Coastal Wild Grapevine Accession (Vitis vinifera L. ssp. sylvestris) Shows Distinct Late and Early Transcriptome Changes under Salt Stress in Comparison to Commercial Rootstock Richter 110
Abstract
1. Introduction
2. Results
2.1. Pre-Harvest Mortality Rates in AS1B and Richter 110
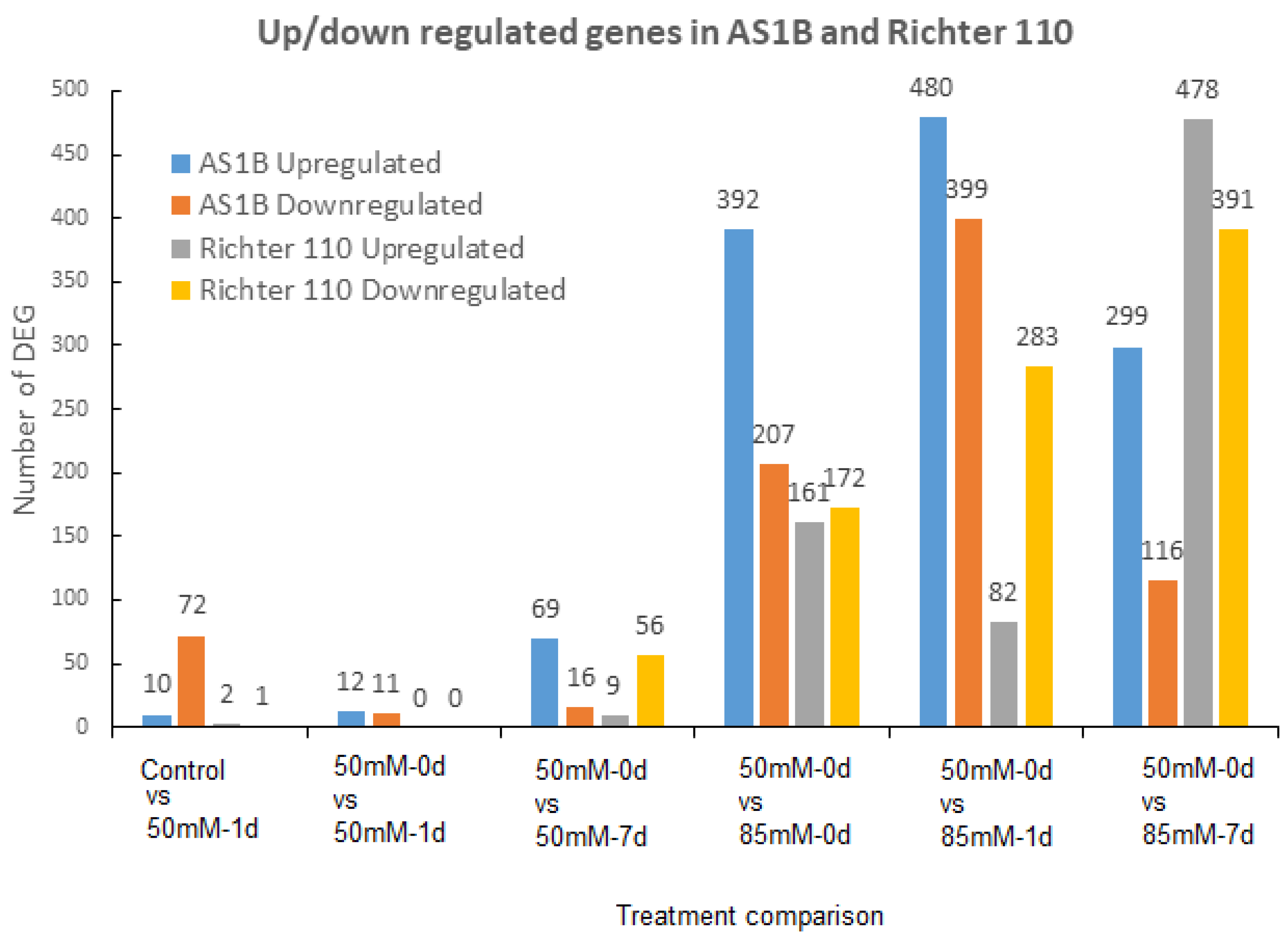
2.2. Differential Gene Expression between AS1B and Ricther 110 under Salt-Stress Conditions
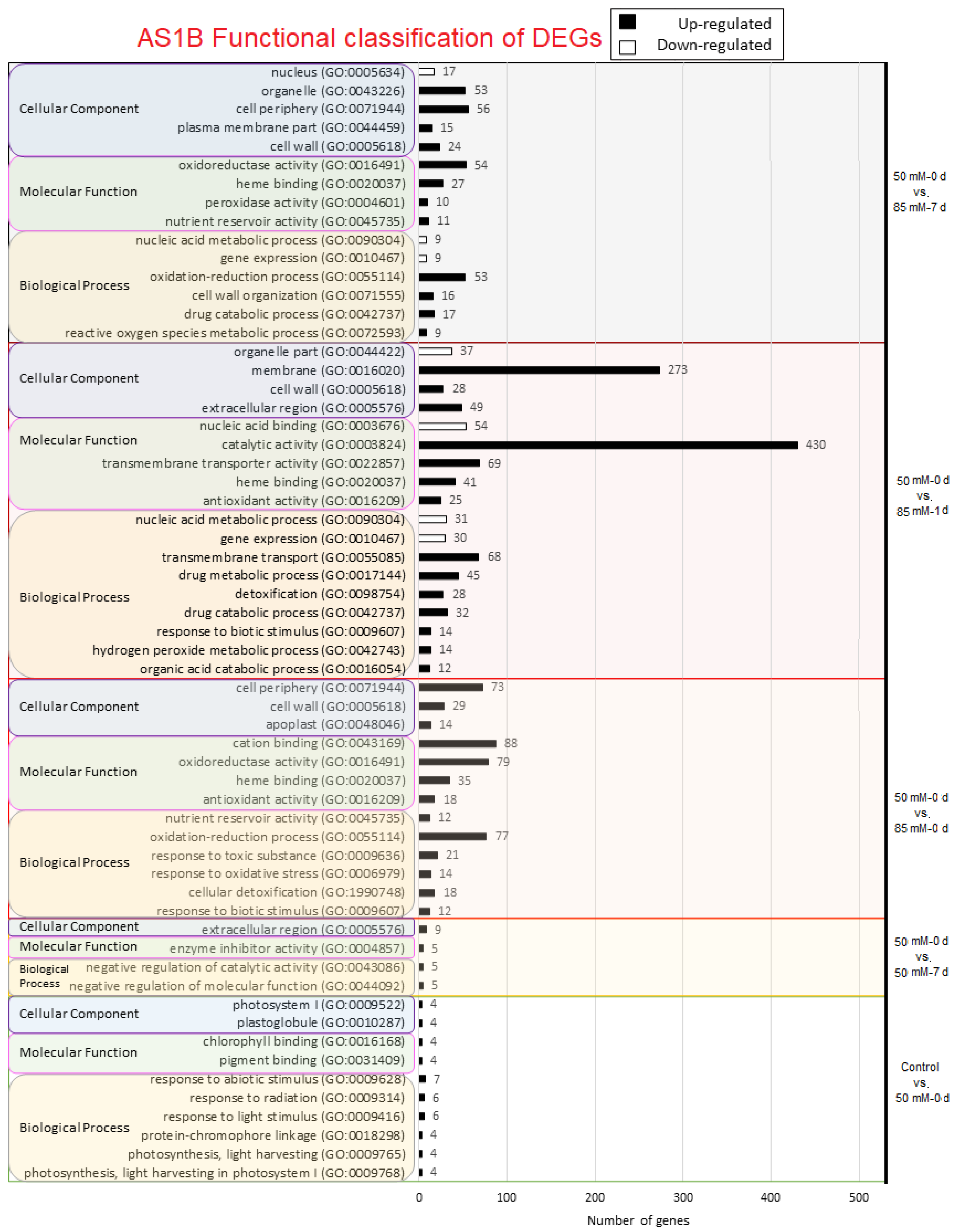
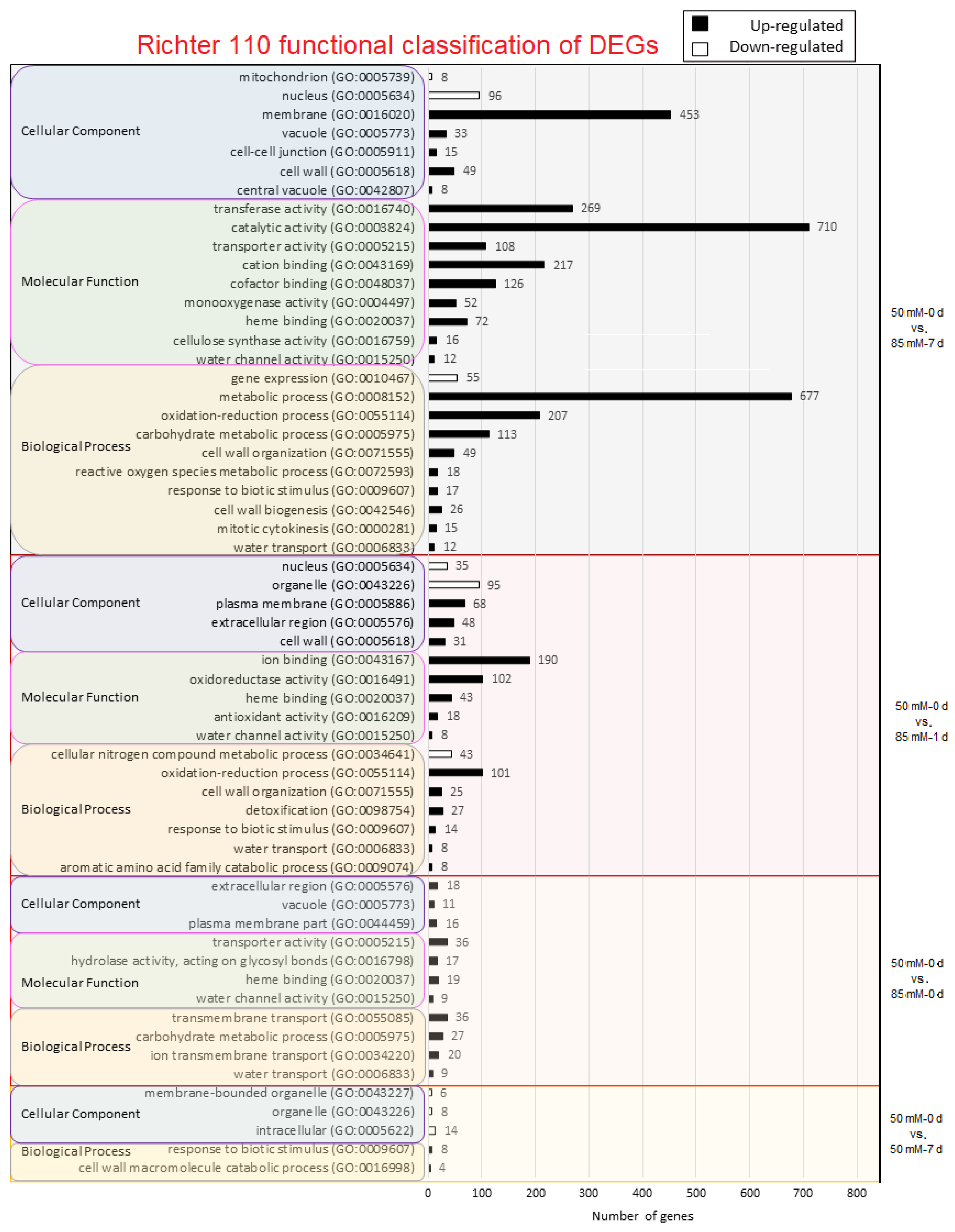
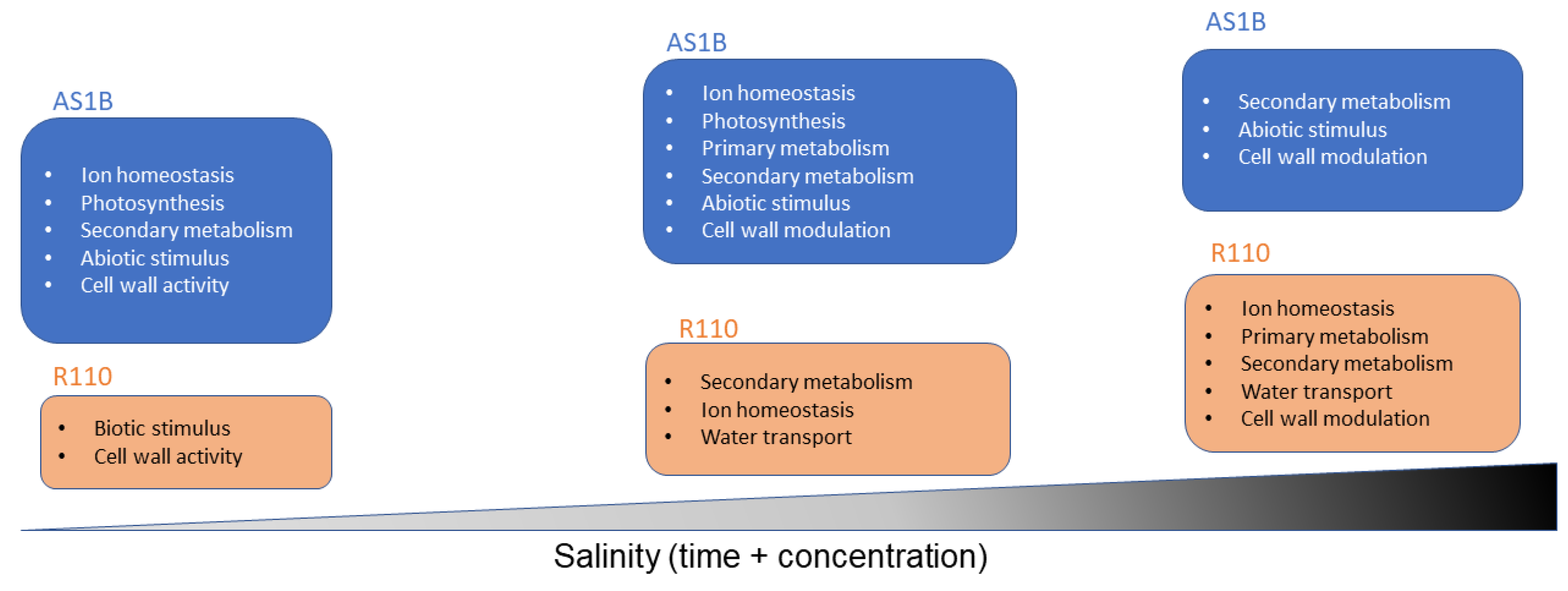
2.3. Gene Ontology Analysis
2.4. Consistent DEGs in AS1B and Richter 110 across Different Treatment Conditions
3. Materials and Methods
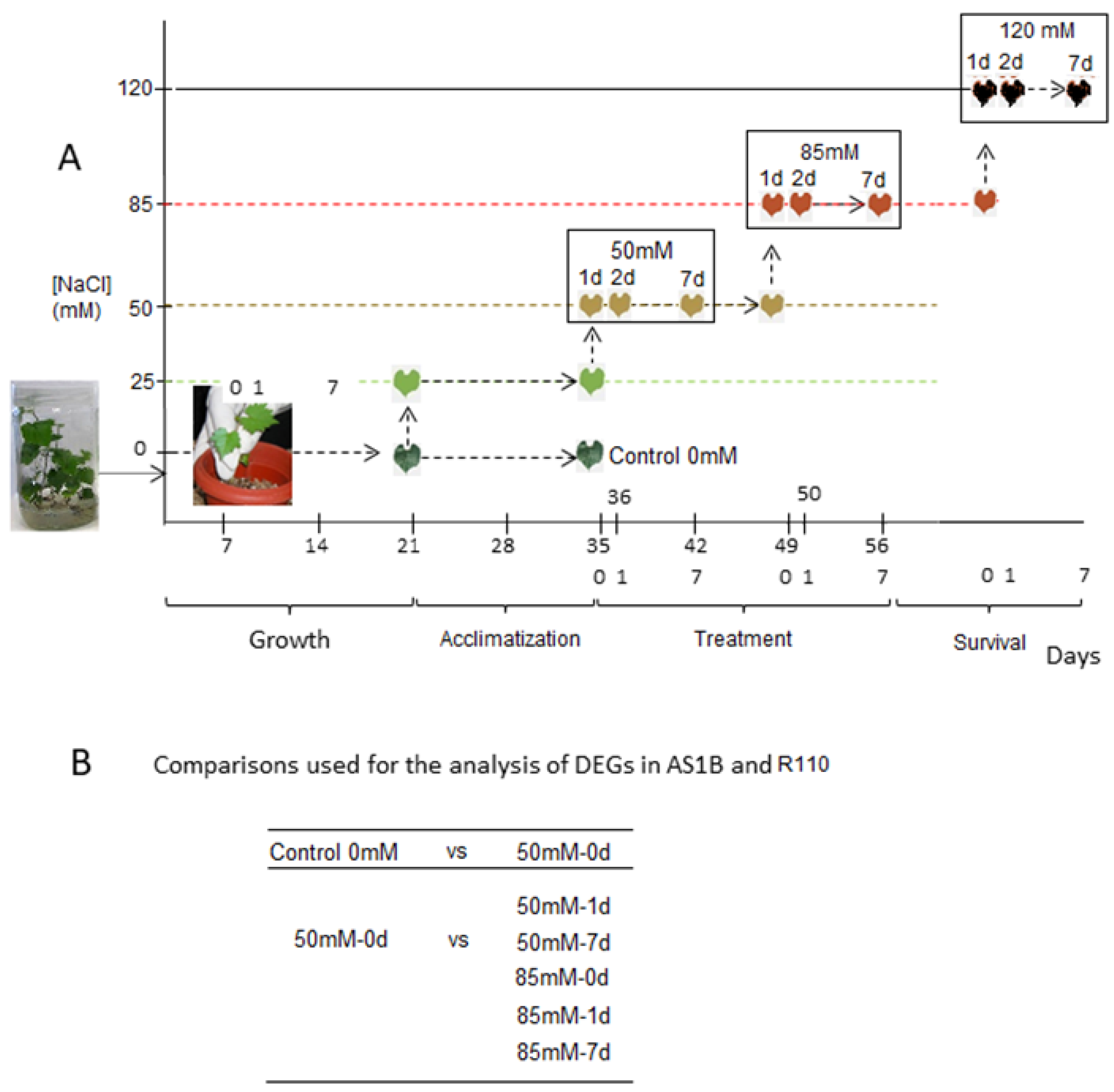
3.1. Preparation of Plant Material and Salt Treatments
3.2. RNA Extraction and Sequencing
3.3. Differential Expression Analysis
3.4. Gene Ontology Analysis
3.5. Gene Validation: RT-qPCR and Linear Regression Analyse
4. Discussion
4.1. Focusing the Comparison of Known Salt Damages and Mechanisms Tolerance in Grapevine
4.2. Distribution of Differentially Expressed Genes from Functional Categories in Both Accessions
4.3. Key Functional Roles in DEGs Present in AS1B’s Early Response to the Initial Saline Input
4.3.1. Control vs. 50 mM-0 d
4.3.2. mM-0 d vs. 50 mM-1 d
4.4. Functional Characterization of Genes with a Continuous Differential Expression from Comparison 3 (50 mM-0 d vs. 50 mM-7 d) until Comparison 6 (50 mM-0 d vs. 85 mM-7 d)
4.4.1. Ion Transport
4.4.2. Secondary Metabolism
4.4.3. Mechanical Adjustment: Cell Wall and Cytoskeleton Dynamics
4.4.4. Hydric Regulation
4.4.5. Regulatory Systems: Hormone Response and Signaling
4.4.6. Biotic Stress
4.4.7. Post-Translational: Protein Stabilization and Ubiquitin-Mediated Degradation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 50 mM-0 d | 50 millimolar of sodium chloride per liter of irrigation solution at day 0 |
| 50 mM-1 d | 50 millimolar of sodium chloride per liter of irrigation solution at day 1 |
| 50 mM-7 d | 50 millimolar of sodium chloride per liter of irrigation solution at day 7 |
| 85 mM-0 d | 85 millimolar of sodium chloride per liter of irrigation solution at day 0 |
| 85 mM-1 d | 85 millimolar of sodium chloride per liter of irrigation solution at day 1 |
| 85 mM-7 d | 85 millimolar of sodium chloride per liter of irrigation solution at day 7 |
| DEA | Differential expression analysis |
| DEG | Differentially expressed gene |
| GO | Gene ontology |
| R110 | Richter 110 |
| RNA-seq | RNA sequencing |
References
- IOV. 2019 Statistical Report on World Vitiviniculture; International Organisation of Vine and Wine Intergovernmental Organisation: Paris, France, 2019; p. 23. [Google Scholar]
- Tate, A.B. Global Warming’s Impact on Wine. J. Wine Res. 2001, 12, 95–109. [Google Scholar] [CrossRef]
- Phogat, V.; Cox, J.W.; Šimůnek, J. Identifying the Future Water and Salinity Risks to Irrigated Viticulture in the Murray-Darling Basin, South Australia. Agric. Water Manag. 2018, 201, 107–117. [Google Scholar] [CrossRef]
- Jones, G.V.; Webb, L.B. Climate Change, Viticulture, and Wine: Challenges and Opportunities. J. Wine Res. 2010, 21, 103–106. [Google Scholar] [CrossRef]
- Hannah, L.; Roehrdanz, P.R.; Ikegami, M.; Shepard, A.V.; Shaw, M.R.; Tabor, G.; Zhi, L.; Marquet, P.A.; Hijmans, R.J. Climate Change, Wine, and Conservation. Proc. Natl. Acad. Sci. USA 2013, 110, 6907–6912. [Google Scholar] [CrossRef]
- Tomaz, A.; Palma, P.; Fialho, S.; Lima, A.; Alvarenga, P.; Potes, M.; Costa, M.J.; Salgado, R. Risk Assessment of Irrigation-Related Soil Salinization and Sodification in Mediterranean Areas. Water 2020, 12, 3569. [Google Scholar] [CrossRef]
- De Loryn, L.C.; Petrie, P.R.; Hasted, A.M.; Johnson, T.E.; Collins, C.; Bastian, S.E.P. Evaluation of Sensory Thresholds and Perception of Sodium Chloride in Grape Juice and Wine. Am. J. Enol. Vitic. 2014, 1, 124–133. [Google Scholar] [CrossRef]
- Walker, R.R.; Blackmore, D.H.; Clingeleffer, P.R.; Holt, H.; Pearson, W.; Francis, I.L. Effect of Rootstock on Yield, Grape Composition and Wine Sensory Attributes of Shiraz Grown in a Moderately Saline Environment. Aust. J. Grape Wine Res. 2019, 25, 414–429. [Google Scholar] [CrossRef]
- Mass, E.V.; Hoffman, G.J. Crop Salt Tolerance-Current Assessment. J. Irrig. Drain. Div. 1977, 103. [Google Scholar] [CrossRef]
- Ahmad, O.A. Research Article Salinity Soil Effects on Yeild, Fruit Quality and Mineral Composition of Superior Seedless Grapevines Grafted on Some Rootstocks. J. Appl. Sci. 2016, 16, 359–371. [Google Scholar] [CrossRef][Green Version]
