ER Stress and the Unfolded Protein Response: Homeostatic Regulation Coordinate Plant Survival and Growth
Abstract
1. Introduction
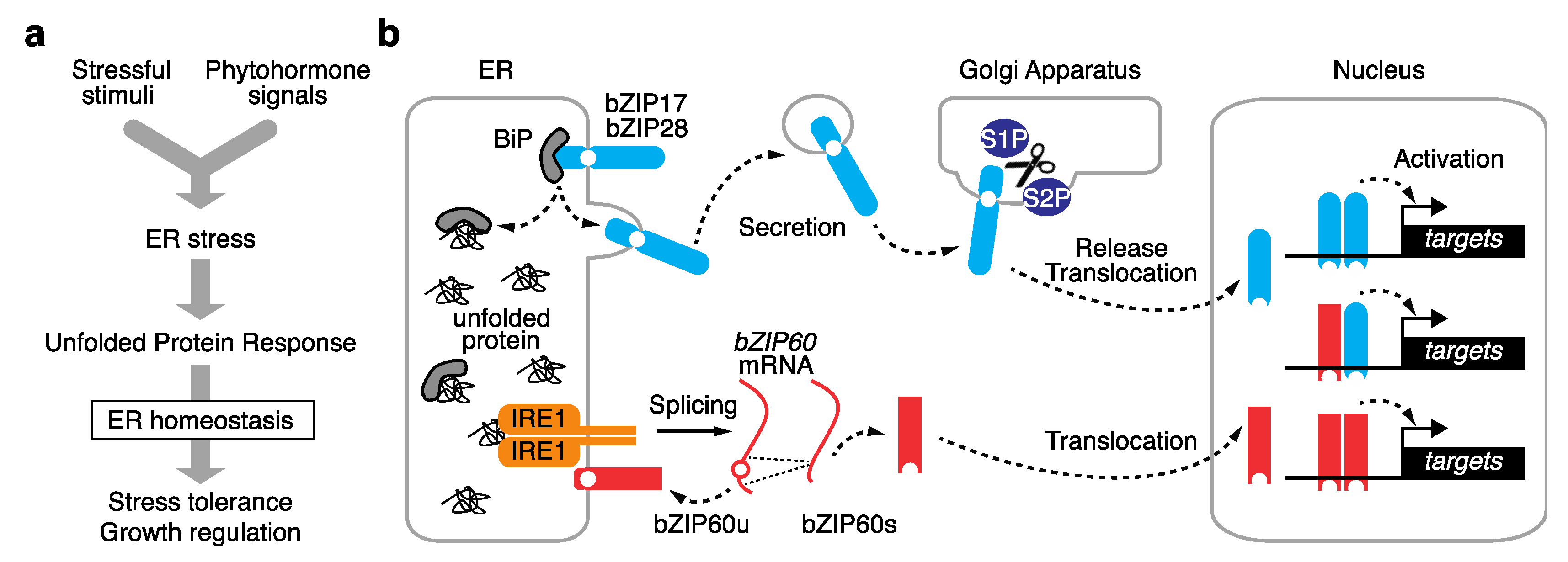
2. Two Distinct Plant UPR Pathways
2.1. Proteolysis-Dependent UPR Pathway
2.2. mRNA Splicing-Dependent UPR Pathway
2.3. Modes of Action of UPR bZIP Dimers
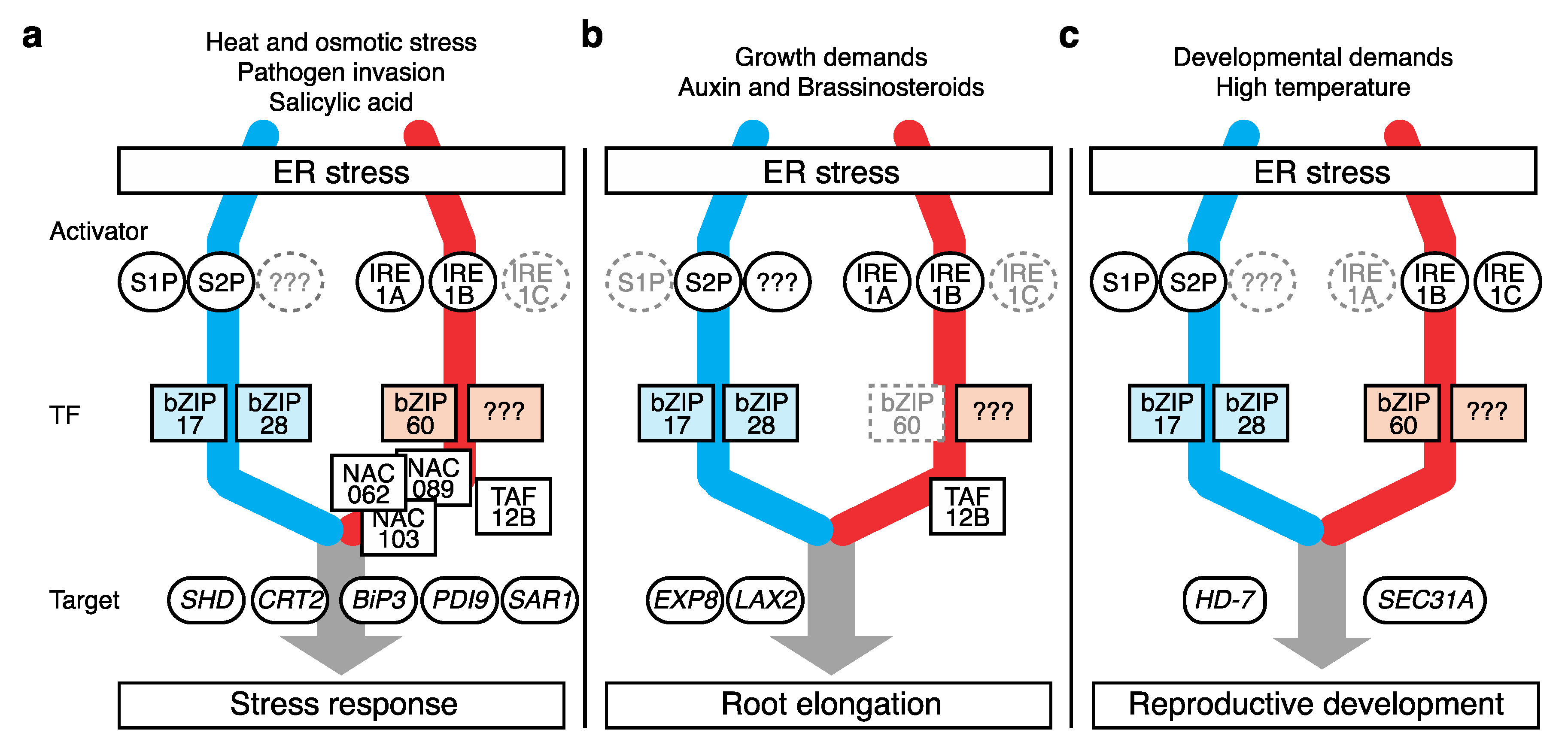
3. UPR Action in Stress Responses
4. Role of the UPR in Root Growth
5. Role of the UPR in Reproductive Development
6. Questions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Saheki, Y.; de Camilli, P. Endoplasmic Reticulum–Plasma Membrane Contact Sites. Annu. Rev. Biochem. 2017, 86, 659–684. [Google Scholar] [CrossRef]
- Anelli, T.; Sitia, R. Protein Quality Control in the Early Secretory Pathway. EMBO J. 2008, 27, 315–327. [Google Scholar] [CrossRef]
- Phillips, B.P.; Gomez-Navarro, N.; Miller, E.A. Protein Quality Control in the Endoplasmic Reticulum. Curr. Opin. Cell Biol. 2020, 65, 96–102. [Google Scholar] [CrossRef] [PubMed]
- Strasser, R. Protein Quality Control in the Endoplasmic Reticulum of Plants. Annu. Rev. Plant Biol. 2018, 69, 147–172. [Google Scholar] [CrossRef]
- Howell, S.H. Endoplasmic Reticulum Stress Responses in Plants. Annu. Rev. Plant Biol. 2013, 64, 477–499. [Google Scholar] [CrossRef]
- Howell, S.H. Evolution of the Unfolded Protein Response in Plants. Plant Cell Environ. 2021, 44, 2625–2635. [Google Scholar] [CrossRef]
- Pastor-Cantizano, N.; Ko, D.K.; Angelos, E.; Pu, Y.; Brandizzi, F. Functional Diversification of ER Stress Responses in Arabidopsis. Trends Biochem. Sci. 2020, 45, 123–136. [Google Scholar] [CrossRef]
- Srivastava, R.; Deng, Y.; Shah, S.; Rao, A.G.; Howell, S.H. BINDING PROTEIN Is a Master Regulator of the Endoplasmic Reticulum Stress Sensor/Transducer BZIP28 in Arabidopsis. Plant Cell 2013, 25, 1416–1429. [Google Scholar] [CrossRef]
- Gallagher, C.M.; Walter, P. Ceapins Inhibit ATF6α Signaling by Selectively Preventing Transport of ATF6α to the Golgi Apparatus during ER Stress. eLife 2016, 5, e11880. [Google Scholar] [CrossRef]
- Oka, O.B.; van Lith, M.; Rudolf, J.; Tungkum, W.; Pringle, M.A.; Bulleid, N.J. ERp18 Regulates Activation of ATF6α during Unfolded Protein Response. EMBO J. 2019, 38, e100990. [Google Scholar] [CrossRef]
