A Tomato EMS-Mutagenized Population Provides New Valuable Resources for Gene Discovery and Breeding of Developmental Traits
Abstract
:1. Introduction
2. Results
2.1. Development of EMS Mutant Collection
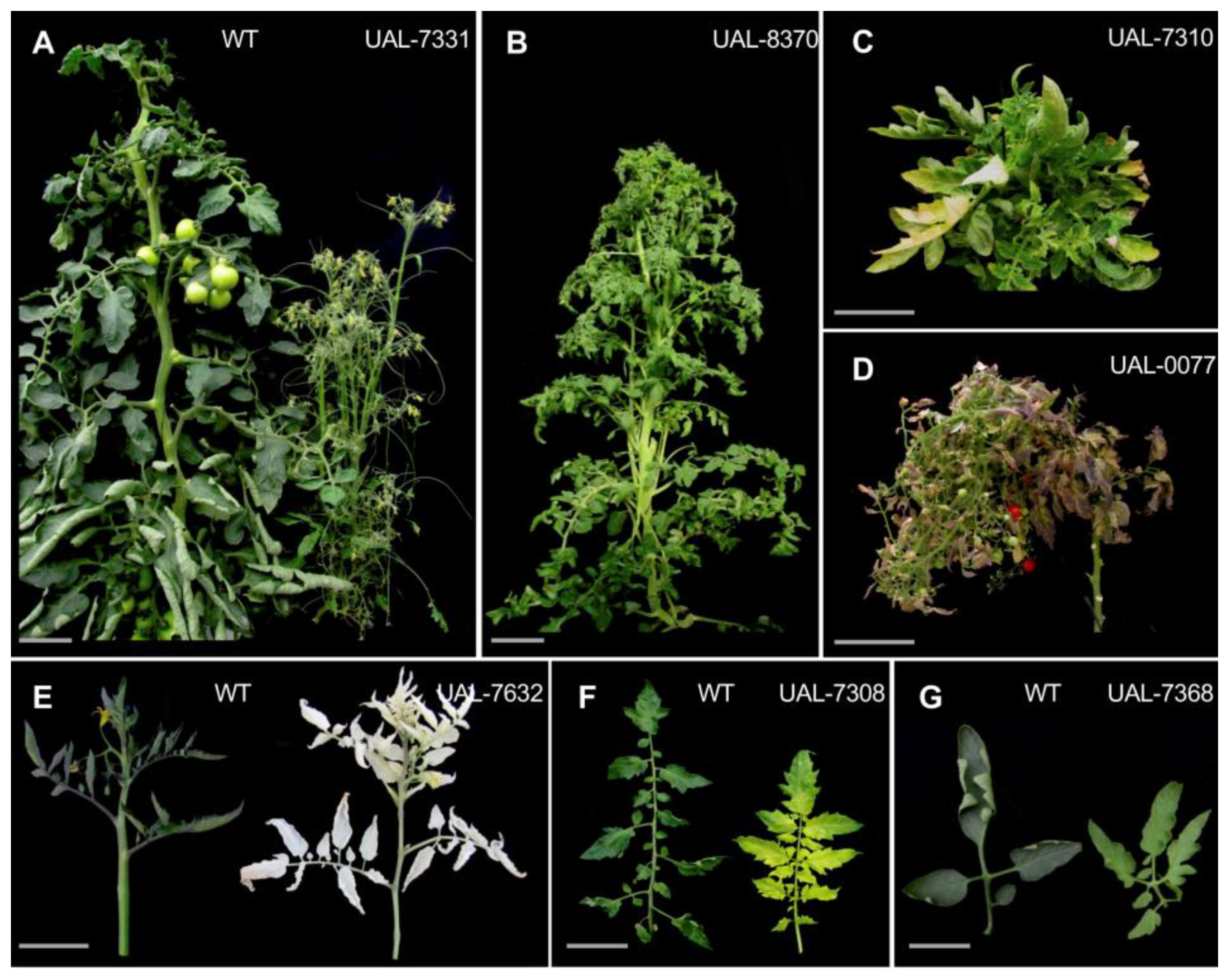
2.2. Phenotypic Screening of EMS Mutant Lines
2.3. Gene Isolation of Causal Mutations by Whole Genome Sequencing Approach
3. Discussion
4. Material and Methods
4.1. Plant Material and Growth Conditions
4.2. EMS Mutagenesis
4.3. Phenotypic Characterization
4.4. Mapping-by-Sequencing of Sin Mutation
4.5. Co-Segregation Test
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gulfishan, M.; Bhat, T.A.; Oves, M. Mutants as a genetic resource for future crop improvement. In Advances in Plant Breeding Strategies: Breeding, Biotechnology and Molecular Tools; Al-Khayri, J., Jain, S., Johnson, D., Eds.; Springer: Cham, Switzerland, 2015; pp. 95–112. [Google Scholar] [CrossRef]
- Jain, S.M. Mutagenesis in crop improvement under the climate change. Rom. Biotechnol. Lett. 2010, 15, 88–106. [Google Scholar]
- Laskar, R.A.; Chaudhary, C.; Khan, S.; Chandra, A. Induction of mutagenized tomato populations for investigation on agronomic traits and mutant phenotyping. J. Saudi Soc. Agric. Sci. 2018, 17, 51–60. [Google Scholar] [CrossRef]
- Capel, C.; Yuste-Lisbona, F.J.; López-Casado, G.; Angosto, T.; Heredia, A.; Cuartero, J.; Fernández-Muñoz, R.; Lozano, R.; Capel, J. QTL mapping of fruit mineral contents provides new chances for molecular breeding of tomato nutritional traits. Theor. Appl. Genet. 2017, 130, 903–913. [Google Scholar] [CrossRef]
- Hobson, G.; Grierson, D. Tomato. In Biochemistry of Fruit Ripening; Seymour, G.B., Taylor, J.E., Tucker, G.A., Eds.; Springer: Dordrecht, The Netherlands, 1993; pp. 405–442. [Google Scholar]
- Kalloo, G. Interspecific and intergeneric hybridization in tomato. In Genetic Improvement of Tomato. Monographs on Theoretical and Applied Genetics; Kalloo, G., Ed.; Springer: Berlin/Heidelberg, Germany, 1991; Volume 14, pp. 73–82. [Google Scholar]
- Arab, L.; Steck, S. Lycopene and cardiovascular disease. Am. J. Clin. Nutr. 2000, 71, 1691S–1695S. [Google Scholar] [CrossRef]
- Pnueli, L.; Carmel-Goren, L.; Hareven, D.; Gutfinger, T.; Alvarez, J.; Ganal, M.; Zamir, D.; Lifschitz, E. The SELF-PRUNING gene of tomato regulates vegetative to reproductive switching of sympodial meristems and is the ortholog of CEN and TFL1. Development 1998, 125, 1979–1989. [Google Scholar] [CrossRef]
- Liu, J.; Van Eck, J.; Cong, B.; Tanksley, S.D. A new class of regulatory genes underlying the cause of pear-shaped tomato fruit. Proc. Natl. Acad. Sci. USA 2002, 99, 13302–13306. [Google Scholar] [CrossRef] [PubMed]
