Development of a Specific Nested PCR Assay for the Detection of 16SrI Group Phytoplasmas Associated with Sisal Purple Leafroll Disease in Sisal Plants and Mealybugs
Abstract
1. Introduction
2. Results
2.1. Design and Screening of Specific Primers
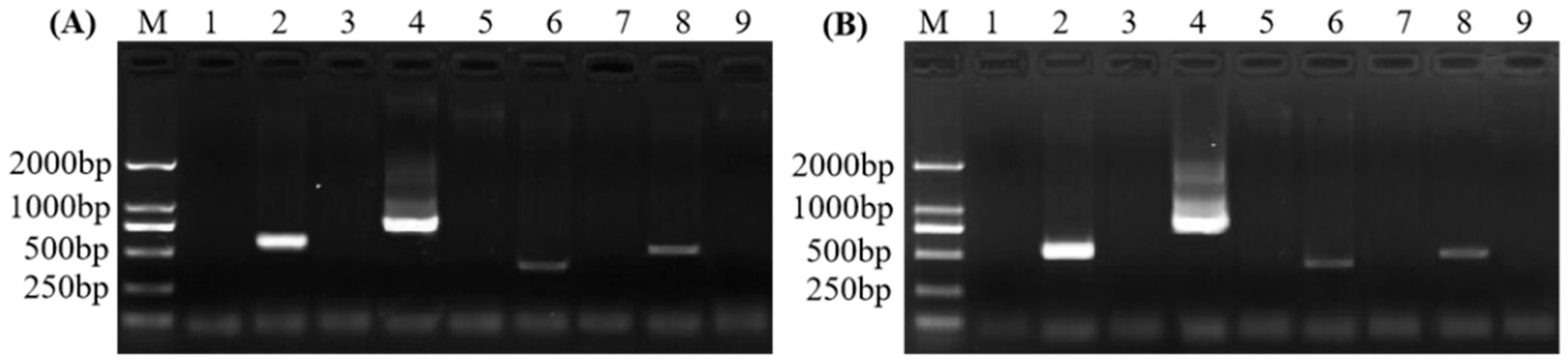
2.2. Optimization of the PCR Detection Method and Analysis of Assay Sensitivity
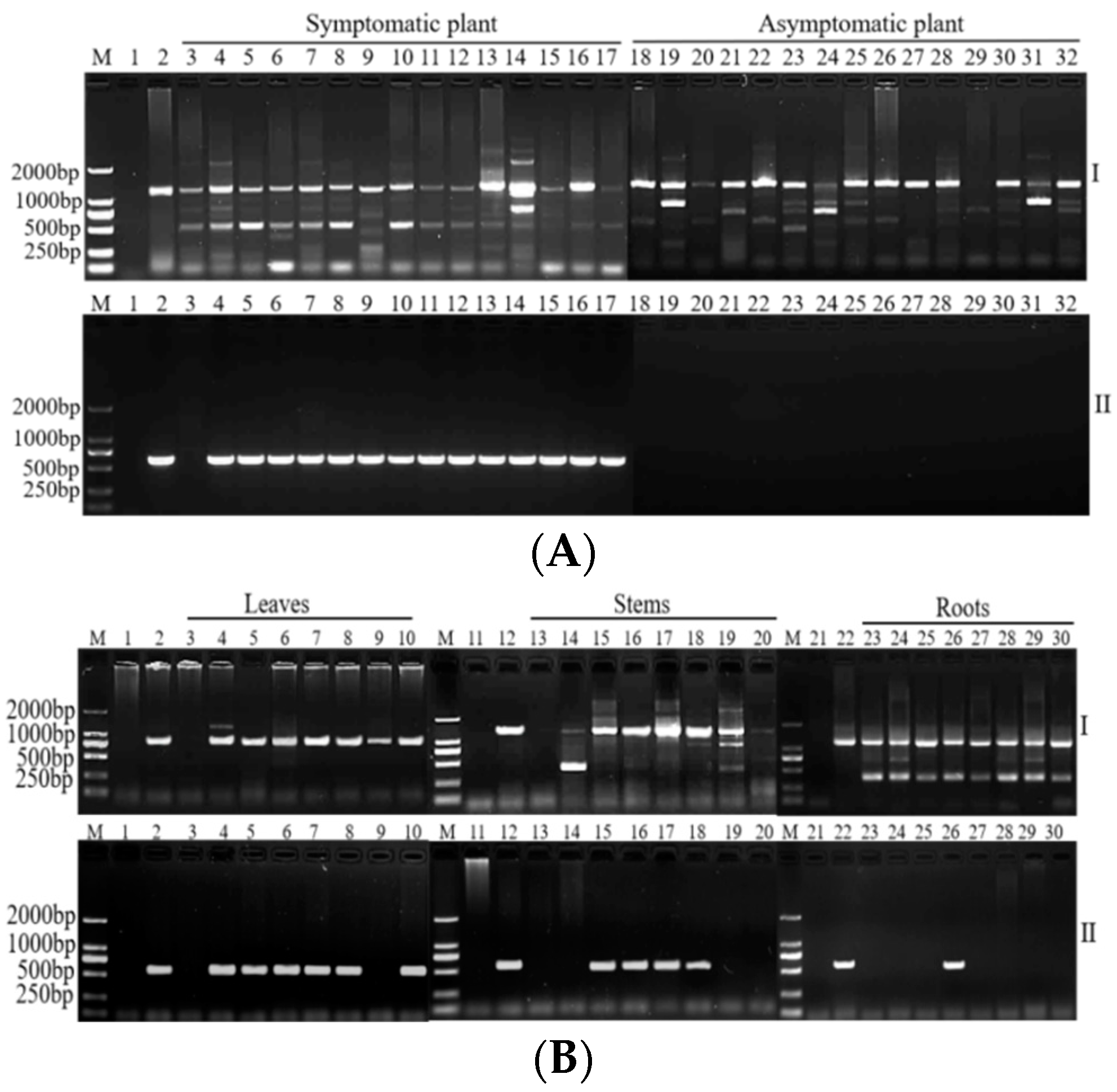
2.3. Evaluation of Specificities
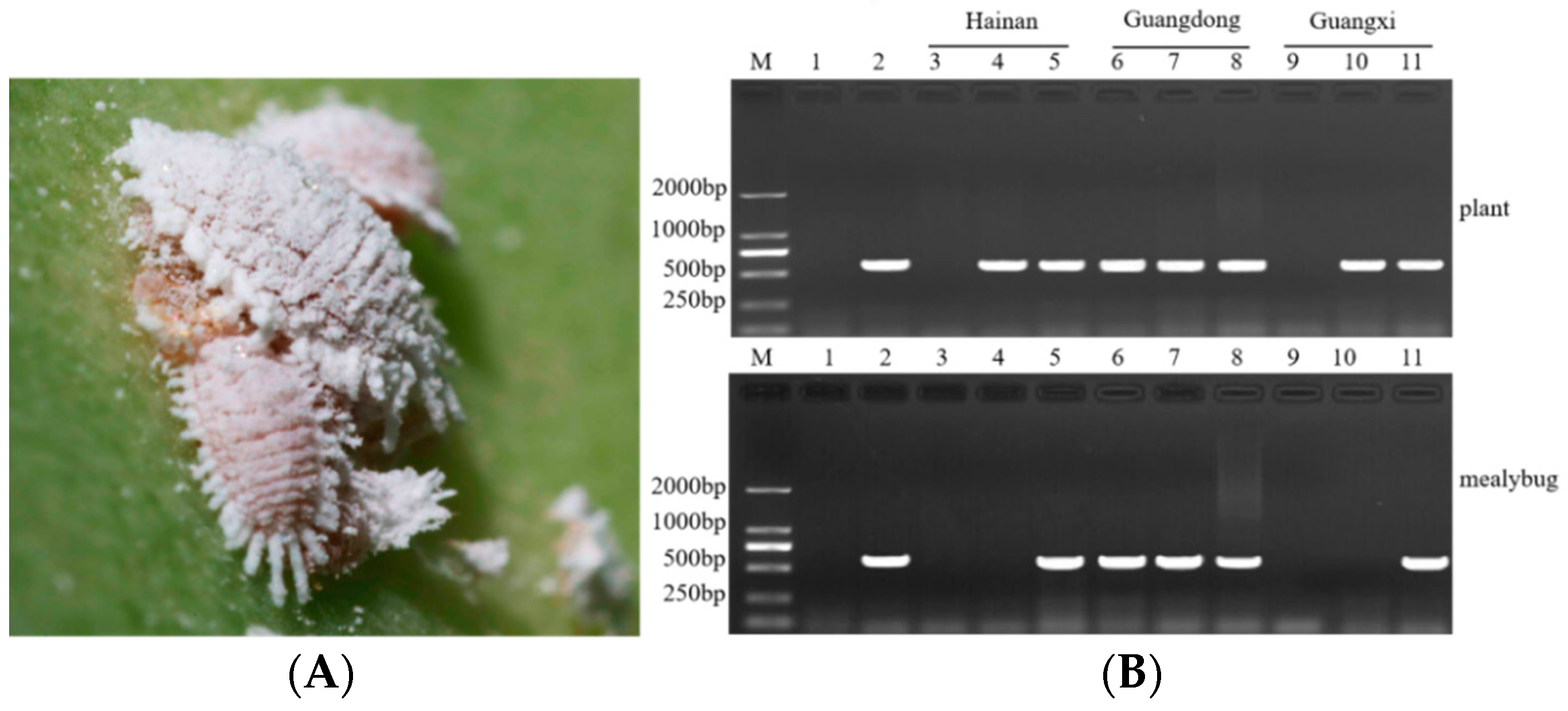
2.4. SPLDaP Detection in Mealybugs
3. Discussion
4. Materials and Methods
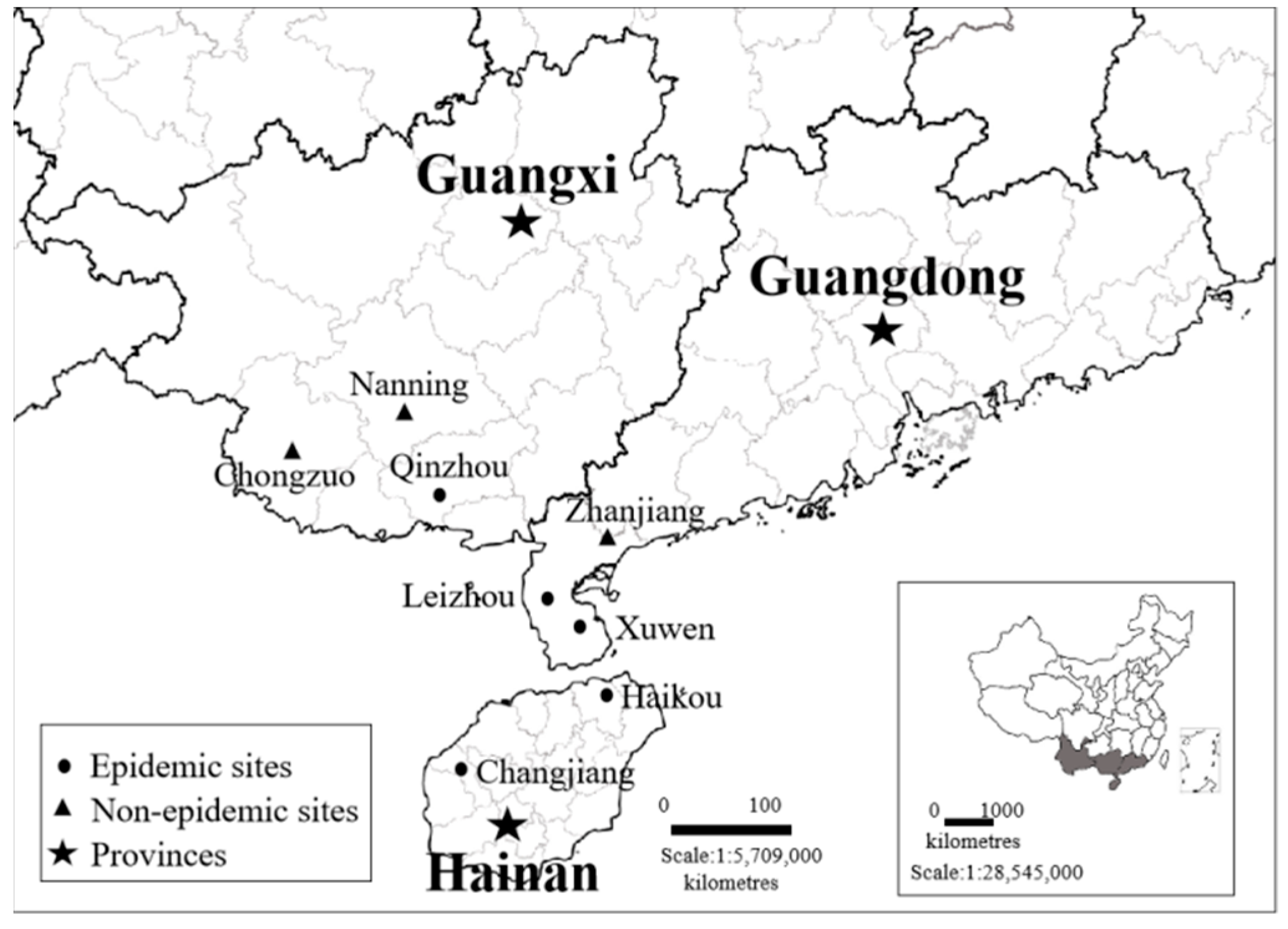
4.1. Plant Samples and Sequence Collection
4.2. Designing and Screening of Specific Primers
4.3. Optimization of the Detection Method and Sensitivity Determination
4.4. Evaluation of Primers Specificity
4.5. Identification of Potential Vectors
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Duarte, E.A.A.; Damasceno, C.L.; De Oliveira, T.A.S.; Oliveira, B.L.D.; Martins, F.M.; Rosa, D.Q.S.J.; De Lima, T.E.F.; Da Silva, R.M.; Kato, R.B.; Bortolini, D.E. Putting the mess in order: Aspergillus welwitschiae (and Not A. niger) is the etiological agent of sisal bole rot disease in Brazil. Front. Microbiol. 2018, 9, 1227. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.M.; Luo, P.; Guo, C.M.; Li, J.Z.; Liu, Q.L.; Chen, H.L.; Zhang, S.Q.; Zheng, J.L.; Jiang, C.J.; Dai, Z.Z.; et al. AFLP analysis and zebra disease resistance identification of 40 sisal genotypes in China. Mol. Biol. Rep. 2012, 39, 6379–6385. [Google Scholar] [CrossRef]
- Xie, H.H.; Long, L.; Huang, S.P.; Mao, L.; Li, J. First report of black spot caused by Neoscytalidium dimidiatum on sisal in Guangxi, China. Plant Dis. 2020, 105, 701. [Google Scholar] [CrossRef] [PubMed]
