Comparative Transcriptomics for Genes Related to Berberine and Berbamine Biosynthesis in Berberidaceae
Abstract
1. Introduction
2. Materials and Methods
2.1. HPLC Analysis
2.2. RNA Extraction, Illumina Sequencing, Data Processing and Assembly
2.3. De Novo Assembly
2.4. DEG
2.5. Functional Annotations and Phylogenetic Analysis
3. Results
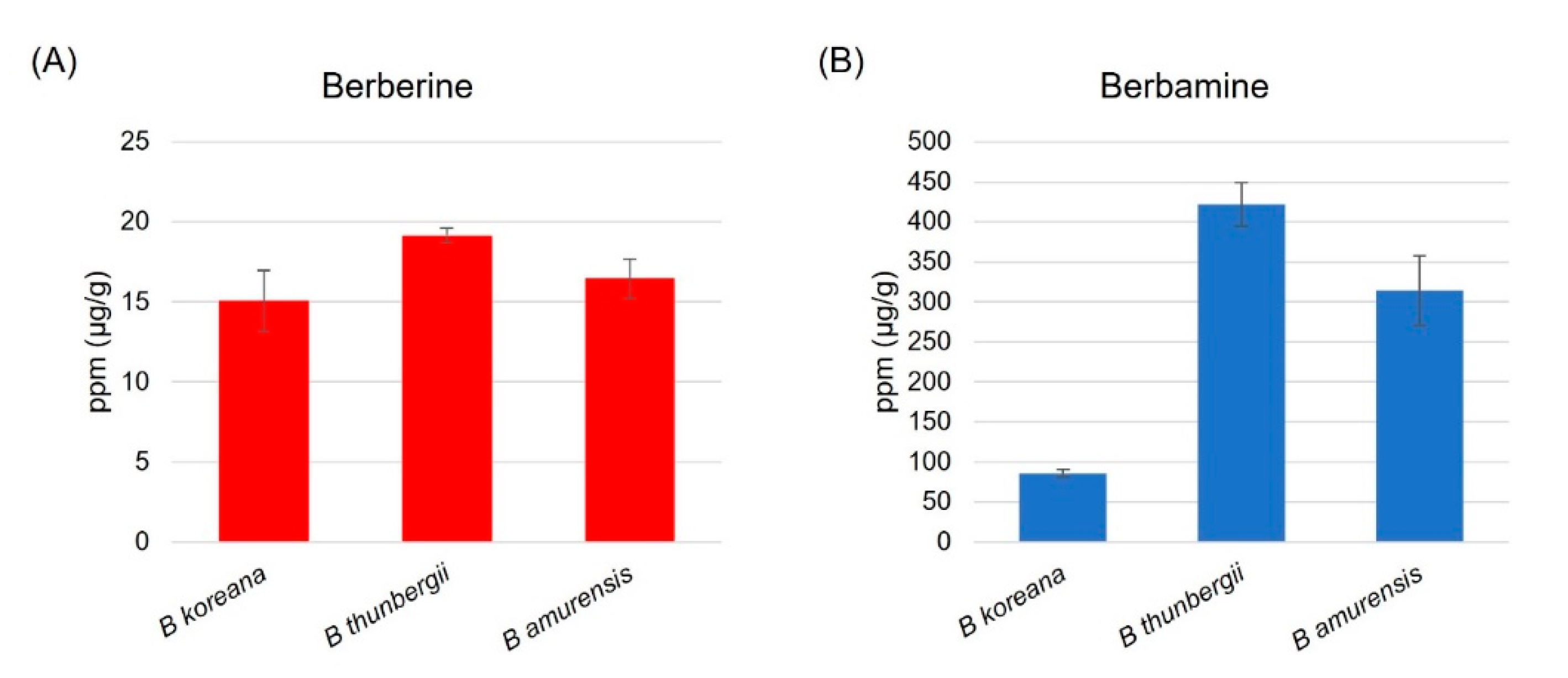
3.1. Contents of Berberine and Berbamine in Leaf Tissues of Berberis Species
3.2. mRNA Sequence Analysis and De Novo Assembly
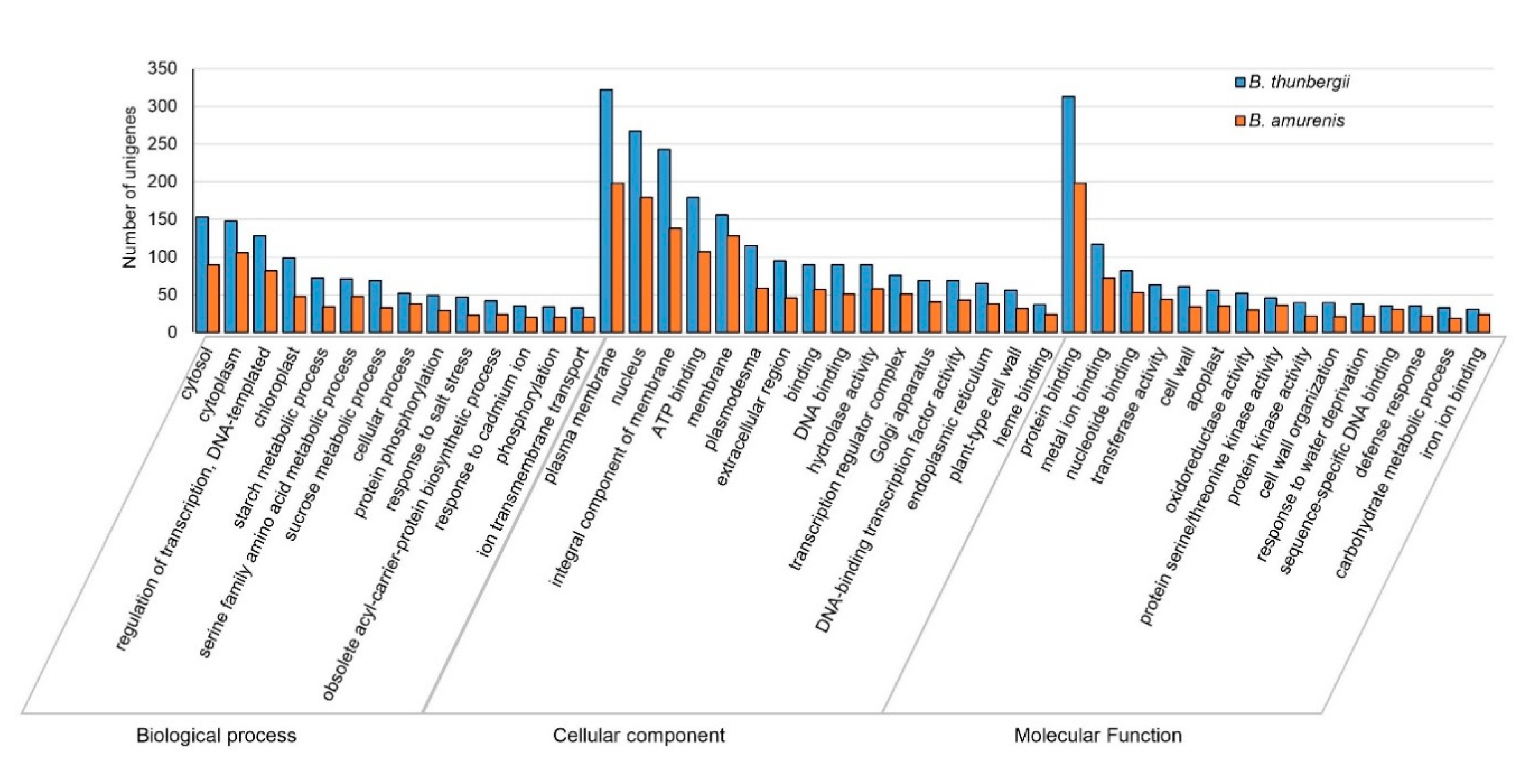
3.3. Functional Annotations and Classifications
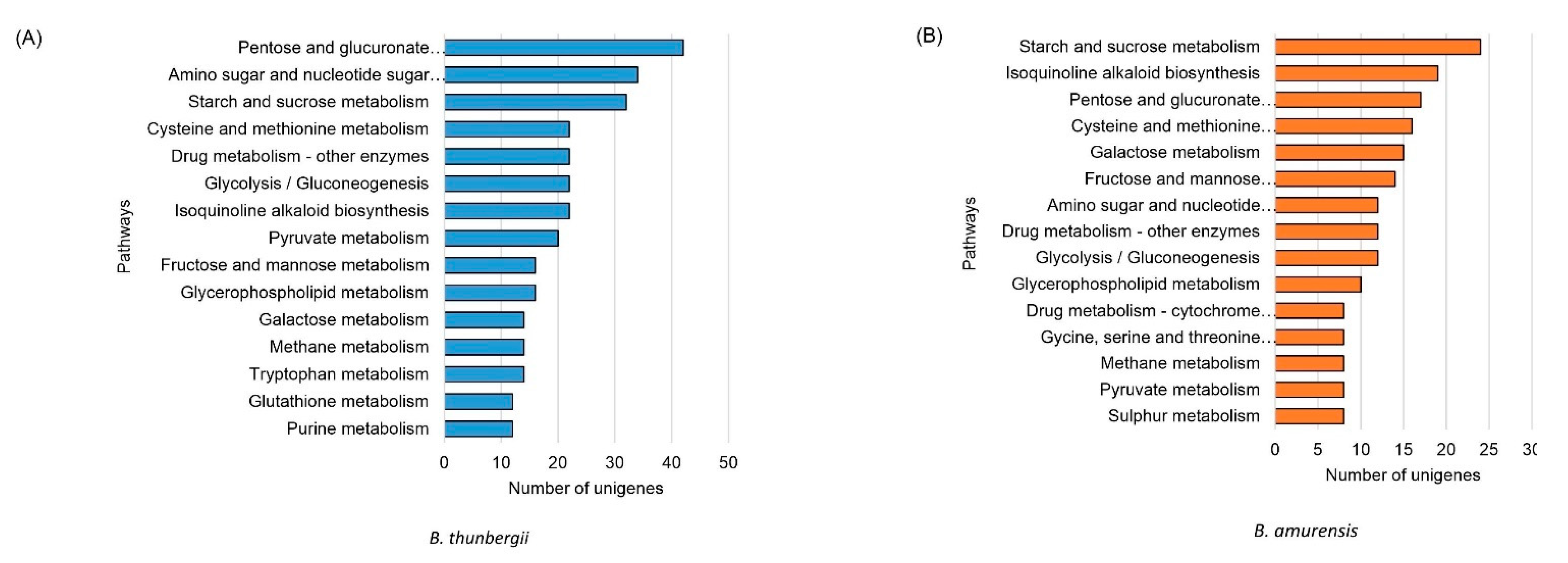
3.4. Differentially Expressed Genes and Classification
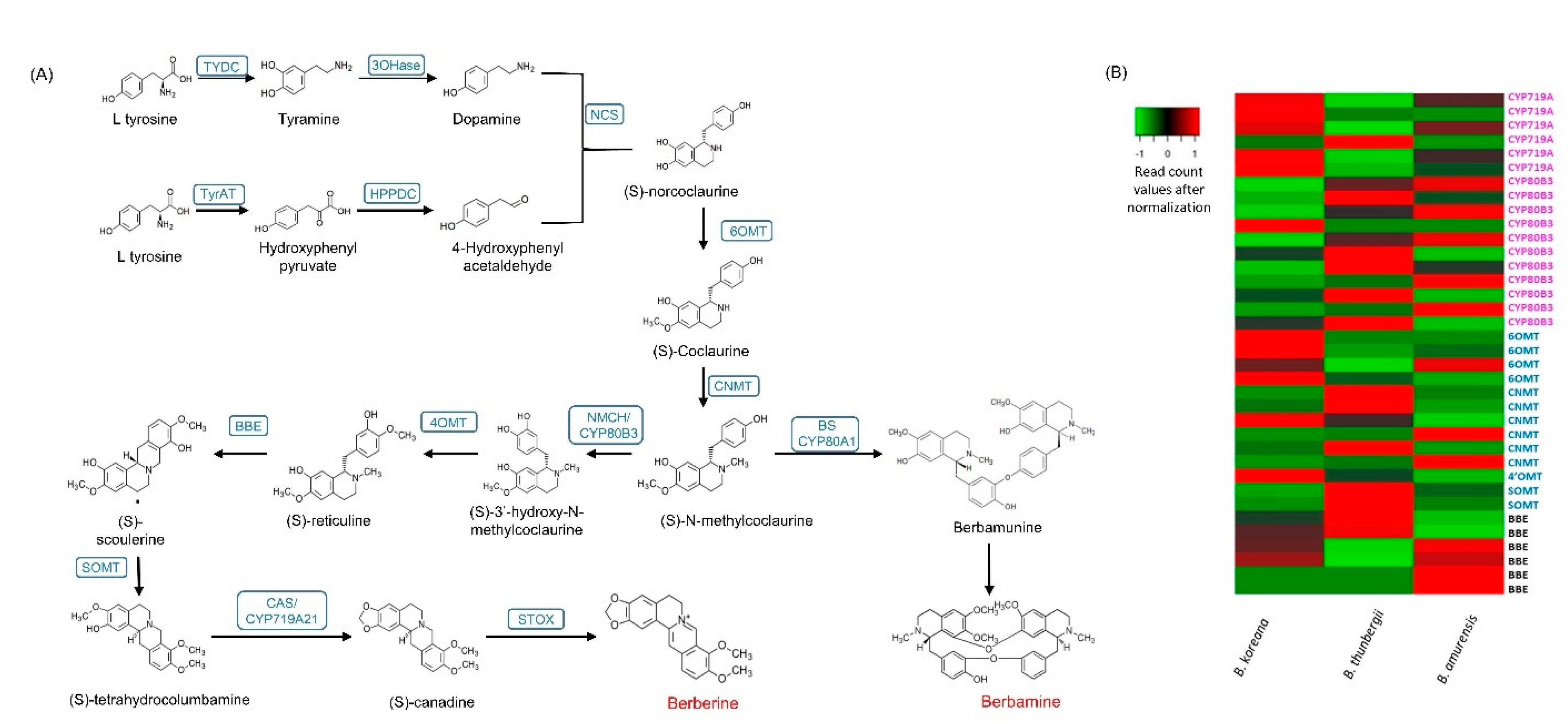
3.5. Genes Encoding Enzymes for BIAs
3.6. CYPs and Methyltransferases among Three Berberis Species
3.7. Phylogenetic Analysis of the Methyltransferases and CYP Oxidases
4. Discussion
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Loconte, H. Berberidaceae. In The Families and Genera of Vascular Plants II. Flowering Plants; Kubitzki, K., Rohwer, J.G., Bittrich, V., Eds.; Springer: Berlin/Heidelberg, Germany; New York, NY, USA, 1993; Volume II, pp. 147–152. [Google Scholar] [CrossRef]
- Christenhusz, M.J.M.; Byng, J.W. The number of known plants species in the world and its annual increase. Phytotaxa 2016, 261, 201–217. [Google Scholar] [CrossRef]
- Bhardwaj, D.; Kaushik, N. Phytochemical and pharmacological studies in genus Berberis. Phytochem. Rev. 2013, 11, 523–542. [Google Scholar] [CrossRef]
- Och, A.; Szewczyk, K.; Pecio, L.; Stochmal, A.; Zaluski, D.; Bogucka-Kocka, A. UPLC-MS/MS Profile of Alkaloids with Cytotoxic Properties of Selected Medicinal Plants of the Berberidaceae and Papaveraceae Families. Oxid. Med. Cell Longev. 2017, 2017, 9369872. [Google Scholar] [CrossRef]