- Mao, L.; Begum, D.; Chuang, H.W.; Budiman, M.A.; Szymkowiak, E.J.; Irish, E.E.; Wing, R.A. JOINTLESS is a MADS-box gene controlling tomato flower abscission zone development. Nature 2000, 406, 910–913. [Google Scholar] [CrossRef]
- Xu, C.; Liberatore, K.L.; MacAlister, C.A.; Huang, Z.; Chu, Y.H.; Jiang, K.; Brooks, C.; Ogawa-Ohnishi, M.; Xiong, G.; Pauly, M.; et al. A cascade of arabinosyltransferases controls shoot meristem size in tomato. Nat. Genet. 2015, 47, 784–792. [Google Scholar] [CrossRef]
- Muños, S.; Ranc, N.; Botton, E.; Bérard, A.; Rolland, S.; Duffé, P.; Carretero, Y.; Le Paslier, M.C.; Delalande, C.; Bouzayen, M.; et al. Increase in tomato locule number is controlled by two single-nucleotide polymorphisms located near WUSCHEL. Plant Phys. 2011, 156, 2244–2254. [Google Scholar] [CrossRef]
- Salinas, M.; Capel, C.; Alba, J.M.; Mora, B.; Cuartero, J.; Fernández-Muñoz, R.; Lozano, R.; Capel, J. Genetic mapping of two QTL from the wild tomato Solanum pimpinellifolium L. controlling resistance against two-spotted spider mite (Tetranychus urticae Koch). Theor. Appl. Genet. 2013, 126, 83–92. [Google Scholar] [CrossRef]
- Kissoudis, C.; Sunarti, S.; van de Wiel, C.; Visser, R.G.; van der Linden, C.G.; Bai, Y. Responses to combined abiotic and biotic stress in tomato are governed by stress intensity and resistance mechanism. J. Exp. Bot. 2016, 67, 5119–5132. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Kissoudis, C.; Yan, Z.; Visser, R.; van der Linden, G. Plant behaviour under combined stress: Tomato responses to combined salinity and pathogen stress. Plant J. 2018, 93, 781–793. [Google Scholar] [CrossRef] [PubMed]
- Tomato Genome Consortium. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 2012, 485, 635–641. [Google Scholar] [CrossRef]
- Hosmani, P.S.; Flores-Gonzalez, M.; van de Geest, H.; Maumus, F.; Bakker, L.V.; Schijlen, E.; van Haarst, J.; Cordewener, J.; Sanchez-Perez, G.; Peters, S.; et al. An improved de novo assembly and annotation of the tomato reference genome using single-molecule sequencing, Hi-C proximity ligation and optical maps. bioRxiv 2019. [Google Scholar] [CrossRef]
- Mathews, H.; Clendennen, S.K.; Caldwell, C.G.; Liu, X.L.; Connors, K.; Matheis, N.; Schuster, D.K.; Menasco, D.J.; Wagoner, W.; Lightner, J.; et al. Activation tagging in tomato identifies a transcriptional regulator of anthocyanin biosynthesis, modification, and transport. Plant Cell 2003, 15, 1689–1703. [Google Scholar] [CrossRef]
- Carter, J.D.; Pereira, A.; Dickerman, A.W.; Veilleux, R.E. An active ac/ds transposon system for activation tagging in tomato cultivar m82 using clonal propagation. Plant Phys. 2013, 162, 145–156. [Google Scholar] [CrossRef]