- Qin, Z.Q.; Qiu, B.L.; Wu, J.H.; Cuthbertson, A.G.S.; Ren, S.X. Effect of temperature on the life history of Dysmicoccus neobrevipes (Hemiptera: Pseudoccocidae): An invasive species of gray pineapple mealybug in South China. Crop Prot. 2013, 45, 141–146. [Google Scholar] [CrossRef]
- Cuervo-Parra, J.A.; Pérez-España, V.H.; López-Pérez, P.A.; Ovando, M.A.M.; Arce-Cervantes, O.; Aparicio-Burgos, J.E.; Romero-Cortes, T. Scyphophorus acupunctatus (Coleoptera: Dryophthoridae): A weevil threatening the production of Agave in Mexico. Fla. Entomol. 2019, 102, 1–9. [Google Scholar]
- Li, G.F.; Wei, M.S.; Ma, J.; Zhu, S.F. First Report of Broad bean wilt virus 2 in Echinacea purpurea in China. Plant Dis. 2012, 96, 1232. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Wang, Y.; Zhao, H.; Zhang, X.; Wang, X.; Li, D.; Yu, J.; Han, C. Brassica yellows virus’ movement protein upregulates anthocyanin accumulation, leading to the development of purple leaf symptoms on Arabidopsis thaliana. Sci. Rep.-UK 2018, 8, 16273. [Google Scholar] [CrossRef]
- Byarugaba, A.A.; Mukasa, S.B.; Barekye, A.; Rubaihayo, P.R. Interactive effects of potato virus Y and potato leafroll virus infection on potato yields in Uganda. Open Agric. 2020, 5, 726–739. [Google Scholar] [CrossRef]
- Sabaté, J.; Laviňa, A.; Batlle, A. First Report of ‘Candidatus Phytoplasma pyri’ causing peach yellow leaf roll (PYLR) in Spain. Plant Dis. 2014, 98, 989. [Google Scholar] [CrossRef]
- Babaei, G.; Esmaeilzadeh-Hosseini, S.A.; Eshaghi, R.; Nikbakht, V. Incidence and molecular characterization of a 16SrI-B phytoplasma strain associated with Vitis vinifera leaf yellowing and reddening in the west of Iran. Can. J. Plant Pathol. 2019, 41, 468–474. [Google Scholar] [CrossRef]