- Iwata, Y.; Koizumi, N. An Arabidopsis Transcription Factor, AtbZIP60, Regulates the Endoplasmic Reticulum Stress Response in a Manner Unique to Plants. Proc. Natl. Acad. Sci. USA 2005, 102, 5280–5285. [Google Scholar] [CrossRef]
- Deng, Y.; Humbert, S.; Liu, J.-X.; Srivastava, R.; Rothstein, S.J.; Howell, S.H. Heat Induces the Splicing by IRE1 of a MRNA Encoding a Transcription Factor Involved in the Unfolded Protein Response in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 7247–7252. [Google Scholar] [CrossRef] [PubMed]
- Nagashima, Y.; Mishiba, K.; Suzuki, E.; Shimada, Y.; Iwata, Y.; Koizumi, N. Arabidopsis IRE1 Catalyses Unconventional Splicing of BZIP60 MRNA to Produce the Active Transcription Factor. Sci. Rep. 2011, 1, 29. [Google Scholar] [CrossRef] [PubMed]
- Koizumi, N.; Martinez, I.M.; Kimata, Y.; Kohno, K.; Sano, H.; Chrispeels, M.J. Molecular Characterization of Two Arabidopsis Ire1 Homologs, Endoplasmic Reticulum-Located Transmembrane Protein Kinases. Plant Physiol. 2001, 127, 949–962. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Brandizzi, F. AtIRE1A/AtIRE1B and AGB1 Independently Control Two Essential Unfolded Protein Response Pathways in Arabidopsis. Plant J. 2012, 69, 266–277. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Srivastava, R.; Howell, S.H. Protein Kinase and Ribonuclease Domains of IRE1 Confer Stress Tolerance, Vegetative Growth, and Reproductive Development in Arabidopsis. Proc. Natl. Acad. Sci. USA 2013, 110, 19633–19638. [Google Scholar] [CrossRef]
- Mishiba, K.; Iwata, Y.; Mochizuki, T.; Matsumura, A.; Nishioka, N.; Hirata, R.; Koizumi, N. Unfolded Protein-Independent IRE1 Activation Contributes to Multifaceted Developmental Processes in Arabidopsis. Life Sci. Alliance 2019, 2, e201900459. [Google Scholar] [CrossRef]
- Pu, Y.; Ruberti, C.; Angelos, E.R.; Brandizzi, F. AtIRE1C, an Unconventional Isoform of the UPR Master Regulator AtIRE1, Is Functionally Associated with AtIRE1B in Arabidopsis Gametogenesis. Plant Direct 2019, 3, e00187. [Google Scholar] [CrossRef]
- Wakasa, Y.; Hayashi, S.; Ozawa, K.; Takaiwa, F. Multiple Roles of the ER Stress Sensor IRE1 Demonstrated by Gene Targeting in Rice. Sci. Rep. 2012, 2, 944. [Google Scholar] [CrossRef]