- Rao, R.R.; Husain, T.; Datt, B.; Garg, A. Revision of the Family Berberidaceae of India—I; Rheedea: Calicut, India, 1998; Volume 1, pp. 1–66. [Google Scholar]
- Rashmi; Rajasekaran, A. The Genus Berberis Linn.: A Review. Parmacogn. Rev. 2008, 2, 369. [Google Scholar]
- Neag, M.A.; Mocan, A.; Echeverria, J.; Pop, R.M.; Bocsan, C.I.; Crisan, G.; Buzoianu, A.D. Berberine: Botanical Occurrence, Traditional Uses, Extraction Methods, and Relevance in Cardiovascular, Metabolic, Hepatic, and Renal Disorders. Front. Pharmacol. 2018, 9, 557. [Google Scholar] [CrossRef]
- Liscombe, D.K.; Macleod, B.P.; Loukanina, N.; Nandi, O.I.; Facchini, P.J. Evidence for the monophyletic evolution of benzylisoquinoline alkaloid biosynthesis in angiosperms. Phytochemistry 2005, 66, 1374–1393. [Google Scholar] [CrossRef]
- Deng, X.; Zhao, L.; Fang, T.; Xiong, Y.; Ogutu, C.; Yang, D.; Vimolmangkang, S.; Liu, Y.; Han, Y. Investigation of benzylisoquinoline alkaloid biosynthetic pathway and its transcriptional regulation in lotus. Hortic. Res. 2018, 5, 29. [Google Scholar] [CrossRef]
- Farrow, S.C.; Hagel, J.M.; Facchini, P.J. Transcript and metabolite profiling in cell cultures of 18 plant species that produce benzylisoquinoline alkaloids. Phytochemistry 2012, 77, 79–88. [Google Scholar] [CrossRef]
- Hagel, J.M.; Morris, J.S.; Lee, E.J.; Desgagne-Penix, I.; Bross, C.D.; Chang, L.; Chen, X.; Farrow, S.C.; Zhang, Y.; Soh, J.; et al. Transcriptome analysis of 20 taxonomically related benzylisoquinoline alkaloid-producing plants. BMC Plant Biol. 2015, 15, 227. [Google Scholar] [CrossRef]
- Mokhber-Dezfuli, N.; Saeidnia, S.; Gohari, A.R.; Kurepaz-Mahmoodabadi, M. Phytochemistry and pharmacology of berberis species. Pharmacogn. Rev. 2014, 8, 8–15. [Google Scholar] [CrossRef]
