Genome-Wide Identification of R2R3-MYB Transcription Factor and Expression Analysis under Abiotic Stress in Rice
Abstract
:1. Introduction
2. Results
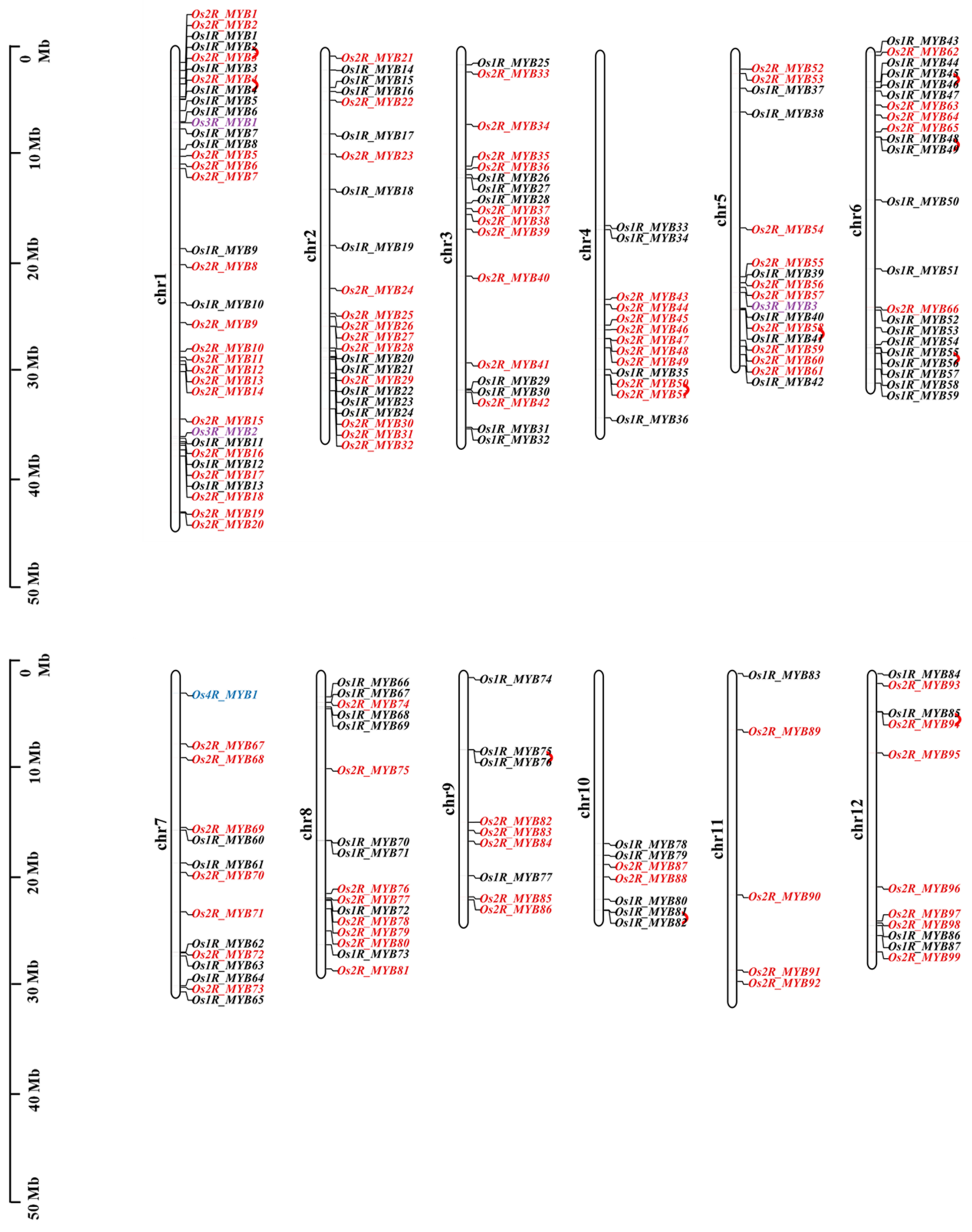
2.1. Genome-Wide Identification and Chromosomal Locations of OsMYBs
2.2. Phylogenetic Analysis of OsMYBs
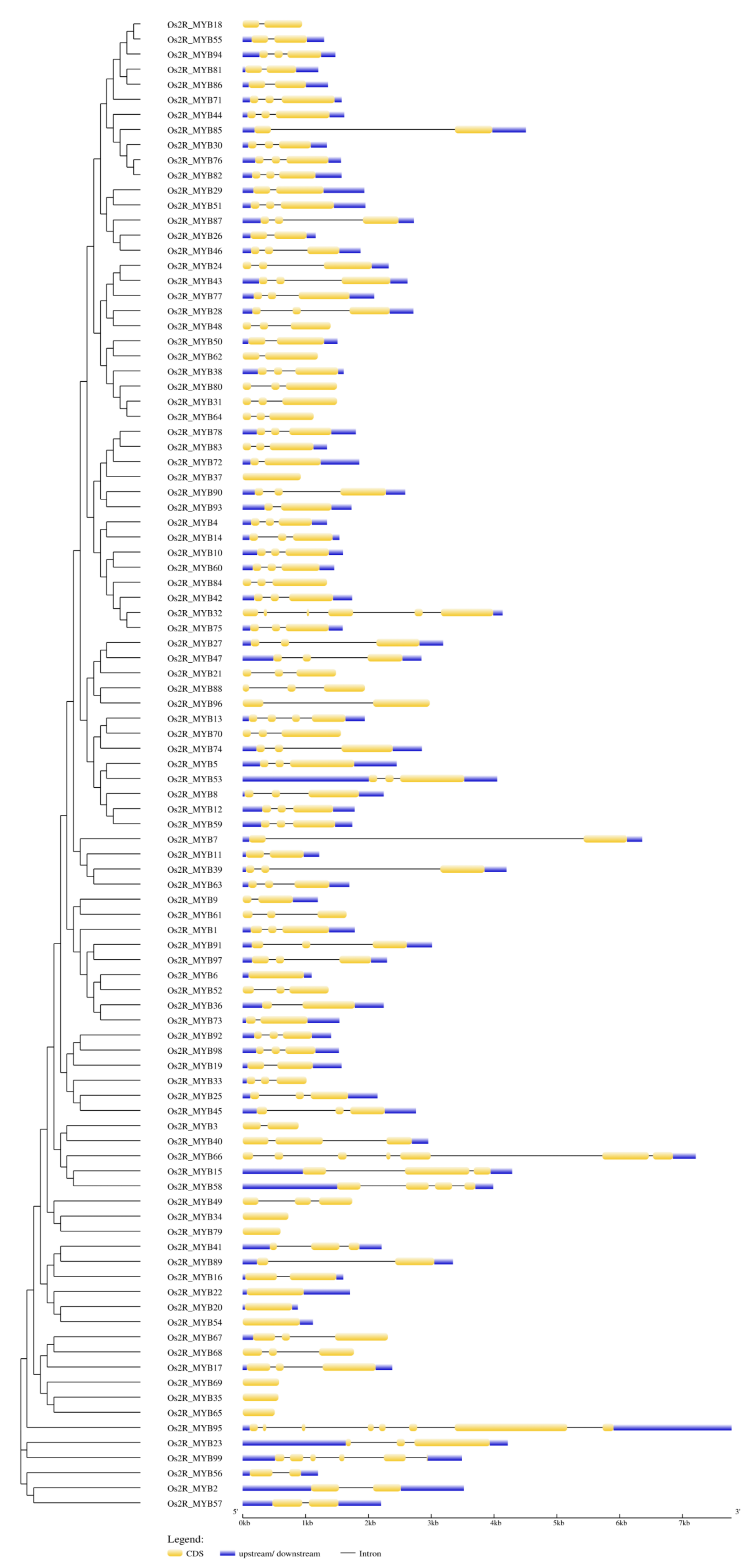
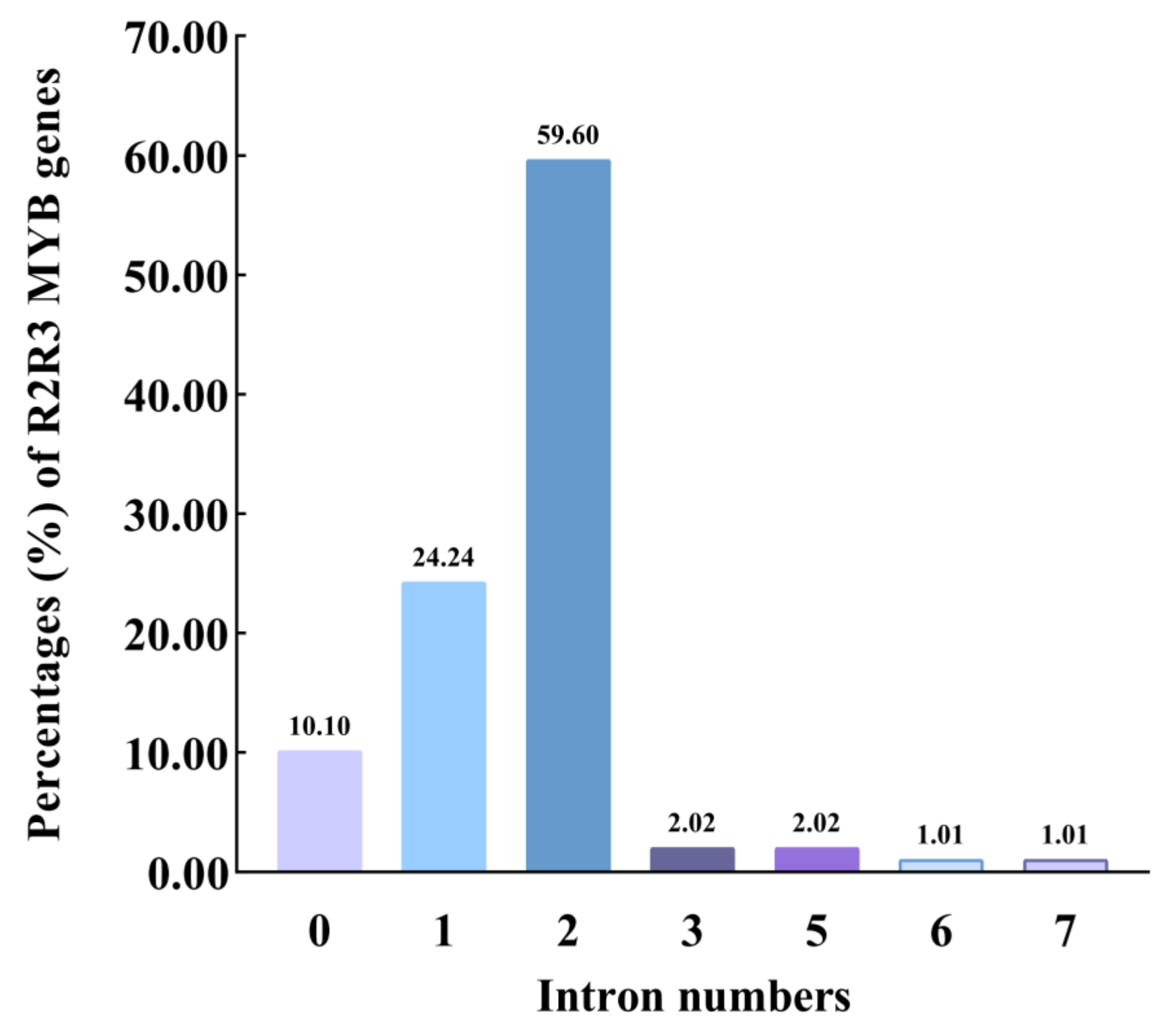
2.3. Structure of Os2R_MYBs Genes
2.4. Analysis of Promoter Cis-Elements
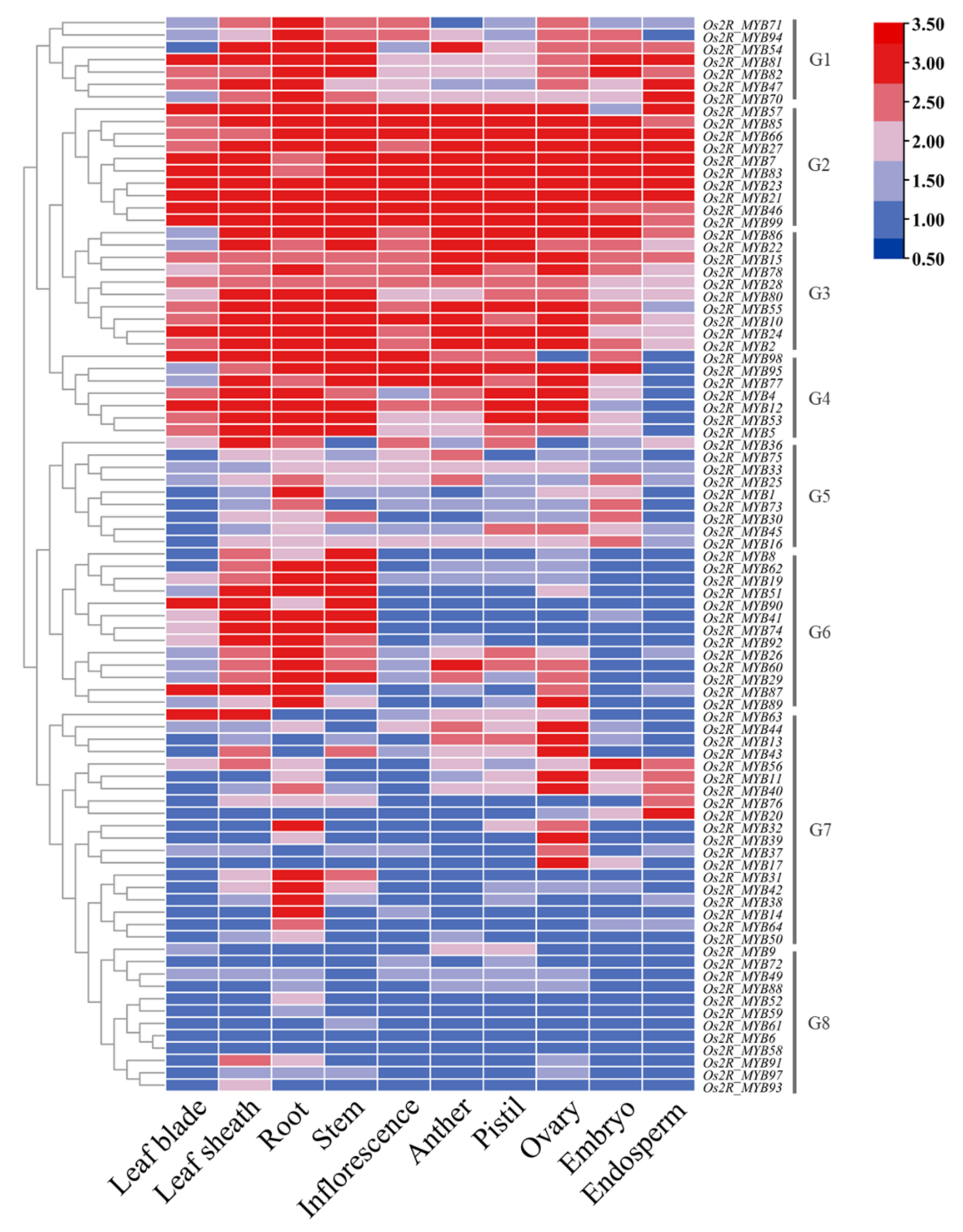
2.5. The Relative Expression Level of Os2R_MYBs in Various Tissues and Growth Stages
2.6. Protein–Protein Interaction Network Analysis
2.7. Gene Expression Levels of Os2R_MYBs under Abiotic Stresses
3. Discussion
4. Materials and Methods
4.1. Identification of MYB Family Members in Rice and Their Chromosomal Locations
4.2. Classification and Phylogenetic Analysis of MYB Family Members
4.3. Structural Analysis of Os2R_MYB Genes
4.4. Analysis of Stress-Related Cis-Elements in Os2R_MYB Promoter Regions
4.5. Analysis of the Relative Expression Level of Os2R_MYBs in Different Tissues and Growth Stages
4.6. Abiotic Stress Treatment of Rice
4.7. Protein–Protein Interaction Network Analysis
4.8. Analysis of Os2R_MYBs Gene Expression in Rice under Abiotic Stress
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Riechmann, J.L.; Heard, J.; Martin, G.; Reuber, L.; Jiang, C.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.J.; Samaha, R.R.; et al. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef] [PubMed]
- Kanei-Ishii, C.; Sarai, A.; Sawazaki, T.; Nakagoshi, H.; He, D.N.; Ogata, K.; Nishimura, Y.; Ishii, S. The tryptophan cluster: A hypothetical structure of the DNA-binding domain of the myb protooncogene product. J. Biol. Chem. 1990, 265, 19990–19995. [Google Scholar] [CrossRef]
- Jia, L.; Clegg, M.T.; Jiang, T. Evolutionary dynamics of the DNA-binding domains in putative R2R3-MYB genes identified from rice subspecies indica and japonica genomes. Plant Physiol. 2004, 134, 575–585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef]