- Yang, Z.-T.; Lu, S.-J.; Wang, M.-J.; Bi, D.-L.; Sun, L.; Zhou, S.-F.; Song, Z.-T.; Liu, J.-X. A Plasma Membrane-Tethered Transcription Factor, NAC062/ANAC062/NTL6, Mediates the Unfolded Protein Response in Arabidopsis. Plant J. Cell Mol. Biol. 2014, 79, 1033–1043. [Google Scholar] [CrossRef]
- Yang, Z.-T.; Wang, M.-J.; Sun, L.; Lu, S.-J.; Bi, D.-L.; Sun, L.; Song, Z.-T.; Zhang, S.-S.; Zhou, S.-F.; Liu, J.-X. The Membrane-Associated Transcription Factor NAC089 Controls ER-Stress-Induced Programmed Cell Death in Plants. PLoS Genet. 2014, 10, e1004243. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Yang, Z.-T.; Song, Z.-T.; Wang, M.-J.; Sun, L.; Lu, S.-J.; Liu, J.-X. The Plant-Specific Transcription Factor Gene NAC103 Is Induced by BZIP60 through a New Cis-Regulatory Element to Modulate the Unfolded Protein Response in Arabidopsis. Plant J. Cell Mol. Biol. 2013, 76, 274–286. [Google Scholar] [CrossRef]
- Kim, J.-S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. ER-Anchored Transcription Factors BZIP17 and BZIP28 Regulate Root Elongation. Plant Physiol. 2018, 176, 2221–2230. [Google Scholar] [CrossRef]
- Dröge-Laser, W.; Snoek, B.L.; Snel, B.; Weiste, C. The Arabidopsis BZIP Transcription Factor Family—An Update. Curr. Opin. Plant Biol. 2018, 45, 36–49. [Google Scholar] [CrossRef]
- Liu, J.-X.; Howell, S.H. BZIP28 and NF-Y Transcription Factors Are Activated by ER Stress and Assemble into a Transcriptional Complex to Regulate Stress Response Genes in Arabidopsis. Plant Cell 2010, 22, 782–796. [Google Scholar] [CrossRef]
- Iwata, Y.; Koizumi, N. Plant Transducers of the Endoplasmic Reticulum Unfolded Protein Response. Trends Plant Sci. 2012, 17, 720–727. [Google Scholar] [CrossRef]
- Iwata, Y.; Fedoroff, N.V.; Koizumi, N. Arabidopsis BZIP60 Is a Proteolysis-Activated Transcription Factor Involved in the Endoplasmic Reticulum Stress Response. Plant Cell 2008, 20, 3107–3121. [Google Scholar] [CrossRef]
- Ruberti, C.; Lai, Y.; Brandizzi, F. Recovery from Temporary Endoplasmic Reticulum Stress in Plants Relies on the Tissue-Specific and Largely Independent Roles of BZIP28 and BZIP60, as Well as an Antagonizing Function of BAX-Inhibitor 1 upon the pro-Adaptive Signaling Mediated by BZIP28. Plant J. 2018, 93, 155–165. [Google Scholar] [CrossRef]