- Chen, W.H.; Pang, J.Y.; Qin, Y.; Peng, Q.; Cai, Z.; Jiang, Z.H. Synthesis of linked berberine dimers and their remarkably enhanced DNA-binding affinities. Bioorg. Med. Chem. Lett. 2005, 15, 2689–2692. [Google Scholar] [CrossRef]
- Srivastava, S.; Srivastava, M.; Misra, A.; Pandey, G.; Rawat, A. A review on biological and chemical diversity in Berberis (Berberidaceae). EXCLI J. 2015, 14, 247–267. [Google Scholar] [CrossRef]
- Sun, Y.; Yao, T.; Li, H.; Peng, Y.; Zheng, J. In vitro and in vivo metabolic activation of berbamine to quinone methide intermediate. J. Biochem. Mol. Toxicol. 2017, 31, e21876. [Google Scholar] [CrossRef] [PubMed]
- Hagel, J.M.; Facchini, P.J. Benzylisoquinoline alkaloid metabolism: A century of discovery and a brave new world. Plant Cell Physiol. 2013, 54, 647–672. [Google Scholar] [CrossRef]
- Meng, Z.; Li, T.; Ma, X.; Wang, X.; Van Ness, C.; Gan, Y.; Zhou, H.; Tang, J.; Lou, G.; Wang, Y.; et al. Berbamine inhibits the growth of liver cancer cells and cancer-initiating cells by targeting Ca(2)(+)/calmodulin-dependent protein kinase II. Mol. Cancer Ther. 2013, 12, 2067–2077. [Google Scholar] [CrossRef]
- Zhao, W.; Bai, B.; Hong, Z.; Zhang, X.; Zhou, B. Berbamine (BBM), a Natural STAT3 Inhibitor, Synergistically Enhances the Antigrowth and Proapoptotic Effects of Sorafenib on Hepatocellular Carcinoma Cells. ACS Omega 2020, 5, 24838–24847. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.B.; Liu, Y.S.; Huang, P.; Ma, Y.S.; Qing, Z.X.; Tang, Q.; Cao, H.F.; Cheng, P.; Zheng, Y.J.; Yuan, Z.J.; et al. The Genome of Medicinal Plant Macleaya cordata Provides New Insights into Benzylisoquinoline Alkaloids Metabolism. Mol. Plant 2017, 10, 975–989. [Google Scholar] [CrossRef]
- He, S.M.; Liang, Y.L.; Cong, K.; Chen, G.; Zhao, X.; Zhao, Q.M.; Zhang, J.J.; Wang, X.; Dong, Y.; Yang, J.L.; et al. Identification and Characterization of Genes Involved in Benzylisoquinoline Alkaloid Biosynthesis in Coptis Species. Front. Plant Sci. 2018, 9, 731. [Google Scholar] [CrossRef]