- Pérez-Martín, F.; Yuste-Lisbona, F.J.; Pineda, B.; Angarita-Díaz, M.P.; García-Sogo, B.; Antón, T.; Sánchez, S.; Giménez, E.; Atarés, A.; Fernández-Lozano, A.; et al. A collection of enhancer trap insertional mutants for functional genomics in tomato. Plant Biotechnol. J. 2017, 15, 1439–1452. [Google Scholar] [CrossRef]
- Li, X.; Song, Y.; Century, K.; Straight, S.; Ronald, P.; Dong, X.; Lassner, M.; Zhang, Y. A fast neutron deletion mutagenesis-based reverse genetics system for plants. Plant J. 2001, 27, 235–242. [Google Scholar] [CrossRef]
- Wu, J.L.; Wu, C.; Lei, C.; Baraoidan, M.; Bordeos, A.; Madamba, M.R.; Ramos-Pamplona, M.; Mauleon, R.; Portugal, A.; Ulat, V.J.; et al. Chemical and irradiation induced mutants of indica rice IR64 for forward and reverse genetics. Plant Mol. Biol. 2005, 59, 85–97. [Google Scholar] [CrossRef]
- Sikora, P.; Chawade, A.; Larsson, M.; Olsson, J.; Olsson, O. Mutagenesis as a tool in plant genetics, functional genomics, and breeding. Int. J. Plant Genom. 2011, 2011, 314829. [Google Scholar] [CrossRef] [Green Version]
- Yuan, L.; Dou, Y.; Kianian, S.F.; Zhang, C.; Holding, D.R. Deletion mutagenesis identifies a haploinsufficient role for γ-zein in opaque2 endosperm modification. Plant Physiol. 2014, 164, 119–130. [Google Scholar] [CrossRef] [PubMed]
- Ries, G.; Heller, W.; Puchta, H.; Sandermann, H.; Seidlitz, H.K.; Hohn, B. Elevated UV-B radiation reduces genome stability in plants. Nature 2000, 406, 98–101. [Google Scholar] [CrossRef] [PubMed]
- Shirasawa, K.; Hirakawa, H.; Nunome, T.; Tabata, S.; Isobe, S. Genome-wide survey of artificial mutations induced by ethyl methanesulfonate and gamma rays in tomato. Plant Biotechnol. J. 2016, 14, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Krieg, D.R. Ethyl methanesulfonate-induced reversion of bacteriophage T4rII mutants. Genetics 1963, 48, 561–580. [Google Scholar] [CrossRef]
- Saito, T.; Ariizumi, T.; Okabe, Y.; Asamizu, E.; Hiwasa-Tanase, K.; Fukuda, N.; Mizoguchi, T.; Yamazaki, Y.; Aoki, K.; Ezura, H. TOMATOMA: A novel tomato mutant database distributing Micro-Tom mutant collections. Plant Cell Physiol. 2011, 52, 283–296. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, R.F.; Campos, M.L.; Pino, L.E.; Crestana, S.L.; Zsögön, A.; Lima, J.E.; Benedito, V.A.; Peres, L.E. Convergence of developmental mutants into a single tomato model system: ‘Micro-Tom’ as an effective toolkit for plant development research. Plant Methods 2011, 7, 18. [Google Scholar] [CrossRef]
- Watanabe, S.; Mizoguchi, T.; Aoki, K.; Kubo, Y.; Mori, H.; Imanishi, S.; Yamazaki, Y.; Shibata, D.; Ezura, H. Ethylmethanesulfonate (EMS) mutagenesis of Solanum lycopersicum cv. Micro-Tom for largescale mutant screens. Plant Biotechnol. 2007, 24, 33–38. [Google Scholar] [CrossRef]