- Downton, W.J.S. Growth and Mineral Composition of the Sultana Grapevine as Influenced by Salinity and Rootstock. Aust. J. Agric. Res. 1985, 36, 425–434. [Google Scholar] [CrossRef]
- Walker, R.R.; Blackmore, D.H.; Clingeleffer, P.R.; Correll, R.L. Rootstock Effects on Salt Tolerance of Irrigated Field-Grown Grapevines (Vitis Vinifera L. Cv. Sultana) 2. Ion Concentrations in Leaves and Juice. Aust. J. Grape Wine Res. 2004, 10, 90–99. [Google Scholar] [CrossRef]
- Stevens, R.; Pitt, T.R.; Dyson, C.; Pech, J.M.; Skewes, M. Salt Tolerant Rootstocks for Long-Term Sustainability in the Limestone Coast. Final Report to Grape and Wine Research & Development Corporation; South Australian Research and Development Institute Sustainable Systems: Urrbrae, SA, Australia, 2011. [Google Scholar]
- Zhou-Tsang, A.; Wu, Y.; Henderson, S.W.; Walker, A.R.; Borneman, A.R.; Walker, R.R.; Gilliham, M. Grapevine Salt Tolerance. Aust. J. Grape Wine Res. 2021, 27, 149–168. [Google Scholar] [CrossRef]
- Schröder, S.; Kortekamp, A.; Heene, E.; Daumann, J.; Valea, I.; Nick, P. Crop Wild Relatives as Genetic Resources–the Case of the European Wild Grape. Can. J. Plant Sci. 2015, 95, 905–912. [Google Scholar] [CrossRef]
- Askri, H.; Daldoul, S.; Ammar, A.B.; Rejeb, S.; Jardak, R.; Rejeb, M.N.; Mliki, A.; Ghorbel, A. Short-Term Response of Wild Grapevines (Vitis Vinifera L. Ssp. Sylvestris) to NaCl Salinity Exposure: Changes of Some Physiological and Molecular Characteristics. Acta Physiol. Plant. 2012, 34, 957–968. [Google Scholar] [CrossRef]
- Popescu, C.F.; Bejan, C.; Dumitrica, R.N.; Dejeu, L.C.; Nedelea, G. Rootstocks and Wild Grapevines Responses to Salinity. Vitis J. Grapevine Res. 2015, 54, 197–201. [Google Scholar]
- Arroyo-García, R.; Cantos, M.; Lara, M.; López, M.Á.; Gallardo, A.; Ocete, C.A.; Pérez, Á.; Bánáti, H.; García, J.L.; Ocete, R. Characterization of the Largest Relic Eurasian Wild Grapevine Reservoir in Southern Iberian Peninsula. Span. J. Agric. Res. 2016, 14, e0708. [Google Scholar] [CrossRef]
- Rozema, J.; Bijwaard, P.; Prast, G.; Broekman, R. Ecophysiological Adaptations of Coastal Halophytes from Foredunes and Salt Marshes. Vegetatio 1985, 62, 499–521. [Google Scholar] [CrossRef]
- Carrasco, D.; Gutierrez, E.; Ocete, R.; Revilla, M.; Arroyo-García, R. Exploitation of the Genetic Diversity of Grapevine (Vitis Vinifera L) in the Adaptation to Salt Stress. In Proceedings of the Annual Meeting COST action INTEGRAPE, Chania, Grece, 25–28 March 2019. [Google Scholar]
- Hoagland, D.; Arnon, D. The Water-Culture Method for Growing Plants without Soil. Circ. Calif. Agric. Exp. Stn. 1950, 347, 32. [Google Scholar]
- Shavrukov, Y. Salt Stress or Salt Shock: Which Genes Are We Studying? J. Exp. Bot. 2013, 64, 119–127. [Google Scholar] [CrossRef]
- Rivasplata-Varillas, P.E. Tolerancia Al NaCl de Plantas Andaluzas de Vid Silvestre (Vitis Vinifera L., Ssp Sylvestris (Gmelin) Hegi); CSIC-Instituto de Recursos Naturales y Agrobiología de Sevilla (IRNAS): Sevilla, Spain, 2005. [Google Scholar]
- Reid, K.E.; Olsson, N.; Schlosser, J.; Peng, F.; Lund, S.T. An Optimized Grapevine RNA Isolation Procedure and Statistical Determination of Reference Genes for Real-Time RT-PCR during Berry Development. BMC Plant Biol. 2006, 6, 27. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data; Babraham Bioinformatics: Cambridge, UK, 2010. [Google Scholar]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A Fast Spliced Aligner with Low Memory Requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Jaillon, O.; Aury, J.M.; Noel, B.; Policriti, A.; Clepet, C.; Casagrande, A.; Choisne, N.; Aubourg, S.; Vitulo, N.; Jubin, C.; et al. The Grapevine Genome Sequence Suggests Ancestral Hexaploidization in Major Angiosperm Phyla. Nature 2007, 449, 463–467. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie Enables Improved Reconstruction of a Transcriptome from RNA-Seq Reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Tarazona, S. Differential Expression in RNA-Seq: A matter of depth. Genome Res. 2011, 21, 2213–2223. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential Expression Analysis for Sequence Count Data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef]
- Blum, M.; Chang, H.-Y.; Chuguransky, S.; Grego, T.; Kandasaamy, S.; Mitchell, A.; Nuka, G.; Paysan-Lafosse, T.; Qureshi, M.; Raj, S.; et al. The InterPro Protein Families and Domains Database: 20 Years On. Nucleic Acids Res. 2021, 49, D344–D354. [Google Scholar] [CrossRef]
