Acetic Acid, Improving Seed Germination and Mitigating Oxidative Stress. J. Zhejiang Univ. Sci. B 2017, 18, 125–137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ek-Ramos, M.J.; Gomez-Flores, R.; Orozco-Flores, A.A.; Rodríguez-Padilla, C.; González-Ochoa, G.; Tamez-Guerra, P. Bioactive Products From Plant-Endophytic Gram-Positive Bacteria. Front. Microbiol. 2019, 10, 463. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.J. Are Microbial Endophytes the ‘Actual’ Producers of Bioactive Antitumor Agents? Trends Cancer 2018, 4, 662–670. [Google Scholar] [CrossRef]
- Newman, D.J. The Impact of Decreasing Biodiversity on Novel Drug Discovery: Is There a Serious Cause for Concern? Expert Opin. Drug Discov. 2019, 14, 521–525. [Google Scholar] [CrossRef]
- Kowalczyk, T.; Wieczfinska, J.; Skała, E.; Śliwiński, T.; Sitarek, P. Transgenesis as a Tool for the Efficient Production of Selected Secondary Metabolites from Plant in Vitro Cultures. Plants 2020, 9, 132. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.; Kanakkanthara, A. Beyond the Paclitaxel and Vinca Alkaloids: Next Generation of Plant-Derived Microtubule-Targeting Agents with Potential Anticancer Activity. Cancers 2020, 12, 1721. [Google Scholar] [CrossRef]
- Newman, D.J.; Cragg, G.M. Plant Endophytes and Epiphytes: Burgeoning Sources of Known and “Unknown” Cytotoxic and Antibiotic Agents? Planta Med. 2020, 86, 891–905. [Google Scholar] [CrossRef] [Green Version]
- Kupchan, S.M.; Komoda, Y.; Court, W.A.; Thomas, G.J.; Smith, R.M.; Karim, A.; Gilmore, C.J.; Haltiwanger, R.C.; Bryan, R.F. Tumor Inhibitors. LXXIII. Maytansine, a Novel Antileukemic Ansa Macrolide from Maytenus Ovatus. J. Am. Chem. Soc. 1972, 94, 1354–1356. [Google Scholar] [CrossRef]
- Cassady, J.M.; Chan, K.K.; Floss, H.G.; Leistner, E. Recent Developments in the Maytansinoid Antitumor Agents. Chem. Pharm. Bull. 2004, 52, 1–26. [Google Scholar] [CrossRef] [Green Version]
- Higashide, E.; Asai, M.; Ootsu, K.; Tanida, S.; Kozai, Y.; Hasegawa, T.; Kishi, T.; Sugino, Y.; Yoneda, M. Ansamitocin, a Group of Novel Maytansinoid Antibiotics with Antitumour Properties from Nocardia. Nature 1977, 270, 721–722. [Google Scholar] [CrossRef] [PubMed]
- Pullen, C.B.; Schmitz, P.; Hoffmann, D.; Meurer, K.; Boettcher, T.; von Bamberg, D.; Pereira, A.M.; de Castro França, S.; Hauser, M.; Geertsema, H.; et al. Occurrence and Non-Detectability of Maytansinoids in Individual Plants of the Genera Maytenus and Putterlickia. Phytochemistry 2003, 62, 377–387. [Google Scholar] [CrossRef]
- Wings, S.; Müller, H.; Berg, G.; Lamshöft, M.; Leistner, E. A Study of the Bacterial Community in the Root System of the Maytansine Containing Plant Putterlickia Verrucosa. Phytochemistry 2013, 91, 158–164. [Google Scholar] [CrossRef] [PubMed]
- Kusari, S.; Lamshöft, M.; Kusari, P.; Gottfried, S.; Zühlke, S.; Louven, K.; Hentschel, U.; Kayser, O.; Spiteller, M. Endophytes Are Hidden Producers of Maytansine in Putterlickia Roots. J. Nat. Prod. 2014, 77, 2577–2584. [Google Scholar] [CrossRef] [Green Version]
- Kusari, P.; Kusari, S.; Eckelmann, D.; Zühlke, S.; Kayser, O.; Spiteller, M. Cross-Species Biosynthesis of Maytansine in Maytenus serrata. RSC Adv. 2016, 6, 10011–10016. [Google Scholar] [CrossRef] [Green Version]