- Gamarra, D.G.; Villar, C.M.; Suarez, G.T.; Esteban, W.D.I.; Contaldo, N.; Lozano, E.C.C.; Bertaccini, A. Diverse phytoplasmas associated with maize bushy stunt disease in Peru. Eur. J. Plant Pathol. 2022, 163, 223–235. [Google Scholar] [CrossRef]
- Muller, A. Health status of sisal plants (Agave sisalana) as related to soils and the mineral composition of their leaves. Chemistry 1964, 15, 129–132. [Google Scholar] [CrossRef]
- Yoneda, K.; Usui, M.; Kubota, S. Effect of nutrient deficiency on growth and flowering of Phalaenopsis. J. Jpn. Soc. Hortic. Sci. 1997, 66, 141–147. [Google Scholar] [CrossRef]
- Ariraman, R.; Kumar, A.P.; Selvakumar, S.; Sowmya, S.R.; Mansingh, M.D. Effect of sulphur nutrition on growth parameters, yield parameters, yield, nutrient uptake, quality and economics of maize: A review. J. Pharmacogn. Phytochem. 2020, 9, 1632–1636. [Google Scholar]
- Lee, I.-M.; Gundersen-Rindal, D.E.; Davis, R.E.; Bottner, K.D.; Marcone, C.; Seemüller, E. ‘Candidatus Phytoplasma asteris’, a novel phytoplasma taxon associated with aster yellows and related diseases. Int. J. Syst. Evol. Microbiol. 2004, 54, 1037–1048. [Google Scholar] [CrossRef]
- Nair, S.; Manimekalai, R. Phytoplasma diseases of plants: Molecular diagnostics and way forward. World J. Microb. Biot. 2021, 6, 102. [Google Scholar] [CrossRef]
- Bertaccini, A.; Paltrinieri, S.; Contaldo, N. Standard detection protocol: PCR and RFLP analyses based on 16S rRNA gene. In Methods in Molecular Biology; Musetti, R., Pagliari, L., Eds.; Humana Press: New York, NY, USA, 2019; pp. 83–95. [Google Scholar]
- Fránová, J.; Bertaccini, A.; Maini, S. Difficulties with conventional phytoplasma diagnostic using PCR/RFLP analyses. Bull. Insectology 2011, 64, S287–S288. [Google Scholar]