- Liu, J.-X.; Srivastava, R.; Howell, S.H. Stress-Induced Expression of an Activated Form of AtbZIP17 Provides Protection from Salt Stress in Arabidopsis. Plant Cell Environ. 2008, 31, 1735–1743. [Google Scholar] [CrossRef]
- Zhang, S.-S.; Yang, H.; Ding, L.; Song, Z.-T.; Ma, H.; Chang, F.; Liu, J.-X. Tissue-Specific Transcriptomics Reveals an Important Role of the Unfolded Protein Response in Maintaining Fertility upon Heat Stress in Arabidopsis. Plant Cell 2017, 29, 1007–1023. [Google Scholar] [CrossRef] [PubMed]
- Cifuentes-Esquivel, N.; Celiz-Balboa, J.; Henriquez-Valencia, C.; Mitina, I.; Arraño-Salinas, P.; Moreno, A.A.; Meneses, C.; Blanco-Herrera, F.; Orellana, A. BZIP17 Regulates the Expression of Genes Related to Seed Storage and Germination, Reducing Seed Susceptibility to Osmotic Stress. J. Cell. Biochem. 2018, 119, 6857–6868. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Wang, M.-J.; Wang, J.-J.; Lu, H.-P.; Liu, J.-X. BZIP17 Regulates Heat Stress Tolerance at Reproductive Stage in Arabidopsis. aBIOTECH 2022, 3, 1–11. [Google Scholar] [CrossRef]
- Zhou, S.-F.; Sun, L.; Valdés, A.E.; Engström, P.; Song, Z.-T.; Lu, S.-J.; Liu, J.-X. Membrane-Associated Transcription Factor Peptidase, Site-2 Protease, Antagonizes ABA Signaling in Arabidopsis. New Phytol. 2015, 208, 188–197. [Google Scholar] [CrossRef]
- Gao, H.; Brandizzi, F.; Benning, C.; Larkin, R.M. A Membrane-Tethered Transcription Factor Defines a Branch of the Heat Stress Response in Arabidopsis Thaliana. Proc. Natl. Acad. Sci. USA 2008, 105, 16398–16403. [Google Scholar] [CrossRef] [PubMed]
- Moreno, A.A.; Mukhtar, M.S.; Blanco, F.; Boatwright, J.L.; Moreno, I.; Jordan, M.R.; Chen, Y.; Brandizzi, F.; Dong, X.; Orellana, A.; et al. IRE1/BZIP60-Mediated Unfolded Protein Response Plays Distinct Roles in Plant Immunity and Abiotic Stress Responses. PLoS ONE 2012, 7, e31944. [Google Scholar] [CrossRef]
- Deng, Y.; Srivastava, R.; Quilichini, T.D.; Dong, H.; Bao, Y.; Horner, H.T.; Howell, S.H. IRE1, a Component of the Unfolded Protein Response Signaling Pathway, Protects Pollen Development in Arabidopsis from Heat Stress. Plant J. 2016, 88, 193–204. [Google Scholar] [CrossRef] [PubMed]
