1. Introduction
The genus
Panax is composed of perennial herbaceous plants whose roots are harvested for medicinal applications, such as improved cognitive functions, lowered blood pressure and stabilized heart rate [
1]. Asian ginseng (
Panax ginseng), Chinese ginseng (
Panax notoginseng) and American ginseng (
Panax quinquefolius) are the most widely cultivated species [
2]. Commercial cultivation requires intensive management and typically lasts for 3 to 4 years with the stems dying back at the end of each growing season [
3]. Ginseng pathogens can cause significant losses with several producing root rots, including
Alternaria panax,
Phytophthora cactorum,
Sclerotinia sclerotiorum and
Cylindrocarpon destructans [
4].
Cylindrocarpon destructans has been divided into
forma specialis based on host with
C. destructans f. sp.
panacis having specificity to ginseng [
5]. Highly aggressive strains to ginseng are
C. destructans f. sp.
panacis, whereas less aggressive strains are other
Cylindrocarpon species [
6].
Cylindrocarpon-like fungi were originally classified into five teleomorphic genera:
Ilyonectria,
Neonectria,
Rugonectria,
Thelonectria and
Campylocarpon [
7]. The
Ilyonectria radicicola species complex was later reclassified into 14 new
Ilyonectria species and 4 previously described ones [
8].
I. mors-panacis,
I. crassa,
I. robusta and
I. panacis were described as being isolated from ginseng species with
I. mors-panacis matching the highly aggressive
C. destructans f. sp.
panacis. A later classification resulted in five species of
Ilyonectria and named
C. destructans as
Ilyonectria destructans as the name
C. destructans was generated prior to
C. radicicola [
9]. However, the classification of Cabral et al. [
8] currently appears to be most used by ginseng researchers. One issue is that earlier studies only referring to
C. destructans make it difficult to compare to more recent studies as it is unknown if the fungus was highly or weakly aggressive to ginseng.
Cylindrocarpon/
Ilyonectria spp. can survive in soil in infected host debris as well as a saprophyte on dead organic matter producing mycelium and conidia, but long-term survival depends on chlamydospores that can remain viable for many years in the soil (
Figure 1) [
10]. The formation of chlamydospores by
Cylindrocarpon/
Ilyonectria spp. has been reported in vitro from conidia and hyphae [
11]. The ability to thrive in low oxygen concentrations also allows
C. destructans to grow in lower soil horizons [
10].
The fungus likely detects a nearby susceptible plant by host signaling compounds exuded from the roots, such as ginsenosides [
12], which triggers germination of chlamydospores, microconidia and macroconidia and then growth toward the host. Host infection normally occurs via wounds [
13], usually starting from the root tips [
14], but not the meristem [
15]. Once it has successfully infected the host, invasion of the root occurs by hyphae growing inter- and intracellularly [
16] resulting in rotting of the tissues (
Figure 1). Eventually, hyphae spread to the host surface to produce micro- and macroconidia that may form chlamydospores in the soil [
17].
Typical symptoms in infected ginseng roots are dry orange-red to black-brown areas with a strong odor [
14,
19]. The root rot can be restricted lesions resulting in plant stunting or more widespread resulting in plant death with shriveled, scaly, blackened roots where only epidermal root tissue remains, which is commonly referred as disappearing root rot (
Figure 1) [
20]. Symptoms in aerial parts can be barely noticeable to red-brown leaves, which can be confused with the symptoms due to
Phytophthora and
Fusarium, to wilting with vascular discoloration of the stem and black-brown lesions at the base of the stem, particularly when roots are highly rotted (
Figure 1) [
3,
21]. Infection by
Cylindrocarpon/
Ilyonectria spp. can result in secondary invasion by non-pathogens, such as
Rhizopus, under wet conditions [
3]. Distribution and spread of the disease can be observed in the gardens as concentric areas of wilted and dead plants that eventually coalesce [
22].
There are many soil fungi other than
Ilyonectria spp. that can cause root rot of ginseng [
23], making it difficult to assign losses only due to
Ilyonectria. Although not impossible to be replanted, losses due to severe
Ilyonectria spp. root rot can reach 100% [
24]. While the cause of replant disease is not fully established, infection by
Ilyonectria spp. have often been associated with replant disease [
8,
25]. Therefore, understanding the molecular and cellular basis of the host response and pathogen virulence is important for improving ginseng production, and this review will examine the relevant literature in that area.
Most of the information about the molecular/cellular interactions between
Cylindrocarpon/
Ilyonectria spp. and ginseng roots is based on studies of infections with restricted black-brown lesions of
P. ginseng [
26,
27,
28,
29,
30,
31,
32], with only a few studies of
P. quinquefolius [
20,
33] and
P. notoginseng [
34]. While a few of these studies have compared host response between aggressive and less aggressive species of
Ilyonectria [
27,
28,
31], none have examined roots described with disappearing root rot symptoms. Thus, it is not clear if the results obtained with restricted
Ilyonectria lesions are comparable to those with disappearing root rot, or how different are the responses of roots to isolates differing in virulence within the same species of
Ilyonectria.