- Zhong, F.; Huang, L.; Qi, L.; Ma, Y.; Yan, Z. Full-length transcriptome analysis of Coptis deltoidea and identification of putative genes involved in benzylisoquinoline alkaloids biosynthesis based on combined sequencing platforms. Plant Mol. Biol. 2020, 102, 477–499. [Google Scholar] [CrossRef]
- Sharma, B.R.; Gautam, L.N.; Adhikari, D.; Karki, R. A Comprehensive Review on Chemical Profiling of Nelumbo nucifera: Potential for Drug Development. Phytother. Res. 2017, 31, 3–26. [Google Scholar] [CrossRef]
- Stadler, R.; Zenk, M.H. The purification and characterization of a unique cytochrome P-450 enzyme from Berberis stolonifera plant cell cultures. J. Biol. Chem. 1993, 268, 823–831. [Google Scholar] [CrossRef]
- Morishige, T.; Tsujita, T.; Yamada, Y.; Sato, F. Molecular characterization of the S-adenosyl-L-methionine:3′-hydroxy-N-methylcoclaurine 4′-O-methyltransferase involved in isoquinoline alkaloid biosynthesis in Coptis japonica. J. Biol. Chem. 2000, 275, 23398–23405. [Google Scholar] [CrossRef] [PubMed]
- Qadir, S.A.; Kwon, M.C.; Han, J.G.; Ha, J.H.; Chung, H.S.; Ahn, J.; Lee, H.Y. Effect of different extraction protocols on anticancer and antioxidant activities of Berberis koreana bark extracts. J. Biosci. Bioeng. 2009, 107, 331–338. [Google Scholar] [CrossRef] [PubMed]
- Yoo, K.Y.; Hwang, I.K.; Lim, B.O.; Kang, T.C.; Kim, D.W.; Kim, S.M.; Lee, H.Y.; Kim, J.D.; Won, M.H. Berberry extract reduces neuronal damage and N-Methyl-D-aspartate receptor 1 immunoreactivity in the gerbil hippocampus after transient forebrain ischemia. Biol. Pharm. Bull. 2006, 29, 623–628. [Google Scholar] [CrossRef]
- Kim, K.H.; Choi, S.U.; Ha, S.K.; Kim, S.Y.; Lee, K.R. Biphenyls from Berberis koreana. J. Nat. Prod. 2009, 72, 2061–2064. [Google Scholar] [CrossRef]