- Zimmermann, L.; Stephens, A.; Nam, S.; Rau, D.; Kübler, J.; Lozajic, M.; Gabler, F.; Söding, J.; Lupas, A.N.; Alva, V. A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at Its Core. J. Mol. Biol. 2018, 430, 2237–2243. [Google Scholar] [CrossRef] [PubMed]
- Wan, Y.; Schwaninger, H.R.; Baldo, A.M.; Labate, J.A.; Zhong, G.Y.; Simon, C.J. A Phylogenetic Analysis of the Grape Genus (Vitis L.) Reveals Broad Reticulation and Concurrent Diversification during Neogene and Quaternary Climate Change. BMC Evol. Biol. 2013, 13, 141. [Google Scholar] [CrossRef] [PubMed]
- Downton, W.J.S. Chloride Accumulation in Different Species of Grapevine. Sci. Hortic. 1977, 7, 249–253. [Google Scholar] [CrossRef]
- Walker, R.; Blackmore, D.H.; Clingeleffer, P.R.; CorrelL, R.L. Rootstock Effects on Salt Tolerance of Irrigated Field-Grown Grapevines (Vitis Vinifera L. Cv. Sultana).: 1. Yield and Vigour Inter-Relationships. Aust. J. Grape Wine Res. 2002, 8, 3–14. [Google Scholar] [CrossRef]
- Donkin, R.; Robinson, S.; Sumby, K.; Harris, V.; McBryde, C.; Jiranek, V. Sodium Chloride in Australian Grape Juice and Its Effect on Alcoholic and Malolactic Fermentation. Am. J. Enol. Vitic. 2010, 61, 392–400. [Google Scholar]
- Munns, R.; Day, D.A.; Fricke, W.; Watt, M.; Arsova, B.; Barkla, B.J.; Bose, J.; Byrt, C.S.; Chen, Z.H.; Foster, K.J.; et al. Energy Costs of Salt Tolerance in Crop Plants. New Phytol. 2020, 225, 1072–1090. [Google Scholar] [CrossRef]
- Munns, R.; Gilliham, M. Salinity Tolerance of Crops-What is the Cost? New Phytol. 2015, 208, 668–673. [Google Scholar] [CrossRef]
- Fozouni, M.; Abbaspour, N.; Baneh, H.D. Leaf Water Potential, Photosynthetic Pigments and Compatible Solutes Alterations in Four Grape Cultivars under Salinity. Vitis J. Grapevine Res. 2012, 51, 147–152. [Google Scholar]
- Osmond, B.; Badger, M.; Maxwell, K.; Björkman, O.; Leegood, R. Too Many Photons: Photorespiration, Photoinhibition and Photooxidation. Trends Plant Sci. 1997, 2, 119–121. [Google Scholar] [CrossRef]
- Downton, W.J.S. Osmotic Adjustment during Water Stress Protects the Photosynthetic Apparatus against Photoinhibition. Plant Sci. Lett. 1983, 30, 137–143. [Google Scholar] [CrossRef]
- Dorji, K. Chloride Accumulation in Grapevine (Vitis) Rootstocks. Ph.D. Thesis, Latrobe University, Melbourne, VIC, Australia, 2015. [Google Scholar]
- Storey, R.; Schachtman, D.P.; Thomas, M.R. Root Structure and Cellular Chloride, Sodium and Potassium Distribution in Salinized Grapevines. Plant Cell Environ. 2003, 26, 789–800. [Google Scholar] [CrossRef]
- Tregeagle, J.M.; Tisdall, J.M.; Tester, M.; Walker, R.R. Cl–Uptake, Transport and Accumulation in Grapevine Rootstocks of Differing Capacity for Cl–-Exclusion. Funct. Plant Biol. 2010, 37, 665–673. [Google Scholar]
- Walker, R.R.; Sedgley, M.; Blesing, M.A.; Douglas, T.J. Anatomy, Ultrastructure and Assimilate Concentrations of Roots of Citrus Genotypes Differing in Ability for Salt Exclusion. J. Exp. Bot. 1984, 35, 1481–1494. [Google Scholar] [CrossRef]
- Premachandra, G.S.; Saneoka, H.; Fujita, K.; Ogata, S. Leaf Water Relations, Osmotic Adjustment, Cell Membrane Stability, Epicuticular Wax Load and Growth as Affected by Increasing Water Deficits in Sorghum. J. Exp. Bot. 1992, 43, 1569–1576. [Google Scholar] [CrossRef]
- Mohammadkhani, N.; Heidari, R.; Abbaspour, N.; Rahmani, F. Growth Responses and Aquaporin Expression in Grape Genotypes under Salinity. Iran. J. Plant Physiol. 2012, 2, 497–508. [Google Scholar]
- Shelden, M.C.; Howitt, S.M.; Kaiser, B.N.; Tyerman, S.D. Identification and Functional Characterisation of Aquaporins in the Grapevine, Vitis Vinifera. Funct. Plant Biol. 2009, 36, 1065–1078. [Google Scholar] [CrossRef]
- Lovisolo, C.; Schubert, A. Effects of Water Stress on Vessel Size and Xylem Hydraulic Conductivity in Vitis Vinifera L. J. Exp. Bot. 1998, 49, 693–700. [Google Scholar] [CrossRef]
- Walker, R.R.; Torokfalvy, E.; Scott, N.S.; Kriedemann, P.E. An Analysis of Photosynthetic Response to Salt Treatment in Vitis Vinifera. Funct. Plant Biol. 1981, 8, 359–374. [Google Scholar] [CrossRef]
- Chalker-Scott, L. Environmental Significance of Anthocyanins in Plant Stress Responses. Photochem. Photobiol. 1999, 70, 1–9. [Google Scholar] [CrossRef]
- Yildirim, Ö.; Aras, S.; Ergül, A. Response of Antioxidant Systems to Short-Term NaCl Stress in Grapevine Rootstock-1616c and Vitis Vinifera L. Cv. Razaki. Acta Biol. Crac. Ser. Bot. 2004, 46, 151–158. [Google Scholar]