- Shikata, M.; Hoshikawa, K.; Ariizumi, T.; Fukuda, N.; Yamazaki, Y.; Ezura, H. TOMATOMA Update: Phenotypic and Metabolite Information in the Micro-Tom Mutant Resource. Plant Cell Physiol. 2016, 57, e11. [Google Scholar] [CrossRef]
- Menda, N.; Semel, Y.; Peled, D.; Eshed, Y.; Zamir, D. In silico screening of a saturated mutation library of tomato. Plant J. 2004, 38, 861–872. [Google Scholar] [CrossRef]
- Okabe, Y.; Asamizu, E.; Saito, T.; Matsukura, C.; Ariizumi, T.; Brès, C.; Rothan, C.; Mizoguchi, T.; Ezura, H. Tomato TILLING technology: Development of a reverse genetics tool for the efficient isolation of mutants from Micro-Tom mutant libraries. Plant Cell Physiol. 2011, 52, 1994–2005. [Google Scholar] [CrossRef] [Green Version]
- Yuste-Lisbona, F.J.; Jiménez-Gómez, J.M.; Capel, C.; Lozano, R. Effective Mapping by Sequencing to Isolate Causal Mutations in the Tomato Genome. In Methods in Molecular Biology; Tripodi, P., Ed.; Humana: New York, NY, USA, 2021; Volume 2264, pp. 89–103. [Google Scholar] [CrossRef]
- Kerr, E.A. Lutescent2, l2. Rep. Tomato Gen. Coop. 1956, 6, 17. [Google Scholar]
- Barry, C.S.; Aldridge, G.M.; Herzog, G.; Ma, Q.; McQuinn, R.P.; Hirschberg, J.; Giovannoni, J.J. Altered chloroplast development and delayed fruit ripening caused by mutations in a zinc metalloprotease at the lutescent2 locus of tomato. Plant Physiol. 2012, 159, 1086–1098. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Yu, H.; Yuan, L.; Li, C.; Ye, J.; Chen, W.; Wang, Y.; Ge, P.; Zhang, J.; Ye, Z.; et al. SlRCM1, which encodes tomato Lutescent1, is required for chlorophyll synthesis and chloroplast development in fruits. Hortic. Res. 2021, 8, 128. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Tyagi, A.K.; Sharma, A.K. Genome-wide analysis of auxin response factor (ARF) gene family from tomato and analysis of their role in flower and fruit development. Mol. Genet. Genom. 2011, 285, 245–260. [Google Scholar] [CrossRef]
- Zouine, M.; Fu, Y.; Chateigner-Boutin, A.L.; Mila, I.; Frasse, P.; Wang, H.; Audran, C.; Roustan, J.P.; Bouzayen, M. Characterization of the tomato ARF gene family uncovers a multi-levels post-transcriptional regulation including alternative splicing. PLoS ONE 2014, 9, e84203. [Google Scholar] [CrossRef]
- Damodharan, S.; Corem, S.; Gupta, S.K.; Arazi, T. Tuning of SlARF10A dosage by sly-miR160a is critical for auxin-mediated compound leaf and flower development. Plant J. 2018, 96, 855–868. [Google Scholar] [CrossRef]
- Gramazio, P.; Pereira-Dias, L.; Vilanova, S.; Prohens, J.; Soler, S.; Esteras, J.; Garmendia, A.; Díez, M.J. Morphoagronomic characterization and whole-genome resequencing of eight highly diverse wild and weedy S. pimpinellifolium and S. lycopersicum var. cerasiforme accessions used for the first interspecific tomato MAGIC population. Hortic. Res. 2020, 7, 174. [Google Scholar] [CrossRef]