- Demeuse, K.L.; Grode, A.S.; Szendrei, Z. Comparing qPCR and nested PCR diagnostic methods for aster yellows phytoplasma in aster vectors. Plant Dis. 2016, 100, 2513–2519. [Google Scholar] [CrossRef]
- Harrison, N.A.; Womack, M.; Carpio, M.L. Detection and characterization of a lethal yellowing (16SrIV) group phytoplasma in Canary Island date palms affected by lethal decline in Texas. Plant Dis. 2002, 86, 676–681. [Google Scholar] [CrossRef]
- Lee, M.E.; Grau, C.R.; Lukaesko, L.A.; Lee, I.M. Identification of aster yellows phytoplasmas in soybean in Wisconsin based on RFLP analysis of PCR-amplified products (16S rDNAs). Can. J. Plant Pathol. 2002, 24, 125–130. [Google Scholar] [CrossRef]
- Malausa, T.; Fenis, A.; Warot, S.; Germain, J.F.; Ris, N.; Prado, E.; Botton, M.; Vanlerberghe-Masutti, F.; Sforza, R.; Cruaud, C.; et al. DNA markers to disentangle complexes of cryptic taxa in mealybugs (Hemiptera: Pseudococcidae). J. Appl. Entomol. 2011, 135, 142–155. [Google Scholar] [CrossRef]
- Valiunas, D.; Jomantiene, R.; Davis, R.E. Evaluation of the DNA-dependent RNA polymerase-subunit gene (rpoB) for phytoplasma classification and phylogeny. Int. J. Syst. Evol. Microbiol. 2013, 63, 3904–3914. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.-M.; Bottner-Parker, K.D.; Zhao, Y.; Davis, R.; Harrison, N. Phylogenetic analysis and delineation of phytoplasmas based on secY gene sequences. Int. J. Syst. Evol. Microbiol. 2010, 60, 2887–2897. [Google Scholar] [CrossRef]
- Marcone, C.; Lee, I.-M.; Davis, R.; Ragozzino, A.; Seemüller, E. Classification of aster yellows-group phytoplasmas based on combined analyses of rRNA and tuf gene sequences. Int. J. Syst. Evol. Microbiol. 2000, 5, 1703–1713. [Google Scholar] [CrossRef] [PubMed]