- Souza, B.J.M.; Lima, P.L.; Chequer, F.M.D.; Duarte-Almeida, J.M. Os Derivados Da Cannabis sativa Têm Potencial Para Limitar a Severidade e a Progressão Da COVID-19? Uma Revisão Da Literatura. Braz. J. Health Pharm. 2020, 2, 83–96. [Google Scholar] [CrossRef]
- Alves, T.B.; Souza-Moreira, T.M.; Valentini, S.R.; Zanelli, C.F.; Furlan, M. Friedelin in Maytenus ilicifolia Is Produced by Friedelin Synthase Isoforms. Molecules 2018, 23, 700. [Google Scholar] [CrossRef] [Green Version]
- Hernandes, C.; Miguita, L.; de Sales, R.O.; Silva, E.D.; Mendonça, P.O.; Lorencini da Silva, B.; Klingbeil, M.D.; Mathor, M.B.; Rangel, E.B.; Marti, L.C.; et al. Anticancer Activities of the Quinone-Methide Triterpenes Maytenin and 22-β-Hydroxymaytenin Obtained from Cultivated Maytenus ilicifolia Roots Associated with Down-Regulation of MiRNA-27a and MiR-20a/MiR-17-5p. Molecules 2020, 25, 760. [Google Scholar] [CrossRef] [Green Version]
- López, M.R.; de León, L.; Moujir, L. Antibacterial Properties of Phenolic Triterpenoids against Staphylococcus epidermidis. Planta Med. 2011, 77, 726–729. [Google Scholar] [CrossRef] [Green Version]
- Rodrigues, V.G.; Duarte, L.P.; Silva, G.D.F.; Silva, F.C.; Góes, J.V.; Takahashi, J.A.; Pimenta, L.P.S.; Vieira Filho, S.A. Evaluation of Antimicrobial Activity and Toxic Potential of Extracts and Triterpenes Isolated from Maytenus imbricata. Quím. Nova 2012, 35, 1375–1380. [Google Scholar] [CrossRef] [Green Version]
- Pitakbut, T.; Spiteller, M.; Kayser, O. In Vitro Production and Exudation of 20-Hydroxymaytenin from Gymnosporia heterophylla (Eckl. and Zeyh.) Loes. Cell Culture Plants 2021, 10, 1493. [Google Scholar] [CrossRef] [PubMed]
- Huitu, Z.; Linzhuan, W.; Aiming, L.; Guizhi, S.; Feng, H.; Qiuping, L.; Yuzhen, W.; Huanzhang, X.; Qunjie, G.; Yiguang, W. PCR Screening of 3-Amino-5-Hydroxybenzoic Acid Synthase Gene Leads to Identification of Ansamycins and AHBA-Related Antibiotic Producers in Actinomycetes. J. Appl. Microbiol. 2009, 106, 755–763. [Google Scholar] [CrossRef] [PubMed]
- Floss, H.G.; Yu, T.-W.; Arakawa, K. The Biosynthesis of 3-Amino-5-Hydroxybenzoic Acid (AHBA), the Precursor of MC7N Units in Ansamycin and Mitomycin Antibiotics: A Review. J. Antibiot. 2011, 64, 35–44. [Google Scholar] [CrossRef] [Green Version]
- Kim, C.Y.; Mitchell, A.J.; Glinkerman, C.M.; Li, F.-S.; Pluskal, T.; Weng, J.-K. The Chloroalkaloid (−)-Acutumine Is Biosynthesized via a Fe(II)- and 2-Oxoglutarate-Dependent Halogenase in Menispermaceae Plants. Nat. Commun. 2020, 11, 1867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kallscheuer, N.; Rast, P.; Jogler, M.; Wiegand, S.; Kohn, T.; Boedeker, C.; Jeske, O.; Heuer, A.; Quast, C.; Glöckner, F.O.; et al. Analysis of Bacterial Communities in a Municipal Duck Pond during a Phytoplankton Bloom and Isolation of Anatilimnocola Aggregata Gen. Nov., Sp. Nov., Lacipirellula Limnantheis Sp. Nov. and Urbifossiella Limnaea Gen. Nov., Sp. Nov. Belonging to the Phylum Planctomycetes. Environ. Microbiol. 2021, 23, 1379–1396. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.; Kim, W.; Hwang, S.; Lee, Y.; Cho, S.; Palsson, B.; Cho, B.-K. Thirty Complete Streptomyces Genome Sequences for Mining Novel Secondary Metabolite Biosynthetic Gene Clusters. Sci. Data 2020, 7, 55. [Google Scholar] [CrossRef] [PubMed]