- Roohanitaziani, R.; de Maagd, R.A.; Lammers, M.; Molthoff, J.; Meijer-Dekens, F.; van Kaauwen, M.P.W.; Finkers, R.; Tikunov, Y.; Visser, R.G.F.; Bovy, A.G. Exploration of a Resequenced Tomato Core Collection for Phenotypic and Genotypic Variation in Plant Growth and Fruit Quality Traits. Genes 2020, 11, 1278. [Google Scholar] [CrossRef]
- Shinozaki, Y.; Nicolas, P.; Fernandez-Pozo, N.; Ma, Q.; Evanich, D.J.; Shi, Y.; Xu, Y.; Zheng, Y.; Snyder, S.I.; Martin, L.; et al. High-resolution spatiotemporal transcriptome mapping of tomato fruit development and ripening. Nat. Commun. 2018, 9, 364. [Google Scholar] [CrossRef]
- Safavi-Rizi, V.; Herde, M.; Stöhr, C. RNA-Seq reveals novel genes and pathways associated with hypoxia duration and tolerance in tomato root. Sci. Rep. 2020, 10, 1692. [Google Scholar] [CrossRef] [Green Version]
- Rick, C.M.; Quiros, C.F.; Lange, W.H.; Stevens, M.A. Monogenic control of resistance in the tomato to the tobacco flea beetle: Probable repellance by foliage volatiles. Euphytica 1976, 25, 521–530. [Google Scholar] [CrossRef]
- Emmanuel, E.; Levy, A.A. Tomato mutants as tools for functional genomics. Curr. Opin. Plant Biol. 2002, 5, 112–117. [Google Scholar] [CrossRef]
- Feldman, A.B.; Murchie, E.H.; Leung, H.; Baraoidan, M.; Coe, R.; Yu, S.M.; Lo, S.F.; Quick, W.P. Increasing leaf vein density by mutagenesis: Laying the foundations for C4 rice. PLoS ONE 2014, 9, e94947. [Google Scholar] [CrossRef]
- Mohapatra, T.; Robin, S.; Sarla, N.; Sheshashayee, M.; Singh, A.K.; Singh, K.; Singh, N.K.; Mithra, A.C.R.; Sharma, R.P. EMS induced mutants of upland rice variety Nagina22: Generation and characterization. Proc. India Natl. Acad. Sci. 2014, 80, 163–172. [Google Scholar] [CrossRef]
- Poli, Y.; Basava, R.K.; Panigrahy, M.; Vinukonda, V.P.; Dokula, N.R.; Voleti, S.R.; Desiraju, S.; Neelamraju, S. Characterization of a Nagina22 rice mutant for heat tolerance and mapping of yield traits. Rice 2013, 6, 36. [Google Scholar] [CrossRef]
- Brunelle, D.C.; Clark, J.K.; Sheridan, W.F. Genetic Screening for EMS-Induced Maize Embryo-Specific Mutants Altered in Embryo Morphogenesis. G3 2017, 7, 3559–3570. [Google Scholar] [CrossRef]
- Heuermann, M.C.; Rosso, M.G.; Mascher, M.; Brandt, R.; Tschiersch, H.; Altschmied, L.; Altmann, T. Combining next-generation sequencing and progeny testing for rapid identification of induced recessive and dominant mutations in maize M2 individuals. Plant J. 2019, 100, 851–862. [Google Scholar] [CrossRef]
- Chandler, J.W. Auxin response factors. Plant Cell Environ. 2016, 39, 1014–1028. [Google Scholar] [CrossRef]