- Amann, M.; Nagakura, N.; Zenk, M.H. Purification and properties of (S)-tetrahydroprotoberberine oxidase from suspension-cultured cells of Berberis wilsoniae. Eur. J. Biochem. 1988, 175, 17–25. [Google Scholar] [CrossRef]
- Gesell, A.; Chavez, M.L.; Kramell, R.; Piotrowski, M.; Macheroux, P.; Kutchan, T.M. Heterologous expression of two FAD-dependent oxidases with (S)-tetrahydroprotoberberine oxidase activity from Arge mone mexicana and Berberis wilsoniae in insect cells. Planta 2011, 233, 1185–1197. [Google Scholar] [CrossRef]
- Roy, N.S.; Choi, I.Y.; Um, T.; Jeon, M.J.; Kim, B.Y.; Kim, Y.D.; Yu, J.K.; Kim, S.; Kim, N.S. Gene Expression and Isoform Identification of PacBio Full-Length cDNA Sequences for Berberine Biosynthesis in Berberis koreana. Plants 2021, 10, 1314. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Cabau, C.; Escudie, F.; Djari, A.; Guiguen, Y.; Bobe, J.; Klopp, C. Compacting and correcting Trinity and Oases RNA-Seq de novo assemblies. PeerJ 2017, 5, e2988. [Google Scholar] [CrossRef]
- Schulz, M.H.; Zerbino, D.R.; Vingron, M.; Birney, E. Oases: Robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics 2012, 28, 1086–1092. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Jeong, S.W.; Lee, W.S.; Park, S.; Kim, Y.H.; Kim, G.S.; Lee, S.J.; Jin, J.S.; Kim, C.Y.; Lee, J.E.; et al. Determination of Polyphenol Components of Korean Prostrate Spurge (Euphorbia supina) by Using Liquid Chromatography-Tandem Mass Spectrometry: Overall Contribution to Antioxidant Activity. J. Anal. Methods Chem. 2014, 2014, 418690. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef] [PubMed]
- Ban, Y.W.; Roy, N.S.; Yang, H.; Choi, H.K.; Kim, J.H.; Babu, P.; Ha, K.S.; Ham, J.K.; Park, K.C.; Choi, I.Y. Comparative transcriptome analysis reveals higher expression of stress and defense responsive genes in dwarf soybeans obtained from the crossing of G. max and G. soja. Genes Genom. 2019, 41, 1315–1327. [Google Scholar] [CrossRef]