- Eckelmann, D.; Kusari, S.; Spiteller, M. Spatial Profiling of Maytansine during the Germination Process of Maytenus senegalensis Seeds. Fitoterapia 2017, 119, 51–56. [Google Scholar] [CrossRef]
- Kanehisa, M. Toward Understanding the Origin and Evolution of Cellular Organisms. Protein Sci. 2019, 28, 1947–1951. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Ishiguro-Watanabe, M.; Tanabe, M. KEGG: Integrating Viruses and Cellular Organisms. Nucleic Acids Res. 2021, 49, D545–D551. [Google Scholar] [CrossRef]
- Thaker, M.N.; Wang, W.; Spanogiannopoulos, P.; Waglechner, N.; King, A.M.; Medina, R.; Wright, G.D. Identifying Producers of Antibacterial Compounds by Screening for Antibiotic Resistance. Nat. Biotechnol. 2013, 31, 922–927. [Google Scholar] [CrossRef]
- Pavarini, D.P.; Selegato, D.M.; Castro-Gamboa, I.; do Sacramento, L.V.S.; Furlan, M. Ecological Insights to Track Cytotoxic Compounds among Maytenus ilicifolia Living Individuals and Clones of an Ex Situ Collection. Molecules 2019, 24, 1160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inácio, M.C.; Paz, T.A.; Pereira, A.M.S.; Furlan, M. Maytenin Plays a Special Role in the Regulation of the Endophytic Bacillus megaterium in Peritassa campestris Adventitious Roots. J. Chem. Ecol. 2019, 45, 789–797. [Google Scholar] [CrossRef] [PubMed]
- Pereira, L.C.; Bertuzzi Pereira, C.; Correia, L.V.; Matera, T.C.; dos Santos, R.F.; de Carvalho, C.; Osipi, E.A.F.; Braccini, A.L. Corn Responsiveness to Azospirillum: Accessing the Effect of Root Exudates on the Bacterial Growth and Its Ability to Fix Nitrogen. Plants 2020, 9, 923. [Google Scholar] [CrossRef] [PubMed]
- Vranova, V.; Rejsek, K.; Skene, K.R.; Janous, D.; Formanek, P. Methods of Collection of Plant Root Exudates in Relation to Plant Metabolism and Purpose: A Review. J. Plant Nutr. Soil Sci. 2013, 176, 175–199. [Google Scholar] [CrossRef]
- Dankwa, A.S.; Machado, R.M.; Perry, J.J. Sources of Food Contamination in a Closed Hydroponic System. Lett. Appl. Microbiol. 2020, 70, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Lee, J. Beneficial Bacteria and Fungi in Hydroponic Systems: Types and Characteristics of Hydroponic Food Production Methods. Sci. Hortic. 2015, 195, 206–215. [Google Scholar] [CrossRef]
- Frank, A.C.; Saldierna Guzmán, J.P.; Shay, J.E. Transmission of Bacterial Endophytes. Microorganisms 2017, 5, 70. [Google Scholar] [CrossRef] [Green Version]
- Saleh, D.; Sharma, M.; Seguin, P.; Jabaji, S. Organic Acids and Root Exudates of Brachypodium distachyon: Effects on Chemotaxis and Biofilm Formation of Endophytic Bacteria. Can. J. Microbiol. 2020, 66, 562–575. [Google Scholar] [CrossRef]
- Xiong, Q.; Liu, D.; Zhang, H.; Dong, X.; Zhang, G.; Liu, Y.; Zhang, R. Quorum Sensing Signal Autoinducer-2 Promotes Root Colonization of Bacillus velezensis SQR9 by Affecting Biofilm Formation and Motility. Appl. Microbiol. Biotechnol. 2020, 104, 7177–7185. [Google Scholar] [CrossRef]