- Soylemezoglu, G.; Demir, K.; Inal, A.; Gunes, A. Effect of Silicon on Antioxidant and Stomatal Response of Two Grapevine (Vitis Vinifera L.) Rootstocks Grown in Boron Toxic, Saline and Boron Toxic-Saline Soil. Sci. Hortic. 2009, 123, 240–246. [Google Scholar] [CrossRef]
- Downton, W.J.S.; Loveys, B.R. Abscisic Acid Content and Osmotic Relations of Salt-Stressed Grapevine Leaves. Funct. Plant Biol. 1981, 8, 443–452. [Google Scholar] [CrossRef]
- Ma, Y.; Wang, L.; Wang, J.; Zhong, Y.; Cheng, Z.M. Isolation and Expression Analysis of Salt Overly Sensitive Gene Family in Grapevine (Vitisvinifera) in Response to Salt and PEG Stress. PLoS ONE 2019, 14, e0212666. [Google Scholar] [CrossRef]
- Dubrovina, A.S.; Kiselev, K.V.; Khristenko, V.S.; Aleynova, O.A. VaCPK21, a Calcium-Dependent Protein Kinase Gene of Wild Grapevine Vitis Amurensis Rupr., Is Involved in Grape Response to Salt Stress. Plant Cell. Tissue Organ Cult. 2016, 124, 137–150. [Google Scholar] [CrossRef]
- Sohrabi, S.; Ebadi, A.; Jalali, S.; Salami, S.A. Enhanced Values of Various Physiological Traits and VvNAC1 Gene Expression Showing Better Salinity Stress Tolerance in Some Grapevine Cultivars as Well as Rootstocks. Sci. Hortic. 2017, 225, 317–326. [Google Scholar] [CrossRef]
- Walker, R.R.; Blackmore, D.H.; Clingeleffer, P.R.; Tarr, C.R. Rootstock Effects on Salt Tolerance of Irrigated Field-Growngrapevines (Vitis Vinifera L. Cv. Sultana) 3. Fresh Fruit Composition and Dried Grape Quality. Aust. J. Grape Wine Res. 2007, 13, 130–141. [Google Scholar] [CrossRef]
- Henderson, S.W.; Baumann, U.; Blackmore, D.H.; Walker, A.R.; Walker, R.R.; Gilliham, M. Shoot Chloride Exclusion and Salt Tolerance in Grapevine is Associated with Differential Ion Transporter Expression in Roots. BMC Plant Biol. 2014, 14, 273. [Google Scholar] [CrossRef]
- Guan, L.; Haider, M.S.; Khan, N.; Nasim, M. Transcriptome Sequence Analysis Elaborates a Complex Defensive Mechanism of Grapevine ( Vitis Vinifera L.) in Response to Salt Stress. Int. J. Mol. Sci. 2018, 19, 4019. [Google Scholar] [CrossRef] [PubMed]
- Das, P.; Majumder, A.L. Transcriptome Analysis of Grapevine under Salinity and Identification of Key Genes Responsible for Salt Tolerance. Funct. Integr. Genom. 2019, 19, 61–73. [Google Scholar] [CrossRef] [PubMed]
- Baggett, J.P.; Habibsadeh, S.; Toups, H.S.; Cochetel, N.; Ghan, R.; Robinson, M.L.; Barrios-Masias, F.H.; Cramer, G.R. Is Foliar Cl- Concentration the Cause of Photosynthetic Decline in Grapevine during Mild Salinity? OENO One 2021, 55, 33–48. [Google Scholar] [CrossRef]
- Ismail, A.; Seo, M.; Takebayashi, Y.; Kamiya, Y.; Eiche, E.; Nick, P. Salt Adaptation Requires Efficient Fine-Tuning of Jasmonate Signalling. Protoplasma 2014, 251, 881–898. [Google Scholar] [CrossRef] [PubMed]
- Mohammadkhani, N.; Abbaspour, N. Effects of Salinity on Antioxidant System in Ten Grape Genotypes. Iran. J. Plant Physiol. 2018, 8, 2247–2255. [Google Scholar] [CrossRef]
- Jacoby, R.P.; Taylor, N.L.; Millar, A.H. The Role of Mitochondrial Respiration in Salinity Tolerance. Trends Plant Sci. 2011, 16, 614–623. [Google Scholar] [CrossRef] [PubMed]
- Jacoby, R.P.; Millar, A.H.; Taylor, N.L. Analysis of the Sodium Chloride-Dependent Respiratory Kinetics of Wheat Mitochondria Reveals Differential Effects on Phosphorylating and Non-Phosphorylating Electron Transport Pathways. Plant Cell Environ. 2016, 39, 823–833. [Google Scholar] [CrossRef] [PubMed]
- Geng, X.; Ye, J.; Yang, X.; Li, S.; Zhang, L.; Song, X. Identification of Proteins Involved in Carbohydrate Metabolism and Energy Metabolism Pathways and Their Regulation of Cytoplasmic Male Sterility in Wheat. Int. J. Mol. Sci. 2018, 19, 324. [Google Scholar] [CrossRef]
- Prinsi, B.; Failla, O.; Scienza, A.; Espen, L. Root Proteomic Analysis of Two Grapevine Rootstock Genotypes Showing Different Susceptibility to Salt Stress. Int. J. Mol. Sci. 2020, 21, 1076. [Google Scholar] [CrossRef]
- Li, W.; Pang, S.; Lu, Z.; Jin, B. Function and Mechanism of WRKY Transcription Factors in Abiotic Stress Responses of Plants. Plants 2020, 9, 1515. [Google Scholar] [CrossRef]
- Li, H.; Xu, Y.; Xiao, Y.; Zhu, Z.; Xie, X.; Zhao, H.; Wang, Y. Expression and Functional Analysis of Two Genes Encoding Transcription Factors, VpWRKY1 and VpWRKY2, Isolated from Chinese Wild Vitis Pseudoreticulata. Planta 2010, 232, 1325–1337. [Google Scholar] [CrossRef]
- Hong, Y.; Zhang, H.; Huang, L.; Li, D.; Song, F. Overexpression of a Stress-Responsive NAC Transcription Factor Gene ONAC022 Improves Drought and Salt Tolerance in Rice. Front. Plant Sci. 2016, 7, 4. [Google Scholar] [CrossRef]