- Kovaka, S.; Zimin, A.V.; Pertea, G.M.; Razaghi, R.; Salzberg, S.L.; Pertea, M. Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol. 2019, 20, 278. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Nguyen, V.K.; Kou, K.G. The biology and total syntheses of bisbenzylisoquinoline alkaloids. Org. Biomol. Chem. 2021, 19, 7535–7543. [Google Scholar] [CrossRef] [PubMed]
- Figueroa-Romero, C.; Hur, J.; Bender, D.E.; Delaney, C.E.; Cataldo, M.D.; Smith, A.L.; Yung, R.; Ruden, D.M.; Callaghan, B.C.; Feldman, E.L. Identification of epigenetically altered genes in sporadic amyotrophic lateral sclerosis. PLoS ONE 2012, 7, e52672. [Google Scholar] [CrossRef] [PubMed]
- Schuler, M.A.; Werck-Reichhart, D. Functional genomics of P450s. Annu. Rev. Plant Biol. 2003, 54, 629–667. [Google Scholar] [CrossRef] [PubMed]
- Menendez-Perdomo, I.M.; Facchini, P.J. Benzylisoquinoline Alkaloids Biosynthesis in Sacred Lotus. Molecules 2018, 23, 2899. [Google Scholar] [CrossRef]
- Weber, C.; Opatz, T. Bisbenzylisoquinoline alkaloids. Alkaloids Chem. Biol. 2019, 81, 1–114. [Google Scholar]
- Kraus, P.F.; Kutchan, T.M. Molecular cloning and heterologous expression of a cDNA encoding berbamunine synthase, a C—O phenol-coupling cytochrome P450 from the higher plant Berberis stolonifera. Proc. Natl. Acad. Sci. USA 1995, 92, 2071–2075. [Google Scholar] [CrossRef]
- Belwal, T.; Bisht, A.; Devkota, H.P.; Ullah, H.; Khan, H.; Pandey, A.; Bhatt, I.D.; Echeverria, J. Phytopharmacology and Clinical Updates of Berberis Species Against Diabetes and Other Metabolic Diseases. Front. Pharmacol. 2020, 11, 41. [Google Scholar] [CrossRef]
- Nelson, D.R.; Schuler, M.A. Cytochrome P450 Genes from the Sacred Lotus Genome. Trop. Plant Biol. 2013, 6, 138–151. [Google Scholar] [CrossRef]