- Forte, F.P.; Schmid, J.; Dijkwel, P.P.; Nagy, I.; Hume, D.E.; Johnson, R.D.; Simpson, W.R.; Monk, S.M.; Zhang, N.; Sehrish, T.; et al. Fungal Endophyte Colonization Patterns Alter Over Time in the Novel Association Between Lolium perenne and Epichloë Endophyte AR37. Front. Plant Sci. 2020, 11, 1566. [Google Scholar] [CrossRef]
- Hornung, A.; Bertazzo, M.; Dziarnowski, A.; Schneider, K.; Welzel, K.; Wohlert, S.-E.; Holzenkämpfer, M.; Nicholson, G.J.; Bechthold, A.; Süssmuth, R.D.; et al. A Genomic Screening Approach to the Structure-Guided Identification of Drug Candidates from Natural Sources. ChemBioChem 2007, 8, 757–766. [Google Scholar] [CrossRef] [PubMed]
- Ramos, A.E.; Muñoz, M.; Moreno-Pérez, D.A.; Patarroyo, M.A. PELMO, an Optimised in-House Cloning Vector. AMB Express 2017, 7, 26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ning, X.; Wang, X.; Wu, Y.; Kang, Q.; Bai, L. Identification and Engineering of Post-PKS Modification Bottlenecks for Ansamitocin P-3 Titer Improvement in Actinosynnema pretiosum Subsp. pretiosum ATCC 31280. Biotechnol. J. 2017, 12, 1700484. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Fan, Y.; Nambou, K.; Wei, L.; Liu, Z.; Imanaka, T.; Hua, Q. Enhancement of Ansamitocin P-3 Production in Actinosynnema pretiosum by a Synergistic Effect of Glycerol and Glucose. J. Ind. Microbiol. Biotechnol. 2014, 41, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 Years of Image Analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Zhi, K.; Mukherji, A.; Gerth, K. Repurposing Antiviral Protease Inhibitors Using Extracellular Vesicles for Potential Therapy of COVID-19. Viruses 2020, 12, 486. [Google Scholar] [CrossRef] [PubMed]
- Carr, N.T. Using Microsoft Excel® to Calculate Descriptive Statistics and Create Graphs. Lang. Assess. Q. 2008, 5, 43–62. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pitakbut, T.; Spiteller, M.; Kayser, O. Genome Mining and Gene Expression Reveal Maytansine Biosynthetic Genes from Endophytic Communities Living inside Gymnosporia heterophylla (Eckl. and Zeyh.) Loes. and the Relationship with the Plant Biosynthetic Gene, Friedelin Synthase. Plants 2022, 11, 321. https://doi.org/10.3390/plants11030321
Pitakbut T, Spiteller M, Kayser O. Genome Mining and Gene Expression Reveal Maytansine Biosynthetic Genes from Endophytic Communities Living inside Gymnosporia heterophylla (Eckl. and Zeyh.) Loes. and the Relationship with the Plant Biosynthetic Gene, Friedelin Synthase. Plants. 2022; 11(3):321. https://doi.org/10.3390/plants11030321
Chicago/Turabian StylePitakbut, Thanet, Michael Spiteller, and Oliver Kayser. 2022. "Genome Mining and Gene Expression Reveal Maytansine Biosynthetic Genes from Endophytic Communities Living inside Gymnosporia heterophylla (Eckl. and Zeyh.) Loes. and the Relationship with the Plant Biosynthetic Gene, Friedelin Synthase" Plants 11, no. 3: 321. https://doi.org/10.3390/plants11030321
APA StylePitakbut, T., Spiteller, M., & Kayser, O. (2022). Genome Mining and Gene Expression Reveal Maytansine Biosynthetic Genes from Endophytic Communities Living inside Gymnosporia heterophylla (Eckl. and Zeyh.) Loes. and the Relationship with the Plant Biosynthetic Gene, Friedelin Synthase. Plants, 11(3), 321. https://doi.org/10.3390/plants11030321