- Martin, C.; Paz-Ares, J. MYB transcription factors in plants. Trends Genet. 1997, 13, 67–73. [Google Scholar] [CrossRef]
- Paz-Ares, J.; Ghosal, D.; Wienand, U.; Peterson, P.A.; Saedler, H. The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. EMBO J. 1987, 6, 3553–3558. [Google Scholar] [CrossRef]
- Tamagnone, L.; Merida, A.; Parr, A.; Mackay, S.; Culianez-Macia, F.A.; Roberts, K.; Martin, C. The AmMYB308 and AmMYB330 transcription factors from antirrhinum regulate phenylpropanoid and lignin biosynthesis in transgenic tobacco. Plant Cell 1998, 10, 135–154. [Google Scholar] [CrossRef] [Green Version]
- Pandey, A.; Misra, P.; Khan, M.P.; Swarnkar, G.; Tewari, M.C.; Bhambhani, S.; Trivedi, R.; Chattopadhyay, N.; Trivedi, P.K. Co-expression of Arabidopsis transcription factor, AtMYB12, and soybean isoflavone synthase, GmIFS1, genes in tobacco leads to enhanced biosynthesis of isoflavones and flavonols resulting in osteoprotective activity. Plant Biotechnol. J. 2014, 12, 69–80. [Google Scholar] [CrossRef]
- Castillejo, C.; Waurich, V.; Wagner, H.; Ramos, R.; Oiza, N.; Muñoz, P.; Triviño, J.C.; Caruana, J.; Liu, Z.; Cobo, N.; et al. Allelic Variation of MYB10 Is the Major Force Controlling Natural Variation in Skin and Flesh Color in Strawberry (Fragaria spp.) Fruit. Plant Cell 2020, 32, 3723–3749. [Google Scholar] [CrossRef]
- Wang, L.; Lu, W.; Ran, L.; Dou, L.; Yao, S.; Hu, J.; Fan, D.; Li, C.; Luo, K. R2R3-MYB transcription factor MYB6 promotes anthocyanin and proanthocyanidin biosynthesis but inhibits secondary cell wall formation in Populus tomentosa. Plant J. 2019, 99, 733–751. [Google Scholar] [CrossRef]
- Dai, X.; Xu, Y.; Ma, Q.; Xu, W.; Wang, T.; Xue, Y.; Chong, K. Overexpression of an R1R2R3 MYB gene, OsMYB3R-2, increases tolerance to freezing, drought, and salt stress in transgenic Arabidopsis. Plant Physiol. 2007, 143, 1739–1751. [Google Scholar] [CrossRef] [Green Version]