Rusty root is another ginseng disease where areas of the root periderm turn red to orange-brown [
20]. While some researchers consider that
I. mors-panacis also causes rusty root [
35], others consider
I. mors-panacis to cause only root rot while rusty root is a result of incompatible interactions between ginseng roots and the weakly aggressive
Ilyonectria species,
I. robusta,
I. crassa and
I. panacis [
36]. However, there are other hypotheses, such as rusty root being related to cellulase and pectinase activities of endophytic bacteria, such as
Lysobacter gummosus and
Pseudomonas marginalis, as well as
C. destructans [
37]. Another hypothesis has related rusty root to nitrate-dependent iron-oxidizing bacteria, such as those in the Acidobacteria and Chloroflexi, with no association found between
Ilyonectria species and the amount of rusty root [
38]. As there are considerable differences in the interpretation about the relationship between
Ilyonectria and rusty root disease, it will not be considered in this review.
2. Triggered Immunity
Plant pathogen infections trigger a wide range of changes in host gene expression. For differentially expressed genes (DEGs) in
P. ginseng roots infected by
C. destructans, it was shown that after infection, there were 538, 513 and 2845 DEGs up-regulated at 0.5, 4- and 12-days post-inoculation (dpi), respectively, with a peak in the level of expression at 0.5 dpi [
29]. There were also 201, 69 and 280 DEGs down-regulated at 0.5, 4 and 12 dpi, respectively, with a peak at 0.5 dpi. Gene ontology of up-regulated DEGs at 0.5 dpi revealed that most of them were classified as a defense response to fungi, such as the ethylene (ET)-regulated transcription factor (TF)
ERF2, and jasmonic acid (JA)-regulated TFs
TIFY10A,
TIFY10B and
MYB108. Gene ontology of down-regulated DEGs at 0.5 dpi showed that most of them were related to basal metabolic processes, such as carbon fixation and photosynthesis, as well as a few related to resistance, such as pathogenesis-related (PR) proteins. A shift from growth, such as basal metabolic responses, to plant defenses during infection is a common observation that pathogen infections slow plant growth, thus reducing the need for assimilates, and shifts resources towards defenses [
39]. The relationship of the defense response to JA was not surprising as root rot pathogens typically induce defenses related to JA rather than salicylic acid (SA) signaling [
40]. It was concluded that the impact of infection on gene expression was more acute at an early stage of infection but broader at later stages. However, it is unknown if the isolate used was that of the more aggressive
I. mors-panacis or less aggressive
I. robusta,
I. crassa,
I. panacis or
I. radicicola, as the authors only referred to the pathogen as
C. destructans.
The same study described the host response as being related to effector triggered immunity (ETI) based on resistance (R) genes that were dramatically induced by the pathogen at 0.5 dpi [
29]. ETI is characterized by a rapid and strong host response, typically a hypersensitive response that stops pathogen growth within 1–3 days [
41]. However, ETI was unlikely to have occurred as there were changes in gene expression even at 12 dpi indicating that the pathogen was continuing to invade the roots. Thus, it is more likely that it was PAMP triggered immunity (PTI) with pathogen associated molecular patterns (PAMPs) triggering a strong host response at 0.5 dpi followed by a decline in gene expression due to effectors suppressing PTI. The broader host response at 12 dpi was possibly due to PTI triggered by PAMPs and damage associated molecular patterns (DAMPs) as the amount of tissue rot expanded.
RNA sequencing of
P. ginseng roots infected with an isolate of the less aggressive
I. robusta examined three time points: 1.5 and 3 dpi described as the initial stage of infection, and 6 dpi described as the adaptive stage of infection [
42]. The largest number of up-regulated and down-regulated DEGs was 4293 and 4537, respectively, at 1.5 dpi. Most of the DEGs for cell-surface pattern-recognition receptors (PRRs),
MYB,
WRKY and ET-related TFs and components of PTI and ETI, such as mitogen-activated protein kinases (MAPKs), that were up-regulated at all three time points, indicating a defense response at all time points. However, there was a peak of expression at 1.5 dpi for DEGs for ET-related TFs and components of PTI and ETI, such as MAPKs, indicating that the pathogen was recognized by 1.5 dpi. At 3 dpi, there was a peak of expression for DEGs for
MYB and
WRKY TFs, indicating that defense signal transduction was induced slightly later.
Since most
WRKY TFs are related to SA-regulated defence responses [
43], and many
MYB TFs are related to JA-regulated defence responses [
44], this indicates that both SA and JA were involved in the host response. Most of the auxin-related DEGs were down-regulated at 1.5 dpi and slightly up-regulated at 3 and 6 dpi showing that there was early plant growth impairment that later returned to near pre-infection levels. It is well established that pathogen infections can impair plant growth as the plant shifts resources from growth to defence [
39]. Similarly, most of ROS-related DEGs were also down-regulated at 1.5 dpi and slightly up-regulated at 3 and 6 dpi, which was believed to reflect a reduced defence response that later returned to near pre-infection levels. The authors concluded that the highest impact of
I. robusta on gene expression was at the initial stage of infection, eventually returning to near pre-infection expression at the adaptive stage of infection [
42].