- Schneider, B.; Gibb, K.S. Sequence and RFLP analysis of the elongation factor Tu gene used in differentiation and classification of phytoplasmas. Microbiology 1997, 143, 3381–3389. [Google Scholar] [CrossRef]
- Dumonceaux, T.J.; Green, M.J.; Hammond, C.; Perez, E.; Olivier, C. Molecular diagnostic tools for detection and differentiation of phytoplasmas based on chaperonin-60 reveal differences in host plant infection patterns. PLoS ONE 2014, 9, e116039. [Google Scholar] [CrossRef]
- van Bel, A.J.; Musetti, R. Sieve-element biology provides leads for research on phytoplasma lifestyle in plant hosts. J. Exp. Bot. 2019, 70, 3737–3755. [Google Scholar] [CrossRef]
- Errea, P.; Aguelo, V.; Hormaza, J.I. Seasonal variations in detection and transmission of pear decline phytoplasma. J. Phytopathol. 2002, 150, 439–443. [Google Scholar] [CrossRef]
- Christensen, N.M.; Nicolaisen, M.; Hansen, M.; Schulz, A. Distribution of phytoplasmas in infected plants as revealed by real-time PCR and bioimaging. Mol. Plant-Microbe Interact. 2004, 17, 1175–1184. [Google Scholar] [CrossRef]
- Baric, S.; Berger, J.; Cainelli, C.; Kerschbamer, C.; Letschka, T.; Dalla, V.J. Seasonal colonization of apple trees by ‘Candidatus Phytoplasma mali’ revealed by a new quantitative TaqMan real-time PCR approach. Eur. J. Plant Pathol. 2011, 129, 455–467. [Google Scholar] [CrossRef]
- Singh, V.K.; Kumar, S.; Lakhanpaul, S. Differential distribution of phytoplasma during phyllody progression in sesame (Sesamum indicum L.) under field conditions—An important consideration for effective sampling of diseased tissue. Crop Prot. 2018, 110, 288–294. [Google Scholar] [CrossRef]