- Xing, C.; Liu, Y.; Zhao, L.; Zhang, S.; Huang, X. A novel MYB transcription factor regulates ascorbic acid synthesis and affects cold tolerance. Plant Cell Environ. 2019, 42, 832–845. [Google Scholar] [CrossRef]
- Agarwal, P.; Mitra, M.; Banerjee, S.; Roy, S. MYB4 transcription factor, a member of R2R3-subfamily of MYB domain protein, regulates cadmium tolerance via enhanced protection against oxidative damage and increases expression of PCS1 and MT1C in Arabidopsis. Plant Sci. 2020, 297, 110501. [Google Scholar] [CrossRef]
- Xiang, X.J.; Sun, L.P.; Yu, P.; Yang, Z.F.; Zhang, P.P.; Zhang, Y.X.; Wu, W.X.; Chen, D.B.; Zhan, X.D.; Khan, R.M.; et al. The MYB transcription factor Baymax1 plays a critical role in rice male fertility. Theor. Appl. Genet. 2021, 134, 453–471. [Google Scholar] [CrossRef]
- Wang, D.; Li, J.; Sun, L.; Hu, Y.; Yu, J.; Wang, C.; Zhang, F.; Hou, H.; Liang, W.; Zhang, D. Two rice MYB transcription factors maintain male fertility in response to photoperiod by modulating sugar partitioning. New Phytol. 2021, 231, 1612–1629. [Google Scholar] [CrossRef]
- Ren, D.; Cui, Y.; Hu, H.; Xu, Q.; Rao, Y.; Yu, X.; Zhang, Y.; Wang, Y.; Peng, Y.; Zeng, D.; et al. AH2 encodes a MYB domain protein that determines hull fate and affects grain yield and quality in rice. Plant J. 2019, 100, 813–824. [Google Scholar] [CrossRef]
- Li, Y.F.; Zeng, X.Q.; Li, Y.; Wang, L.; Zhuang, H.; Wang, Y.; Tang, J.; Wang, H.L.; Xiong, M.; Yang, F.Y.; et al. MULTI-FLORET SPIKELET 2, a MYB Transcription Factor, Determines Spikelet Meristem Fate and Floral Organ Identity in Rice. Plant Physiol. 2020, 184, 988–1003. [Google Scholar] [CrossRef]
- Zhu, N.; Cheng, S.; Liu, X.; Du, H.; Dai, M.; Zhou, D.X.; Yang, W.; Zhao, Y. The R2R3-type MYB gene OsMYB91 has a function in coordinating plant growth and salt stress tolerance in rice. Plant Sci. 2015, 236, 146–156. [Google Scholar] [CrossRef]
- Yin, X.; Cui, Y.; Wang, M.; Xia, X. Overexpression of a novel MYB-related transcription factor, OsMYBR1, confers improved drought tolerance and decreased ABA sensitivity in rice. Biochem. Biophys. Res. Commun. 2017, 490, 1355–1361. [Google Scholar] [CrossRef]