| Raw Data | Filtered Data | |||||
|---|---|---|---|---|---|---|
| Species | Reads | Total Length | >Q20 (%) | Reads | Total Length | Rate (%) |
| B. koreana | 35,998,374 | 5,435,754,474 | 97.41 | 33,381,816 | 4,733,258,684 | 92.7 |
| B. amurensis | 50,706,530 | 7,656,686,030 | 96.75 | 46,670,652 | 6,449,979,707 | 92 |
| B. thunbergii | 80,966,878 | 12,225,998,578 | 96.92 | 74,940,106 | 10,354,513,883 | 92.6 |
| Oases (v0.2.09) | CD-Hit | Trans Decoder | Bwa | ||
|---|---|---|---|---|---|
| File | Oases | all_dgb | all_contigs | transdecoder.cds | coding_transcripts_fpkm |
| Total contigs | 1,221,878 | 553,383 | 285,922 | 65,268 | 42,278 |
| Total length | 450,175,132 | 251,071,869 | 186,123,210 | 34,735,974 | 44,989,957 |
| Min. length | 100 | 50 | 200 | 297 | 244 |
| Avg. length | 368.4 | 453.7 | 651 | 532.2 | 1064.1 |
| Max. length | 4395 | 4395 | 5356 | 3627 | 5354 |
| Name | Total Reads | Mapped Reads | Mapped Rate (%) | Prop Paired Read | Prop Paired Mapped Rate (%) |
|---|---|---|---|---|---|
| B. koreana | 33,381,816 | 18,809,373 | 56.3 | 16,617,264 | 49.8 |
| B. amurensis | 30,875,256 | 15,672,027 | 50.8 | 13,667,334 | 44.3 |
| B. thunbergii | 74,940,106 | 33,975,391 | 45.3 | 29,602,862 | 39.5 |
| Control | Target | Number of Transcripts Compared | p-Value < 0.05 & log2FC abs1 | ||
|---|---|---|---|---|---|
| All | Upregulated | Downregulated | |||
| B. koreana | B. amurensis | 39,521 | 3014 | 486 | 2528 |
| B. koreana | B. thunbergii | 40,036 | 4494 | 847 | 3334 |
| Enzyme | Abbreviations | B. koreana * | B. thunbergii * | B. amurensis * |
|---|---|---|---|---|
| Tyrosine decarboxylase | TYDC | 425.10 (8) ** | 158.35 | 217.39 |
| Tyrosine aminotransferase | TyrAT | 1068.84 (4) | 2015.10 | 1719.99 |
| (S)-norcoclaurine synthase | NCS | 1880.62 (8) | 514.78 | 1341.78 |
| (RS)-norcoclaurine 6-O-methyltransferase | 6OMT | 20.02 (2) | 7.29 | 54.43 |