- Alshareef, N.O.; Wang, J.Y.; Ali, S.; Al-Babili, S.; Tester, M.; Schmöckel, S.M. Overexpression of the NAC Transcription Factor JUNGBRUNNEN1 (JUB1) Increases Salinity Tolerance in Tomato. Plant Physiol. Biochem. 2019, 140, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Wu, Z.; Gu, H.; Cheng, D.; Guo, X.; Li, L.; Shi, C.; Xu, G.; Gu, S.; Abid, M.; et al. AvNAC030, a NAC Domain Transcription Factor, Enhances Salt Stress Tolerance in Kiwifruit. Int. J. Mol. Sci. 2021, 22, 11897. [Google Scholar] [CrossRef]
- Aydemir, B.C.; Özmen, C.Y.; Kibar, U.; Mutaf, F.; Büyük, P.B.; Bakır, M.; Ergül, A. Salt Stress Induces Endoplasmic Reticulum Stress-Responsive Genes in a Grapevine Rootstock. PLoS ONE 2020, 15, e0236424. [Google Scholar]
- He, X.-J.; Zhang, Z.-G.; Yan, D.-Q.; Zhang, J.-S.; Chen, S.-Y. A Salt-Responsive Receptor-like Kinase Gene Regulated by the Ethylene Signaling Pathway Encodes a Plasma Membrane Serine/Threonine Kinase. Theor. Appl. Genet. 2004, 109, 377–383. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.-L.; Yu, Q.-Y.; Tang, L.-L.; Ji, W.; Bai, X.; Cai, H.; Liu, X.-F.; Ding, X.-D.; Zhu, Y.-M. GsSRK, a G-Type Lectin S-Receptor-like Serine/Threonine Protein Kinase, Is a Positive Regulator of Plant Tolerance to Salt Stress. J. Plant Physiol. 2013, 170, 505–515. [Google Scholar] [CrossRef]
- Ge, R.-C.; Chen, G.-P.; Zhao, B.-C.; Shen, Y.-Z.; Huang, Z.-J. Cloning and Functional Characterization of a Wheat Serine/Threonine Kinase Gene (TaSTK) Related to Salt-Resistance. Plant Sci. 2007, 173, 55–60. [Google Scholar] [CrossRef]
- De Barajas-Lopez, J.D.; Moreno, J.R.; Gamez-Arjona, F.M.; Pardo, J.M.; Punkkinen, M.; Zhu, J.-K.; Quintero, F.J.; Fujii, H. Upstream Kinases of Plant SnRKs Are Involved in Salt Stress Tolerance. Plant J. 2018, 93, 107–118. [Google Scholar] [CrossRef] [PubMed]
- JiangTao, G.; TuanJie, Z.; Yan, L.; JunYi, G. Cloning and expression analysis of an adenylate kinase gene GmADK in soybean. Acta Agron. Sin. 2013, 39, 1739–1745. [Google Scholar] [CrossRef]
- Wera, S.; Hemmings, B.A. Serine/Threonine Protein Phosphatases. Biochem. J. 1995, 311 Pt 1, 17–29. [Google Scholar] [CrossRef]
- Liu, J.; Lin, Q.F.; Qi, S.L.; Feng, X.J.; Han, H.L.; Xu, T.; Hua, X.J. The F-Box Protein EST1 Modulates Salt Tolerance in Arabidopsis by Regulating Plasma Membrane Na+/H+ Antiport Activity. J. Plant Physiol. 2020, 251, 153217. [Google Scholar] [CrossRef]
- Yu, Y.; Wang, P.; Bai, Y.; Wang, Y.; Wan, H.; Liu, C.; Ni, Z. The Soybean F-Box Protein GmFBX176 Regulates ABA-Mediated Responses to Drought and Salt Stress. Environ. Exp. Bot. 2020, 176, 104056. [Google Scholar] [CrossRef]
- Zhu, J.K. Regulation of Ion Homeostasis under Salt Stress. Curr. Opin. Plant Biol. 2003, 6, 441–445. [Google Scholar] [CrossRef]
- Sánchez-Fernández, R.; Davies, T.G.E.; Coleman, J.O.D.; Rea, P.A. The Arabidopsis Thaliana ABC Protein Superfamily, a Complete Inventory. J. Biol. Chem. 2001, 276, 30231–30244. [Google Scholar] [CrossRef]
- Nour-Eldin, H.H.; Andersen, T.G.; Burow, M.; Madsen, S.R.; Jørgensen, M.E.; Olsen, C.E.; Dreyer, I.; Hedrich, R.; Geiger, D.; Halkier, B.A. NRT/PTR Transporters Are Essential for Translocation of Glucosinolate Defence Compounds to Seeds. Nature 2012, 488, 531–534. [Google Scholar] [CrossRef]
- Kim, D.; Jin, J.; Alejandro, S.; Martinoia, E.; Lee, Y. Overexpression of AtABCG36 Improves Drought and Salt Stress Resistance in Arabidopsis. Physiol. Plant. 2010, 139, 170–180. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Henderson, S.W.; Walker, R.R.; Gilliham, M. Root-Specific Expression of Vitis Vinifera VviNPF2.2 Modulates Shoot Anion Concentration in Transgenic Arabidopsis. Front. Plant Sci. 2022, 13, 863971. [Google Scholar] [CrossRef]
- Shani, U.; Ben-Gal, A. Long-Term Response of Grapevines to Salinity: Osmotic Effects and Ion Toxicity. Am. J. Enol. Vitic. 2005, 56, 148–154. [Google Scholar]
- Yu, S.; Zhang, X.; Guan, Q.; Takano, T.; Liu, S. Expression of a Carbonic Anhydrase Gene Is Induced by Environmental Stresses in Rice (Oryza Sativa L.). Biotechnol. Lett. 2007, 29, 89–94. [Google Scholar] [CrossRef] [PubMed]
- DiMario, R.J.; Clayton, H.; Mukherjee, A.; Ludwig, M.; Moroney, J. V Plant Carbonic Anhydrases: Structures, Locations, Evolution, and Physiological Roles. Mol. Plant 2017, 10, 30–46. [Google Scholar] [CrossRef] [PubMed]
- Ribba, T.; Garrido-Vargas, F.; O’Brien, J.A. Auxin-Mediated Responses under Salt Stress: From Developmental Regulation to Biotechnological Applications. J. Exp. Bot. 2020, 71, 3843–3853. [Google Scholar] [CrossRef] [PubMed]