- Su, C.F.; Wang, Y.C.; Hsieh, T.H.; Lu, C.A.; Tseng, T.H.; Yu, S.M. A novel MYBS3-dependent pathway confers cold tolerance in rice. Plant Physiol. 2010, 153, 145–158. [Google Scholar] [CrossRef] [Green Version]
- Adil, M.F.; Sehar, S.; Chen, S.; Lwalaba, J.L.W.; Jilani, G.; Chen, Z.H.; Shamsi, I.H. Stress signaling convergence and nutrient crosstalk determine zinc-mediated amelioration against cadmium toxicity in rice. Ecotoxicol. Environ. Saf. 2021, 230, 113128. [Google Scholar] [CrossRef]
- Smita, S.; Katiyar, A.; Chinnusamy, V.; Pandey, D.M.; Bansal, K.C. Transcriptional Regulatory Network Analysis of MYB Transcription Factor Family Genes in Rice. Front. Plant Sci. 2015, 6, 1157. [Google Scholar] [CrossRef] [Green Version]
- Chaney, R.L.; Reeves, P.G.; Ryan, J.A.; Simmons, R.W.; Welch, R.M.; Angle, J.S. An improved understanding of soil Cd risk to humans and low cost methods to phytoextract Cd from contaminated soils to prevent soil Cd risks. BioMetals 2004, 17, 549–553. [Google Scholar] [CrossRef]
- Feng, Q.; Zhang, Y.; Hao, P.; Wang, S.; Fu, G.; Huang, Y.; Li, Y.; Zhu, J.; Liu, Y.; Hu, X.; et al. Sequence and analysis of rice chromosome 4. Nature 2002, 420, 316–320. [Google Scholar] [CrossRef]
- Yu, J.; Hu, S.; Wang, J.; Wong, G.K.; Li, S.; Liu, B.; Deng, Y.; Dai, L.; Zhou, Y.; Zhang, X.; et al. A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 2002, 296, 79–92. [Google Scholar] [CrossRef]
- Katiyar, A.; Smita, S.; Lenka, S.K.; Rajwanshi, R.; Chinnusamy, V.; Bansal, K.C. Genome-wide classification and expression analysis of MYB transcription factor families in rice and Arabidopsis. BMC Genom. 2012, 13, 544. [Google Scholar] [CrossRef] [Green Version]
- Ogata, K.; Kanei-Ishii, C.; Sasaki, M.; Hatanaka, H.; Nagadoi, A.; Enari, M.; Nakamura, H.; Nishimura, Y.; Ishii, S.; Sarai, A. The cavity in the hydrophobic core of Myb DNA-binding domain is reserved for DNA recognition and trans-activation. Nat. Struct. Biol. 1996, 3, 178–187. [Google Scholar] [CrossRef]