- Gayral, M.; Arias Gaguancela, O.; Vasquez, E.; Herath, V.; Flores, F.J.; Dickman, M.B.; Verchot, J. Multiple ER-to-Nucleus Stress Signaling Pathways Are Activated during Plantago Asiatica Mosaic Virus and Turnip Mosaic Virus Infection in Arabidopsis thaliana. Plant J. 2020, 103, 1233–1245. [Google Scholar] [CrossRef]
- Liu, J.-X.; Srivastava, R.; Che, P.; Howell, S.H. Salt Stress Responses in Arabidopsis Utilize a Signal Transduction Pathway Related to Endoplasmic Reticulum Stress Signaling: Salt Stress Elicits ER Stress Response. Plant J. 2007, 51, 897–909. [Google Scholar] [CrossRef]
- Zhang, L.; Chen, H.; Brandizzi, F.; Verchot, J.; Wang, A. The UPR Branch IRE1-BZIP60 in Plants Plays an Essential Role in Viral Infection and Is Complementary to the Only UPR Pathway in Yeast. PLoS Genet. 2015, 11, e1005164. [Google Scholar] [CrossRef]
- Zhou, J.-M.; Zhang, Y. Plant Immunity: Danger Perception and Signaling. Cell 2020, 181, 978–989. [Google Scholar] [CrossRef]
- Nagashima, Y.; Iwata, Y.; Ashida, M.; Mishiba, K.; Koizumi, N. Exogenous Salicylic Acid Activates Two Signaling Arms of the Unfolded Protein Response in Arabidopsis. Plant Cell Physiol. 2014, 55, 1772–1778. [Google Scholar] [CrossRef]
- Lu, D.-P.; Christopher, D.A. Endoplasmic Reticulum Stress Activates the Expression of a Sub-Group of Protein Disulfide Isomerase Genes and AtbZIP60 Modulates the Response in Arabidopsis Thaliana. Mol. Genet. Genom. 2008, 280, 199–210. [Google Scholar] [CrossRef]
- Hayashi, S.; Wakasa, Y.; Takaiwa, F. Functional Integration between Defence and IRE1-Mediated ER Stress Response in Rice. Sci. Rep. 2012, 2, 670. [Google Scholar] [CrossRef]
- Kim, J.-S.; Sakamoto, Y.; Takahashi, F.; Shibata, M.; Urano, K.; Matsunaga, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Arabidopsis TBP-ASSOCIATED FACTOR 12 Ortholog NOBIRO6 Controls Root Elongation with Unfolded Protein Response Cofactor Activity. Proc. Natl. Acad. Sci. USA 2022, 119, e2120219119. [Google Scholar] [CrossRef]
- Liu, J.-X.; Srivastava, R.; Che, P.; Howell, S.H. An Endoplasmic Reticulum Stress Response in Arabidopsis Is Mediated by Proteolytic Processing and Nuclear Relocation of a Membrane-Associated Transcription Factor, BZIP28. Plant Cell 2007, 19, 4111–4119. [Google Scholar] [CrossRef]
- Iwata, Y.; Ashida, M.; Hasegawa, C.; Tabara, K.; Mishiba, K.; Koizumi, N. Activation of the Arabidopsis Membrane-Bound Transcription Factor BZIP28 Is Mediated by Site-2 Protease, but Not Site-1 Protease. Plant J. 2017, 91, 408–415. [Google Scholar] [CrossRef]