- Kaya, C.; Ashraf, M.; Dikilitas, M.; Tuna, A.L. Alleviation of Salt Stress-Induced Adverse Effects on Maize Plants by Exogenous Application of Indoleacetic Acid (IAA) and Inorganic Nutrients-A Field Trial. Aust. J. Crop Sci. 2013, 7, 249–254. [Google Scholar]
- Abdel Latef, A.A.; Tahjib-Ul-Arif, M.; Rhaman, M.S. Exogenous Auxin-Mediated Salt Stress Alleviation in Faba Bean (Vicia Faba L.). Agronomy 2021, 11, 547. [Google Scholar] [CrossRef]
- Feng, Y.; Lichao, Q.; Candida, N.; Qiaohong, D.; Daniel, K.; Kathryn, L.; Xiaoqian, L.; Changqing, L.; Hui, L.; Congcong, H.; et al. FERONIA Receptor Kinase Pathway Suppresses Abscisic Acid Signaling in Arabidopsis by Activating ABI2 Phosphatase. Proc. Natl. Acad. Sci. USA 2012, 109, 14693–14698. [Google Scholar] [CrossRef]
- Tenhaken, R. Cell Wall Remodeling under Abiotic Stress. Front. Plant Sci. 2015, 5, 771. [Google Scholar] [CrossRef]
- Hanana, M.; Cagnac, O.; Yamaguchi, T.; Hamdi, S.; Ghorbel, A.; Blumwald, E. Molecular Biology and Transport Properties of Grapevine Na+/H+ Antiporter BT-Biosaline Agriculture and High Salinity Tolerance; Abdelly, C., Öztürk, M., Ashraf, M., Grignon, C., Eds.; Birkhäuser Basel: Basel, Switzerland, 2008; pp. 305–315. [Google Scholar]
- Rodríguez-Rosales, M.P.; Gálvez, F.J.; Huertas, R.; Aranda, M.N.; Baghour, M.; Cagnac, O.; Venema, K. Plant NHX Cation/Proton Antiporters. Plant Signal. Behav. 2009, 4, 265–276. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Wang, J.; Zhong, Y.; Geng, F.; Cramer, G.R.; Cheng, Z.-M.M. Subfunctionalization of Cation/Proton Antiporter 1 Genes in Grapevine in Response to Salt Stress in Different Organs. Hortic. Res. 2015, 2, 15031. [Google Scholar] [CrossRef] [PubMed]
- Chanroj, S.; Wang, G.; Venema, K.; Zhang, M.W.; Delwiche, C.F.; Sze, H. Conserved and Diversified Gene Families of Monovalent Cation/H+ Antiporters from Algae to Flowering Plants. Front. Plant Sci. 2012, 3, 25. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Wang, J.; Zhong, Y.; Cramer, G.; Cheng, M. Genome-Wide Analysis of the Cation/Proton Antiporter (CPA) Super Family Genes in Grapevine (Vitis Vinifera L.). Plant Omics 2015, 8, 300–311. [Google Scholar]
- Kholodova, V.; Volkov, K.; Kuznetsov, V. Plants Under Heavy Metal Stress in Saline Environments. In Soil Heavy Metals, Soil Biology; Springer: Berlin/Heidelberg, Germany, 2010; Volume 19, pp. 163–183. ISBN 978-3-642-02435. [Google Scholar]
- Denancé, N.; Szurek, B.; Noël, L.D. Emerging Functions of Nodulin-like Proteins in Non-Nodulating Plant Species. Plant Cell Physiol. 2014, 55, 469–474. [Google Scholar] [CrossRef] [PubMed]
- Berglund, T.; Ohlsson, A.B. Defensive and Secondary Metabolism in Plant Tissue Cultures, with Special Reference to Nicotinamide, Glutathione and Oxidative Stress. Plant Cell Tissue Organ Cult. 1995, 43, 137–145. [Google Scholar] [CrossRef]
- Abogadallah, G.M. Antioxidative Defense under Salt Stress. Plant Signal. Behav. 2010, 5, 369–374. [Google Scholar] [CrossRef]
- Akram, N.A.; Shafiq, F.; Ashraf, M. Ascorbic Acid-a Potential Oxidant Scavenger and Its Role in Plant Development and Abiotic Stress Tolerance. Front. Plant Sci. 2017, 8, 613. [Google Scholar] [CrossRef]
- Carvalho, L.C.; Vidigal, P.; Amancio, S. Oxidative Stress Homeostasis in Grapevine (Vitis Vinifera L.). Front. Environ. Sci. 2015, 3, 20. [Google Scholar] [CrossRef]
- Jambunathan, N. Determination and Detection of Reactive Oxygen Species (ROS), Lipid Peroxidation, and Electrolyte Leakage in Plants. In Plant Stress Tolerance; Humana: Totowa, NJ, USA, 2010; pp. 291–297. [Google Scholar]
- Wang, C.; Zhang, L.-J.; Huang, R.-D. Cytoskeleton and Plant Salt Stress Tolerance. Plant Signal. Behav. 2011, 6, 29–31. [Google Scholar] [CrossRef]
- Davin, L.B.; Lewis, N.G. Dirigent Proteins and Dirigent Sites Explain the Mystery of Specificity of Radical Precursor Coupling in Lignan and Lignin Biosynthesis. Plant Physiol. 2000, 123, 453–462. [Google Scholar] [CrossRef] [PubMed]
- Moura, J.C.M.S.; Bonine, C.A.V.; de Oliveira Fernandes Viana, J.; Dornelas, M.C.; Mazzafera, P. Abiotic and Biotic Stresses and Changes in the Lignin Content and Composition in Plants. J. Integr. Plant Biol. 2010, 52, 360–376. [Google Scholar] [CrossRef] [PubMed]
- Ito, S.; Suzuki, Y.; Miyamoto, K.; Ueda, J.; Yamaguchi, I. AtFLA11, a Fasciclin-Like Arabinogalactan-Protein, Specifically Localized in Screlenchyma Cells. Biosci. Biotechnol. Biochem. 2005, 69, 1963–1969. [Google Scholar] [CrossRef] [PubMed]