- Wakasa, Y.; Yasuda, H.; Oono, Y.; Kawakatsu, T.; Hirose, S.; Takahashi, H.; Hayashi, S.; Yang, L.; Takaiwa, F. Expression of ER quality control-related genes in response to changes in BiP1 levels in developing rice endosperm. Plant J. 2011, 65, 675–689. [Google Scholar] [CrossRef]
- Gu, M.; Zhang, J.; Li, H.; Meng, D.; Li, R.; Dai, X.; Wang, S.; Liu, W.; Qu, H.; Xu, G. Maintenance of phosphate homeostasis and root development are coordinately regulated by MYB1, an R2R3-type MYB transcription factor in rice. J. Exp. Bot. 2017, 68, 3603–3615. [Google Scholar] [CrossRef] [Green Version]
- Yang, W.T.; Baek, D.; Yun, D.J.; Hwang, W.H.; Park, D.S.; Nam, M.H.; Chung, E.S.; Chung, Y.S.; Yi, Y.B.; Kim, D.H. Overexpression of OsMYB4P, an R2R3-type MYB transcriptional activator, increases phosphate acquisition in rice. Plant Physiol. Biochem. 2014, 80, 259–267. [Google Scholar] [CrossRef]
- Yang, A.; Dai, X.; Zhang, W.H. A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J. Exp. Bot. 2012, 63, 2541–2556. [Google Scholar] [CrossRef] [PubMed]
- Feller, A.; Machemer, K.; Braun, E.L.; Grotewold, E. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 2011, 66, 94–116. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Li, E.; Porth, I.; Chen, J.G.; Mansfield, S.D.; Douglas, C.J. Regulation of secondary cell wall biosynthesis by poplar R2R3 MYB transcription factor PtrMYB152 in Arabidopsis. Sci. Rep. 2014, 4, 5054. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, B.; Song, Z.; Li, C.; Jiang, J.; Zhou, Y.; Wang, R.; Wang, Q.; Ni, C.; Liang, Q.; Chen, H.; et al. RSM1, an Arabidopsis MYB protein, interacts with HY5/HYH to modulate seed germination and seedling development in response to abscisic acid and salinity. PLoS Genet. 2018, 14, e1007839. [Google Scholar] [CrossRef]
- Ma, D.; Reichelt, M.; Yoshida, K.; Gershenzon, J.; Constabel, C.P. Two R2R3-MYB proteins are broad repressors of flavonoid and phenylpropanoid metabolism in poplar. Plant J. 2018, 96, 949–965. [Google Scholar] [CrossRef] [Green Version]