- Che, P.; Bussell, J.D.; Zhou, W.; Estavillo, G.M.; Pogson, B.J.; Smith, S.M. Signaling from the Endoplasmic Reticulum Activates Brassinosteroid Signaling and Promotes Acclimation to Stress in Arabidopsis. Sci. Signal. 2010, 3, ra69. [Google Scholar] [CrossRef]
- Bao, Y.; Bassham, D.C.; Howell, S.H. A Functional Unfolded Protein Response Is Required for Normal Vegetative Development. Plant Physiol. 2019, 179, 1834–1843. [Google Scholar] [CrossRef]
- Angelos, E.; Brandizzi, F. The UPR Regulator IRE1 Promotes Balanced Organ Development by Restricting TOR-Dependent Control of Cellular Differentiation in Arabidopsis. Plant J. 2022, 109, 1229–1248. [Google Scholar] [CrossRef]
- Wang, D.; Weaver, N.D.; Kesarwani, M.; Dong, X. Induction of Protein Secretory Pathway Is Required for Systemic Acquired Resistance. Science 2005, 308, 1036–1040. [Google Scholar] [CrossRef]
- Zeier, J. Metabolic Regulation of Systemic Acquired Resistance. Curr. Opin. Plant Biol. 2021, 62, 102050. [Google Scholar] [CrossRef] [PubMed]
- Humbert, S.; Zhong, S.; Deng, Y.; Howell, S.H.; Rothstein, S.J. Alteration of the BZIP60/IRE1 Pathway Affects Plant Response to ER Stress in Arabidopsis thaliana. PLoS ONE 2012, 7, e39023. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Aung, K.; Rolčík, J.; Walicki, K.; Friml, J.; Brandizzi, F. Inter-Regulation of the Unfolded Protein Response and Auxin Signaling. Plant J. Cell Mol. Biol. 2014, 77, 97–107. [Google Scholar] [CrossRef]
- Robles, L.M.; Wampole, J.S.; Christians, M.J.; Larsen, P.B. Arabidopsis Enhanced Ethylene Response 4 Encodes an EIN3-Interacting TFIID Transcription Factor Required for Proper Ethylene Response, Including ERF1 Induction. J. Exp. Bot. 2007, 58, 2627–2639. [Google Scholar] [CrossRef] [PubMed]
- Kubo, M.; Furuta, K.; Demura, T.; Fukuda, H.; Liu, Y.-G.; Shibata, D.; Kakimoto, T. The CKH1/EER4 Gene Encoding a TAF12-Like Protein Negatively Regulates Cytokinin Sensitivity in Arabidopsis Thaliana. Plant Cell Physiol. 2011, 52, 629–637. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, M.; Nozaki, M.; Iwata, Y.; Koizumi, N.; Sato, Y. THESEUS1 Is Involved in Tunicamycin-Induced Root Growth Inhibition, Ectopic Lignin Deposition, and Cell Wall Damage-Induced Unfolded Protein Response. Plant Biotechnol. 