- Hendelman, A.; Buxdorf, K.; Stav, R.; Kravchik, M.; Arazi, T. Inhibition of lamina outgrowth following Solanum lycopersicum AUXIN RESPONSE FACTOR 10 (SlARF10) derepression. Plant Mol. Biol. 2012, 78, 561–576. [Google Scholar] [CrossRef]
- Wu, L.; Tian, Z.; Zhang, J. Functional Dissection of Auxin Response Factors in Regulating Tomato Leaf Shape Development. Front. Plant Sci. 2018, 9, 957. [Google Scholar] [CrossRef] [Green Version]
- Yuan, Y.; Mei, L.; Wu, M.; Wei, W.; Shan, W.; Gong, Z.; Zhang, Q.; Yang, F.; Yan, F.; Zhang, Q.; et al. SlARF10, an auxin response factor, is involved in chlorophyll and sugar accumulation during tomato fruit development. J. Exp. Bot. 2018, 69, 5507–5518. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, R. La Mutagénesis como Herramienta de Genómica Funcional en Tomate: Caracterización de los Mutantes Succulent stamens2 y hairplus. Ph.D. Thesis, University of Almería, Almeria, Spain, 9 June 2020. [Google Scholar]
- Fonseca, R.; Capel, C.; Yuste-Lisbona, F.J.; Quispe, J.L.; Gómez-Martín, C.; Lebrón, R.; Hackenberg, M.; Oliver, J.L.; Angosto, T.; Lozano, R.; et al. Functional characterization of the tomato HAIRPLUS gene reveals the implication of the epigenome in the control of glandular trichome formation. Hort. Res. 2022, 9, uhab015. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.; Yu, T.; Yang, Q.; Li, C.; Xiong, C.; Gao, S.; Xie, Q.; Zheng, F.; Li, H.; Tian, Z.; et al. Hair, encoding a single C2H2 zinc-finger protein, regulates multicellular trichome formation in tomato. Plant J. 2018, 96, 90–102. [Google Scholar] [CrossRef] [PubMed]
- Garcia, V.; Bres, C.; Just, D.; Fernandez, L.; Tai, F.W.; Mauxion, J.P.; Le Paslier, M.C.; Bérard, A.; Brunel, D.; Aoki, K.; et al. Rapid identification of causal mutations in tomato EMS populations via mapping-by-sequencing. Nat. Protoc. 2016, 11, 2401–2418. [Google Scholar] [CrossRef] [PubMed]
- Lozano, R.; Angosto, T.; Gomez, P.; Payan, C.; Capel, J.; Huijser, P.; Salinas, J.; Martinez-Zapater, J.M. Tomato flower abnormalities induced by low temperatures are associated with changes of expression of MADS-Box genes. Plant Phys. 1998, 117, 91–100. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- DePristo, M.A.; Banks, E.; Poplin, R.; Garimella, K.V.; Maguire, J.R.; Hartl, C.; Philippakis, A.A.; del Angel, G.; Rivas, M.A.; Hanna, M.; et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2010, 43, 491–498. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. 1000 Genomes Project Analysis Group. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2011. [Google Scholar]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from next-generation sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef]