- Bian, S.; Jin, D.; Sun, G.; Shan, B.; Zhou, H.; Wang, J.; Zhai, L.; Li, X. Characterization of the soybean R2R3-MYB transcription factor GmMYB81 and its functional roles under abiotic stresses. Gene 2020, 753, 144803. [Google Scholar] [CrossRef]
- Li, T.; Zhang, X.-Y.; Huang, Y.; Xu, Z.-S.; Wang, F.; Xiong, A.-S. An R2R3-MYB transcription factor, SlMYB28, involved in the regulation of TYLCV infection in tomato. Sci. Hortic. 2018, 237, 192–200. [Google Scholar] [CrossRef]
- Li, Q.; Lu, L.; Liu, H.; Bai, X.; Zhou, X.; Wu, B.; Yuan, M.; Yang, L.; Xing, Y. A minor QTL, SG3, encoding an R2R3-MYB protein, negatively controls grain length in rice. Theor. Appl. Genet. 2020, 133, 2387–2399. [Google Scholar] [CrossRef]
- Hong, L.; Niu, F.; Lin, Y.; Wang, S.; Chen, L.; Jiang, L. MYB106 is a negative regulator and a substrate for CRL3(BPM) E3 ligase in regulating flowering time in Arabidopsis thaliana. J. Integr. Plant Biol. 2021, 63, 1104–1119. [Google Scholar] [CrossRef]
- Fu, S.; Yun, T.; Ma, D.; Zheng, B.; Jiang, D.; He, Y. Thylakoid Transit Peptide Is Related to the Expression and Localization of NdhB Subunits in Soybean. Phyton Int. J. Exp. Bot. 2021, 90, 99–110. [Google Scholar] [CrossRef]
- Mandaokar, A.; Thines, B.; Shin, B.; Lange, B.M.; Choi, G.; Koo, Y.J.; Yoo, Y.J.; Choi, Y.D.; Choi, G.; Browse, J. Transcriptional regulators of stamen development in Arabidopsis identified by transcriptional profiling. Plant J. 2006, 46, 984–1008. [Google Scholar] [CrossRef]
- Lu, J.Y.; Xiong, S.X.; Yin, W.; Teng, X.D.; Lou, Y.; Zhu, J.; Zhang, C.; Gu, J.N.; Wilson, Z.A.; Yang, Z.N. MS1, a direct target of MS188, regulates the expression of key sporophytic pollen coat protein genes in Arabidopsis. J. Exp. Bot. 2020, 71, 4877–4889. [Google Scholar] [CrossRef]
- Han, Y.; Zhou, S.D.; Fan, J.J.; Zhou, L.; Shi, Q.S.; Zhang, Y.F.; Liu, X.L.; Chen, X.; Zhu, J.; Yang, Z.N. OsMS188 Is a Key Regulator of Tapetum Development and Sporopollenin Synthesis in Rice. Rice 2021, 14, 4. [Google Scholar] [CrossRef]