- Wormit, A.; Usadel, B. The Multifaceted Role of Pectin Methylesterase Inhibitors (PMEIs). Int. J. Mol. Sci. 2018, 19, 2878. [Google Scholar] [CrossRef]
- Park, J.; Cui, Y.; Kang, B.-H. AtPGL3 is an Arabidopsis BURP Domain Protein That Is Localized to the Cell Wall and Promotes Cell Enlargement. Front. Plant Sci. 2015, 6, 412. [Google Scholar] [CrossRef] [PubMed]
- Lovisolo, C.; Lavoie-Lamoureux, A.; Tramontini, S.; Ferrandino, A. Grapevine Adaptations to Water Stress: New Perspectives about Soil/Plant Interactions. Theor. Exp. Plant Physiol. 2016, 28, 53–66. [Google Scholar] [CrossRef]
- Tyerman, S.D.; Niemietz, C.M.; Bramley, H. Plant Aquaporins: Multifunctional Water and Solute Channels with Expanding Roles. Plant. Cell Environ. 2002, 25, 173–194. [Google Scholar]
- Byrt, C.S.; Zhao, M.; Kourghi, M.; Bose, J.; Henderson, S.W.; Qiu, J.; Gilliham, M.; Schultz, C.; Schwarz, M.; Ramesh, S.A.; et al. Non-Selective Cation Channel Activity of Aquaporin AtPIP2;1 Regulated by Ca2+ and PH. Plant. Cell Environ. 2017, 40, 802–815. [Google Scholar] [CrossRef]
- Vandeleur, R.K.; Mayo, G.; Shelden, M.C.; Gilliham, M.; Kaiser, B.N.; Tyerman, S.D. The Role of Plasma Membrane Intrinsic Protein Aquaporins in Water Transport through Roots: Diurnal and Drought Stress Responses Reveal Different Strategies between Isohydric and Anisohydric Cultivars of Grapevine. Plant Physiol. 2009, 149, 445–460. [Google Scholar] [CrossRef]
- Galmés, J.; Pou, A.; Alsina, M.M.; Tomàs, M.; Medrano, H.; Flexas, J. Aquaporin Expression in Response to Different Water Stress Intensities and Recovery in Richter-110 (Vitis Sp.): Relationship with Ecophysiological Status. Planta 2007, 226, 671–681. [Google Scholar] [CrossRef]
- Hundertmark, M.; Hincha, D.K. LEA (Late Embryogenesis Abundant) Proteins and Their Encoding Genes in Arabidopsis Thaliana. BMC Genom. 2008, 9, 118. [Google Scholar] [CrossRef] [PubMed]
- Rocheta, M.; Coito, J.L.; Ramos, M.J.N.; Carvalho, L.; Becker, J.D.; Carbonell-Bejerano, P.; Amâncio, S. Transcriptomic Comparison between Two Vitis Vinifera L. Varieties (Trincadeira and Touriga Nacional) in Abiotic Stress Conditions. BMC Plant Biol. 2016, 16, 224. [Google Scholar] [CrossRef] [PubMed]
- Cramer, G.R.; Ergul, A.; Grimplet, J.; Tillett, R.L.; Tattersall, E.A.R.; Bohlman, M.C.; Vincent, D.; Sonderegger, J.; Evans, J.; Osborne, C.; et al. Water and Salinity Stress in Grapevines: Early and Late Changes in Transcript and Metabolite Profiles. Funct. Integr. Genom. 2007, 7, 111–134. [Google Scholar] [CrossRef]
- Braun, H.; Czihal, A.; Shutov, A.D.; Baumlein, H. A Vicilin-like Seed Protein of Cycads: Similarity to Sucrose-Binding Proteins. Plant Mol. Biol. 1996, 31, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Anil Kumar, S.; Hima Kumari, P.; Shravan Kumar, G.; Mohanalatha, C.; Kavi Kishor, P.B. Osmotin: A Plant Sentinel and a Possible Agonist of Mammalian Adiponectin. Front. Plant Sci. 2015, 6, 163. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Poovaiah, B.W. Calcium/Calmodulin-Mediated Signal Network in Plants. Trends Plant Sci. 2003, 8, 505–512. [Google Scholar] [CrossRef] [PubMed]
- Feuillet, C.; Schachermayr, G.; Keller, B. Molecular Cloning of a New Receptor-like Kinase Gene Encoded at the Lr10 Disease Resistance Locus of Wheat. Plant J. 1997, 11, 45–52. [Google Scholar] [CrossRef]
- Sharma, A.; Shahzad, B.; Kumar, V.; Kohli, S.K.; Sidhu, G.P.S.; Bali, A.S.; Handa, N.; Kapoor, D.; Bhardwaj, R.; Zheng, B. Phytohormones Regulate Accumulation of Osmolytes under Abiotic Stress. Biomolecules 2019, 9, 285. [Google Scholar] [CrossRef]
- Guy, E.; Lautier, M.; Chabannes, M.; Roux, B.; Lauber, E.; Arlat, M.; Noël, L.D. XopAC-Triggered Immunity against Xanthomonas Depends on Arabidopsis Receptor-like Cytoplasmic Kinase Genes PBL2 and RIPK. PLoS ONE 2013, 8, e73469. [Google Scholar] [CrossRef]
- Návarová, H.; Bernsdorff, F.; Döring, A.-C.; Zeier, J. Pipecolic Acid, an Endogenous Mediator of Defense Amplification and Priming, Is a Critical Regulator of Inducible Plant Immunity. Plant Cell 2012, 24, 5123–5141. [Google Scholar] [CrossRef]
- Borsani, O.; Valpuesta, V.; Botella, M.A. Evidence for a Role of Salicylic Acid in the Oxidative Damage Generated by NaCl and Osmotic Stress in Arabidopsis Seedlings. Plant Physiol. 2001, 126, 1024–1030. [Google Scholar] [CrossRef] [PubMed]