| (S)-coclaurine N-methyltransferase | CNMT | 268.97 (4) | 226.20 | 109.22 |
| (S)-N-methylcoclaurine 3′-hydroxylase/N-methylcoclaurine 3′-monooxygenase | NMCH/CYP80B3 | 1485.02 (6) | 3959.62 | 4927.05 |
| 3′-hydroxy-N-methylcoclaurine 4′-O-methyltransferase | 4′OMT | 18.13 (2) | 43.63 | 82.71 |
| Berberine bridge enzyme (reticuline oxidase) | BBE | 40.32 (1) | 13.67 | 26.16 |
| (S)-scoulerine 9-O-methyltransferase | SOMT | 51.68 (1) | 123.71 | 42.06 |
| Canadine synthase enzyme | CYP719A21 | 851.83 (6) | 343.40 | 589.24 |
| Berbamunine synthase *** | CYP80A1 | - (1) | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roy, N.S.; Park, N.-I.; Kim, N.-S.; Park, Y.; Kim, B.-Y.; Kim, Y.-D.; Yu, J.-K.; Kim, Y.-I.; Um, T.; Kim, S.; et al. Comparative Transcriptomics for Genes Related to Berberine and Berbamine Biosynthesis in Berberidaceae. Plants 2022, 11, 2676. https://doi.org/10.3390/plants11202676
Roy NS, Park N-I, Kim N-S, Park Y, Kim B-Y, Kim Y-D, Yu J-K, Kim Y-I, Um T, Kim S, et al. Comparative Transcriptomics for Genes Related to Berberine and Berbamine Biosynthesis in Berberidaceae. Plants. 2022; 11(20):2676. https://doi.org/10.3390/plants11202676
Chicago/Turabian StyleRoy, Neha Samir, Nam-Il Park, Nam-Soo Kim, Yeri Park, Bo-Yun Kim, Young-Dong Kim, Ju-Kyung Yu, Yong-In Kim, Taeyoung Um, Soonok Kim, and et al. 2022. "Comparative Transcriptomics for Genes Related to Berberine and Berbamine Biosynthesis in Berberidaceae" Plants 11, no. 20: 2676. https://doi.org/10.3390/plants11202676
APA StyleRoy, N. S., Park, N.-I., Kim, N.-S., Park, Y., Kim, B.-Y., Kim, Y.-D., Yu, J.-K., Kim, Y.-I., Um, T., Kim, S., & Choi, I.-Y. (2022). Comparative Transcriptomics for Genes Related to Berberine and Berbamine Biosynthesis in Berberidaceae. Plants, 11(20), 2676. https://doi.org/10.3390/plants11202676