2022, 39, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Maruyama, D.; Sugiyama, T.; Endo, T.; Nishikawa, S. Multiple BiP Genes of Arabidopsis Thaliana Are Required for Male Gametogenesis and Pollen Competitiveness. Plant Cell Physiol. 2014, 55, 801–810. [Google Scholar] [CrossRef]
- Maruyama, D.; Endo, T.; Nishikawa, S. BiP3 Supports the Early Stages of Female Gametogenesis in the Absence of BiP1 and BiP2 in Arabidopsis Thaliana. Plant Signal. Behav. 2015, 10, 6. [Google Scholar] [CrossRef]
- Wakasa, Y.; Yasuda, H.; Oono, Y.; Kawakatsu, T.; Hirose, S.; Takahashi, H.; Hayashi, S.; Yang, L.; Takaiwa, F. Expression of ER Quality Control-Related Genes in Response to Changes in BiP1 Levels in Developing Rice Endosperm. Plant J. 2011, 65, 675–689. [Google Scholar] [CrossRef]
- Yang, W.; Xu, P.; Zhang, J.; Zhang, S.; Li, Z.; Yang, K.; Chang, X.; Li, Y. OsbZIP60-Mediated Unfolded Protein Response Regulates Grain Chalkiness in Rice. J. Genet. Genom. 2022, 49, 414–426. [Google Scholar] [CrossRef]
- Nawkar, G.M.; Kang, C.H.; Maibam, P.; Park, J.H.; Jung, Y.J.; Chae, H.B.; Chi, Y.H.; Jung, I.J.; Kim, W.Y.; Yun, D.-J.; et al. HY5, a Positive Regulator of Light Signaling, Negatively Controls the Unfolded Protein Response in Arabidopsis. Proc. Natl. Acad. Sci. USA 2017, 114, 2084–2089. [Google Scholar] [CrossRef]
- Ko, D.K.; Brandizzi, F. Transcriptional Competition Shapes Proteotoxic ER Stress Resolution. Nat. Plants 2022, 8, 481–490. [Google Scholar] [CrossRef]
- Song, Z.-T.; Sun, L.; Lu, S.-J.; Tian, Y.; Ding, Y.; Liu, J.-X. Transcription Factor Interaction with COMPASS-like Complex Regulates Histone H3K4 Trimethylation for Specific Gene Expression in Plants. Proc. Natl. Acad. Sci. USA 2015, 112, 2900–2905. [Google Scholar] [CrossRef]
- Liu, Y.; Lv, Y.; Wei, A.; Guo, M.; Li, Y.; Wang, J.; Wang, X.; Bao, Y. Unfolded Protein Response in Balancing Plant Growth and Stress Tolerance. Front. Plant Sci. 2022, 13, 1019414. [Google Scholar] [CrossRef]
| Knockout Mutant | Viability | Stress Tolerance | Primary Root Growth (% of Wild Type) | Other Characteristics |
|---|---|---|---|---|
| bzip17 | n.s. | Reduced tolerance to heat [32] and salinity [31] | 100% [16,23] | Ectopic expression enhanced salinity tolerance [29] |