- Gocal, G.F.; Sheldon, C.C.; Gubler, F.; Moritz, T.; Bagnall, D.J.; MacMillan, C.P.; Li, S.F.; Parish, R.W.; Dennis, E.S.; Weigel, D.; et al. GAMYB-like genes, flowering, and gibberellin signaling in Arabidopsis. Plant Physiol. 2001, 127, 1682–1693. [Google Scholar] [CrossRef]
- Shumayla; Mendu, V.; Singh, K.; Upadhyay, S.K. Insight into the Roles of Proline-Rich Extensin-like Receptor Protein Kinases of Bread Wheat (Triticum aestivum L.). Life 2022, 12, 941. [Google Scholar] [CrossRef]
- Rathour, M.; Shumayla; Alok, A.; Upadhyay, S.K. Investigation of Roles of TaTALE Genes during Development and Stress Response in Bread Wheat. Plants 2022, 11, 587. [Google Scholar] [CrossRef]
- Huang, D.; Sun, Y.; Ma, Z.; Ke, M.; Cui, Y.; Chen, Z.; Chen, C.; Ji, C.; Tran, T.M.; Yang, L. Salicylic acid-mediated plasmodesmal closure via Remorin-dependent lipid organization. Proc. Natl. Acad. Sci. USA 2019, 116, 21274–21284. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Xie, X.; Tao, S.; Zhou, K.; Yu, Y.; Lu, Z.; Jiang, D.; Zheng, B.; He, Y. Plasmodesmata play a critical role in promoting the germination of floral buds in Ilex verticillata. Plant Growth Regul. 2020, 91, 349–357. [Google Scholar] [CrossRef]
- Kawahara, Y.; de la Bastide, M.; Hamilton, J.P.; Kanamori, H.; McCombie, W.R.; Ouyang, S.; Schwartz, D.C.; Tanaka, T.; Wu, J.; Zhou, S.; et al. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 2013, 6, 4. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Niu, B.; Gao, Y.; Fu, L.; Li, W. CD-HIT Suite: A web server for clustering and comparing biological sequences. Bioinformatics 2010, 26, 680–682. [Google Scholar] [CrossRef]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar] [CrossRef]
- Sievers, F.; Higgins, D.G. Clustal omega. Curr. Protoc. Bioinform. 2014, 48, 3.13.11–3.13.16. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [Green Version]
- Sakai, H.; Lee, S.S.; Tanaka, T.; Numa, H.; Kim, J.; Kawahara, Y.; Wakimoto, H.; Yang, C.C.; Iwamoto, M.; Abe, T.; et al. Rice Annotation Project Database (RAP-DB): An integrative and interactive database for rice genomics. Plant Cell Physiol. 2013, 54, e6. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [Green Version]