- Wright, A.A.; Shires, M.K.; Molnar, C.; Bishop, G.; Johnson, A.M.; Frias, C.; Harper, S.J. Titer and distribution of Candidatus Phytoplasma pruni in Prunus avium. Phytopathology 2022, 112, 1406–1412. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.H.; Lin, L.; Ren, S.X.; Qin, Z.Q. New pest on sisal in Leizhou Peninsula a briefing about gray pineapple mealybug, Dysmicoccus neobrevipes Beardsley. Guangdong Agric. Sci. 2008, 4, 47–48. [Google Scholar]
- Qin, Z.; Wu, J.; Qiu, B.; Ali, S.; Cuthbertson, A.G. The impact of Cryptolaemus montrouzieri mulsant (Coleoptera: Coccinellidae) on control of Dysmicoccus neobrevipes Beardsley (Hemiptera: Pseudococcidae). Insects 2019, 10, 131. [Google Scholar] [CrossRef]
- Sether, D.M.; Ullman, D.E.; Hu, J.S. Transmission of pineapple mealybug wilt-associated virus by two species of mealybug (Dysmicoccus spp.). Phytopathology 1998, 88, 1224–1230. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.W.; Rowhani, A.; Golino, D.A.; Daane, K.M.; Almeida, R.P.P. Mealybug transmission of grapevine leafroll viruses: An analysis of virus–vector specificity. Phytopathology 2010, 100, 830–834. [Google Scholar] [CrossRef]
- Jelkmann, W.; Fechtner, B.; Agranovsky, A.A. Complete genome structure and phylogenetic analysis of little cherry virus, a mealybug-transmissible closterovirus. J. Gen. Virol. 1997, 78, 2067–2071. [Google Scholar] [CrossRef]
- Herrbach, E.; Maguet, J.L.; Hommay, G. Virus transmission by mealybugs and soft scales (Hemiptera, Coccoidea). In Vector-Mediated Transmission of Plant Pathogens; Brown, J., Ed.; APS Press: Paul, MN, USA, 2016; pp. 147–161. [Google Scholar]