| bzip28 | n.s. | Reduced tolerance to heat [34] | 100% [16,23] | - |
| bzip1728 | n.s. | - | 10% [23,35] | Shoot growth defect recovered by grafting to wild-type roots [35] |
| bzip60 | n.s. | Reduced tolerance to heat [36], viral infection [37] | 100% [16] | - |
| bzip1760 | n.s. | Reduced tolerance to viral infection [38] | 100% [16,23] | - |
| bzip2860 | n.s. | Reduced tolerance to heat [30] | 100% [16,23] | - |
| bzip172860 | Lethal [23] | n.a. | n.a. | - |
| s1p | n.s. | Reduced tolerance to salinity [39] | 100% [39] | - |
| s2p | n.s. | Reduced tolerance to heat [36], drought [40] | 40% [40] | - |
| s1p2p | n.s. | n.a. | 40% [23] | - |
| ire1a | n.s. | Reduced tolerance to pathogen [41] | 100% [16] | - |
| ire1b | One lethal mutant [15,42] | n.s. | 100% [16] | - |
| ire1c | n.s. | n.a. | 100% [18] | - |
| ire1ab | n.s. | Reduced tolerance to heat [36], viral infection [37] | 60% [43] | - |
| ire1bc | Decreased pollen viability [17] | n.s. | 100% [17] | - |
| ire1abc | Lethal [17,18] | n.a. | n.a. | - |
| bzip17ire1a | n.s. | n.s. | 100% [44] | - |
| bzip17ire1b | n.s. | n.a. | 60% [44] | - |
| bzip17ire1ab | n.s. | n.a. | 10% [44] | Delayed flowering [44] |
| bzip28ire1ab | Lethal [16] | n.a. | n.a. | - |
| bzip60ire1ab | n.s. | n.a. | 60% [16] | Similar genetic defects to ire1ab |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.-S.; Mochida, K.; Shinozaki, K. ER Stress and the Unfolded Protein Response: Homeostatic Regulation Coordinate Plant Survival and Growth. Plants 2022, 11, 3197. https://doi.org/10.3390/plants11233197
Kim J-S, Mochida K, Shinozaki K. ER Stress and the Unfolded Protein Response: Homeostatic Regulation Coordinate Plant Survival and Growth. Plants. 2022; 11(23):3197. https://doi.org/10.3390/plants11233197
Chicago/Turabian StyleKim, June-Sik, Keiichi Mochida, and Kazuo Shinozaki. 2022. "ER Stress and the Unfolded Protein Response: Homeostatic Regulation Coordinate Plant Survival and Growth" Plants 11, no. 23: 3197. https://doi.org/10.3390/plants11233197
APA StyleKim, J.-S., Mochida, K., & Shinozaki, K. (2022). ER Stress and the Unfolded Protein Response: Homeostatic Regulation Coordinate Plant Survival and Growth. Plants, 11(23), 3197. https://doi.org/10.3390/plants11233197