- Nishiyama, R.; Le, D.T.; Watanabe, Y.; Matsui, A.; Tanaka, M.; Seki, M.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Tran, L.-S.P. Transcriptome Analyses of a Salt-Tolerant Cytokinin-Deficient Mutant Reveal Differential Regulation of Salt Stress Response by Cytokinin Deficiency. PLoS ONE 2012, 7, e32124. [Google Scholar] [CrossRef] [PubMed]
- Moons, A.; Prinsen, E.; Bauw, G.; Van Montagu, M. Antagonistic Effects of Abscisic Acid and Jasmonates on Salt Stress-Inducible Transcripts in Rice Roots. Plant Cell 1997, 9, 2243–2259. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Qin, Y.; Han, J.; Zhang, L.; Xu, X.; Liu, X.; Wang, C.; Liu, X. Expression Analysis of Innate Immunity Related Genes in the True/Field Blast Resistance Gene-Mediated Defence Response. Biotechnol. Biotechnol. Equip. 2014, 28, 999–1007. [Google Scholar] [CrossRef]
- Degaris, K.A.; Walker, R.R.; Loveys, B.R.; Tyerman, S.D. Exogenous Application of Abscisic Acid to Root Systems of Grapevines with or without Salinity Influences Water Relations and Ion Allocation. Aust. J. Grape Wine Res. 2017, 23, 66–76. [Google Scholar] [CrossRef]
- Orsini, F.; Cascone, P.; De Pascale, S.; Barbieri, G.; Corrado, G.; Rao, R.; Maggio, A. Systemin-Dependent Salinity Tolerance in Tomato: Evidence of Specific Convergence of Abiotic and Biotic Stress Responses. Physiol. Plant. 2010, 138, 10–21. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, N.; Rivero, R.M.; Shulaev, V.; Blumwald, E.; Mittler, R. Abiotic and Biotic Stress Combinations. New Phytol. 2014, 203, 32–43. [Google Scholar] [CrossRef]
- Walker, R.R.; Blackmore, D.H.; Clingeleffer, P.R. Impact of Rootstock on Yield and Ion Concentrations in Petioles, Juice and Wine of Shiraz and Chardonnay in Different Viticultural Environments with Different Irrigation Water Salinity. Aust. J. Grape Wine Res. 2010, 16, 243–257. [Google Scholar] [CrossRef]
- Cameron, A. Rootstock and Clonal Suitability for the Murray Valley Winegrape Region; Murray Valley Wine Growers’ INC: Mildura, VIC, Australia, 2015. [Google Scholar]
- Lowe, K.M.; Walker, M.A. Genetic Linkage Map of the Interspecific Grape Rootstock Cross Ramsey (Vitis champinii) × Riparia Gloire (Vitis riparia). Theor. Appl. Genet. 2006, 112, 1582–1592. [Google Scholar] [CrossRef]
- Hartl, F.U. Molecular Chaperones in Cellular Protein Folding. Nature 1996, 381, 571–579. [Google Scholar] [CrossRef] [PubMed]
- Lyzenga, W.J.; Stone, S.L. Abiotic Stress Tolerance Mediated by Protein Ubiquitination. J. Exp. Bot. 2011, 63, 599–616. [Google Scholar] [CrossRef] [PubMed]
- Sharma, B.; Joshi, D.; Yadav, P.K.; Gupta, A.K.; Bhatt, T.K. Role of Ubiquitin-Mediated Degradation System in Plant Biology. Front. Plant Sci. 2016, 7, 806. [Google Scholar] [CrossRef] [PubMed]
- Koizumi, S.; Irie, T.; Hirayama, S.; Sakurai, Y.; Yashiroda, H.; Naguro, I.; Ichijo, H.; Hamazaki, J.; Murata, S. The Aspartyl Protease DDI2 Activates Nrf1 to Compensate for Proteasome Dysfunction. Elife 2016, 5, e18357. [Google Scholar] [CrossRef] [PubMed]
- Rose, G.D.; Wolfenden, R. Hydrogen Bonding, Hydrophobicity, Packing, and Protein Folding. Annu. Rev. Biophys. Biomol. Struct. 1993, 22, 381–415. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carrasco, D.; Zhou-Tsang, A.; Rodriguez-Izquierdo, A.; Ocete, R.; Revilla, M.A.; Arroyo-García, R. Coastal Wild Grapevine Accession (Vitis vinifera L. ssp. sylvestris) Shows Distinct Late and Early Transcriptome Changes under Salt Stress in Comparison to Commercial Rootstock Richter 110. Plants 2022, 11, 2688. https://doi.org/10.3390/plants11202688
Carrasco D, Zhou-Tsang A, Rodriguez-Izquierdo A, Ocete R, Revilla MA, Arroyo-García R. Coastal Wild Grapevine Accession (Vitis vinifera L. ssp. sylvestris) Shows Distinct Late and Early Transcriptome Changes under Salt Stress in Comparison to Commercial Rootstock Richter 110. Plants. 2022; 11(20):2688. https://doi.org/10.3390/plants11202688
Chicago/Turabian StyleCarrasco, David, Andres Zhou-Tsang, Alberto Rodriguez-Izquierdo, Rafael Ocete, María Angeles Revilla, and Rosa Arroyo-García. 2022. "Coastal Wild Grapevine Accession (Vitis vinifera L. ssp. sylvestris) Shows Distinct Late and Early Transcriptome Changes under Salt Stress in Comparison to Commercial Rootstock Richter 110" Plants 11, no. 20: 2688. https://doi.org/10.3390/plants11202688
APA StyleCarrasco, D., Zhou-Tsang, A., Rodriguez-Izquierdo, A., Ocete, R., Revilla, M. A., & Arroyo-García, R. (2022). Coastal Wild Grapevine Accession (Vitis vinifera L. ssp. sylvestris) Shows Distinct Late and Early Transcriptome Changes under Salt Stress in Comparison to Commercial Rootstock Richter 110. Plants, 11(20), 2688. https://doi.org/10.3390/plants11202688