- Přibylová, J.; Špak, J.; Fránová, J.; Petrzik, K. Association of aster yellows subgroup 16SrI-B phytoplasmas with a disease of Rehmannia glutinosa var. purpurea. Plant Pathol. 2001, 50, 776–781. [Google Scholar] [CrossRef]
- Zhao, Y.; Wei, W.; Lee, I.-M.; Shao, J.; Suo, X.; Davis, R.E. Construction of an interactive online phytoplasma classification tool, iPhyClassifier, and its application in analysis of the peach X-disease phytoplasma group (16SrIII). Int. J. Syst. Evol. Microbiol. 2009, 59, 2582–2593. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, G.; Wu, W.; Tan, S.; Liang, Y.; He, C.; Chen, H.; Huang, X.; Yi, K. Development of a Specific Nested PCR Assay for the Detection of 16SrI Group Phytoplasmas Associated with Sisal Purple Leafroll Disease in Sisal Plants and Mealybugs. Plants 2022, 11, 2817. https://doi.org/10.3390/plants11212817
Wang G, Wu W, Tan S, Liang Y, He C, Chen H, Huang X, Yi K. Development of a Specific Nested PCR Assay for the Detection of 16SrI Group Phytoplasmas Associated with Sisal Purple Leafroll Disease in Sisal Plants and Mealybugs. Plants. 2022; 11(21):2817. https://doi.org/10.3390/plants11212817
Chicago/Turabian StyleWang, Guihua, Weihuai Wu, Shibei Tan, Yanqiong Liang, Chunping He, Helong Chen, Xing Huang, and Kexian Yi. 2022. "Development of a Specific Nested PCR Assay for the Detection of 16SrI Group Phytoplasmas Associated with Sisal Purple Leafroll Disease in Sisal Plants and Mealybugs" Plants 11, no. 21: 2817. https://doi.org/10.3390/plants11212817
APA StyleWang, G., Wu, W., Tan, S., Liang, Y., He, C., Chen, H., Huang, X., & Yi, K. (2022). Development of a Specific Nested PCR Assay for the Detection of 16SrI Group Phytoplasmas Associated with Sisal Purple Leafroll Disease in Sisal Plants and Mealybugs. Plants, 11(21), 2817. https://doi.org/10.3390/plants11212817