| Phenotypic Class | Category | Dominants | Recessives | Complex Inheritance a | Total | Frequency (%) |
|---|---|---|---|---|---|---|
| I | Seedling lethality and albinism | 0 | 243 | 10 | 253 | 9.04 |
| II | Root development | 5 | 177 | 13 | 195 | 6.96 |
| III | Plant size, architecture, and branching | 74 | 1038 | 84 | 1196 | 42.71 |
| IV | Leaf morphology and color | 45 | 21 | 9 | 75 | 2.68 |
| V | Shoot apical and leaf senescence | 15 | 17 | 3 | 35 | 1.25 |
| VI | Flowering time | 0 | 2 | 2 | 4 | 0.14 |
| VII | Inflorescence architecture | 11 | 139 | 33 | 183 | 6.54 |
| VIII | Flower morphology and color | 17 | 182 | 21 | 220 | 7.86 |
| IX | Flower abscission zone | 2 | 3 | 0 | 5 | 0.18 |
| X | Fruit setting | 20 | 35 | 12 | 67 | 2.39 |
| XI | Fruit morphology/color | 14 | 197 | 18 | 229 | 8.18 |
| XII | Parthenocarpy (seedless fruits) | 0 | 218 | 21 | 239 | 8.54 |
| XIII | Fruit ripening | 11 | 55 | 12 | 78 | 2.79 |
| XIV | Cuticle/cracked fruit | 8 | 10 | 3 | 21 | 0.75 |
| TOTAL | 222 | 2337 | 241 | 2800 | 100 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fonseca, R.; Capel, C.; Nieto-Canseco, R.; Ortiz-Atienza, A.; Bretones, S.; López-Fábregas, J.D.; Quevedo-Colmena, A.S.; Lebrón, R.; Barragán-Lozano, T.; Villalobos-Ramírez, V.; et al. A Tomato EMS-Mutagenized Population Provides New Valuable Resources for Gene Discovery and Breeding of Developmental Traits. Plants 2022, 11, 2453. https://doi.org/10.3390/plants11192453
Fonseca R, Capel C, Nieto-Canseco R, Ortiz-Atienza A, Bretones S, López-Fábregas JD, Quevedo-Colmena AS, Lebrón R, Barragán-Lozano T, Villalobos-Ramírez V, et al. A Tomato EMS-Mutagenized Population Provides New Valuable Resources for Gene Discovery and Breeding of Developmental Traits. Plants. 2022; 11(19):2453. https://doi.org/10.3390/plants11192453
Chicago/Turabian StyleFonseca, Rocío, Carmen Capel, Roberto Nieto-Canseco, Ana Ortiz-Atienza, Sandra Bretones, Juan D. López-Fábregas, Abraham S. Quevedo-Colmena, Ricardo Lebrón, Teresa Barragán-Lozano, Víctor Villalobos-Ramírez, and et al. 2022. "A Tomato EMS-Mutagenized Population Provides New Valuable Resources for Gene Discovery and Breeding of Developmental Traits" Plants 11, no. 19: 2453. https://doi.org/10.3390/plants11192453
APA StyleFonseca, R., Capel, C., Nieto-Canseco, R., Ortiz-Atienza, A., Bretones, S., López-Fábregas, J. D., Quevedo-Colmena, A. S., Lebrón, R., Barragán-Lozano, T., Villalobos-Ramírez, V., Yuste-Lisbona, F. J., Angosto, T., Capel, J., & Lozano, R. (2022). A Tomato EMS-Mutagenized Population Provides New Valuable Resources for Gene Discovery and Breeding of Developmental Traits. Plants, 11(19), 2453. https://doi.org/10.3390/plants11192453