| Type | Cis-Elements | Gene Numbers | Percentages |
|---|---|---|---|
| Stress-related | C-box | 1 | 1.0% |
| ACE | 13 | 13.1% | |
| 3-AF1 binding site | 6 | 6.1% | |
| AAAC-motif | 1 | 1.0% | |
| Sp1 | 43 | 43.4% | |
| MRE | 24 | 24.2% | |
| GT1-motif | 59 | 59.6% | |
| G-box | 90 | 90.9% | |
| WUN-motif | 6 | 6.1% | |
| LTR | 44 | 44.4% | |
| MBS | 58 | 58.6% | |
| TC-rich repeats | 33 | 33.3% | |
| Phytohormone-related | SARE | 3 | 3.0% |
| TCA-element | 44 | 44.4% | |
| TATC-box | 16 | 16.2% | |
| P-box | 23 | 23.2% | |
| GARE-motif | 20 | 20.2% | |
| ABRE | 91 | 91.9% | |
| AuxRR-core | 18 | 18.2% | |
| TGA-element | 34 | 34.3% | |
| TGACG-motif | 79 | 79.8% | |
| CGTCA-motif | 79 | 79.8% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kang, L.; Teng, Y.; Cen, Q.; Fang, Y.; Tian, Q.; Zhang, X.; Wang, H.; Zhang, X.; Xue, D. Genome-Wide Identification of R2R3-MYB Transcription Factor and Expression Analysis under Abiotic Stress in Rice. Plants 2022, 11, 1928. https://doi.org/10.3390/plants11151928
Kang L, Teng Y, Cen Q, Fang Y, Tian Q, Zhang X, Wang H, Zhang X, Xue D. Genome-Wide Identification of R2R3-MYB Transcription Factor and Expression Analysis under Abiotic Stress in Rice. Plants. 2022; 11(15):1928. https://doi.org/10.3390/plants11151928
Chicago/Turabian StyleKang, Lihua, Yangyang Teng, Qiwen Cen, Yunxia Fang, Quanxiang Tian, Xiaoqin Zhang, Hua Wang, Xian Zhang, and Dawei Xue. 2022. "Genome-Wide Identification of R2R3-MYB Transcription Factor and Expression Analysis under Abiotic Stress in Rice" Plants 11, no. 15: 1928. https://doi.org/10.3390/plants11151928
APA StyleKang, L., Teng, Y., Cen, Q., Fang, Y., Tian, Q., Zhang, X., Wang, H., Zhang, X., & Xue, D. (2022). Genome-Wide Identification of R2R3-MYB Transcription Factor and Expression Analysis under Abiotic Stress in Rice. Plants, 11(15), 1928. https://doi.org/10.3390/plants11151928