Overall, it appears that
Cylindrocarpon/
Ilyonectria spp. infections can rapidly trigger a rapid host response, likely related to PTI, with later broader scale changes that perhaps are also due to PTI from damaged host cells as the pathogen continues to invade the root. Host basal metabolism and growth shifts towards plant defenses with PRRs, phosphorylation cascade, ROS and activation of TFs related to the defense hormones, SA, ET and JA. There is no clear evidence for ETI with rapid host cell death and cessation of pathogen growth, although ETI may be related to rusty root symptoms [
36], as the superficial damage in rusty root could be related to the hypersensitive response.
3. Ginsenosides
Infections by
Cylindrocarpon/
Ilyonectria spp. of ginseng roots can trigger the production of ginsenosides, which are saponins with antifungal activity. Growth of
C. destructans was significantly inhibited by 80% when protopanaxatriol (PPT) ginsenosides were added to V8 media but was significantly enhanced by almost 130% when protopanaxadiol (PPD) ginsenosides were added to the media [
45]. Although growth of
I. robusta was reduced by 20% when PPT ginsenoside was added to V8 media, there was no significant difference observed with the addition of PPD ginsenoside [
27]. In contrast, growth of
I. mors-panacis was reduced by 45% and 40% when PPT or PPD ginsenoside, respectively, were added to V8 media, and growth of
I. leucospermi was reduced by 15% and 10% when PPT or PPD ginsenoside, respectively, was added to V8 media. Although all isolates showed growth inhibition by some form of ginsenoside, different species and perhaps even isolates can differ greatly in their sensitivity to ginsenosides. As both PPD and PPT ginsenosides accumulate in ginseng roots and can have fungitoxic activity against some
Ilyonectria sp., ginsenoside biosynthesis during the infection process could therefore contribute to pathogen resistance.
Changes in ginsenoside root content were related to differences in aggressiveness of
Ilyonectria sp. possibly due to triggering or suppression of resistance or the ability of the fungi to degrade ginsenosides in
P. ginseng roots grown in pots [
27]. The disease severity indices of roots infected with
I. mors-panacis was 4.99 at 30 dpi, whereas it was 1.95 and 1.85 at 30 dpi with
I. robusta and
I. leucospermi, respectively, showing the difference in aggressiveness. Infection by
I. mors-panacis decreased PPT ginsenosides in roots by 30.0% and PPD ginsenosides by 26.8% at 30 dpi, compared to the water control. However, infection by
I. robusta and
I. leucospermi significantly increased PPT ginsenosides by 20.0% and 10.0%, and PPD ginsenosides significantly increased by 43.9% and 46.3% at 30 dpi, respectively, compared to the water control. The decrease in ginsenoside root content was not associated with
I. mors-panacis degrading ginsenosides as none of the isolates could degrade ginsenosides based on an in vitro fungitoxicity assay. It appeared to be due to suppression of triggering ginsenoside root content by the more aggressive isolate, which was not true for the less aggressive isolates. It appeared that
I. mors-panacis may have been more aggressive due to effectors that were better able to suppress PTI than those of less aggressive
I. robusta and
I. leucospermi.
JA can increase ginsenoside root content, such as exogenous JA increasing ginsenoside content by 5-fold in ginseng roots [
46]. Based on the relationship between ginsenosides and JA, the expression of several TFs potentially associated with JA and SA regulation was examined to determine if they were involved in ginsenoside biosynthesis in
P. ginseng roots infected by
I. mors-panacis [
26]. Infection of ginseng roots by
I. mors-panacis decreased PPT ginsenoside (Rg1 + Re + Rf) by 68.2% and PPD ginsenoside (Rb1 + Rc + Rb2 + Rd) by 70.0% at 30 dpi. At 4 dpi, only the JA/SA-regulated
PgWRKY22 was slightly induced, whereas at 8 dpi,
PgWRKY22 and the SA-regulated
PgMYC2b were suppressed and the JA-regulated
PgMYB3 was slightly induced. At 16 dpi,
PgWRKY22,
PgMYB3, PgMYC2b and
PgMYC2a were all strongly suppressed. Resistance to
I. mors-panacis was increased by treating roots with silicon, which was associated with higher ginsenoside content. Infected silicon treated roots had a weak induction of
PgMYC2b at 4 dpi, a strong induction of
PgMYB3 and
PgWRKY22 at 8 dpi, and a strong induction of
PgMYC2a at 16 dpi. However,
PgMYC2b,
PgMYB3 and
PgWRKY22 were still suppressed at 16 dpi. Thus, it appears that
I. mors-panacis can trigger a weak early induction of two of the four TFs, but it is able to later suppress the response based on the later suppression in expression of all four TFs and ginsenoside root content, perhaps due to effector-triggered suppression of PTI. Silicon-induced resistance could result in higher expression of both SA and JA-regulated TFs and higher ginsenoside root content. The results with non-treated and silicon-treated roots show a connection between resistance to
I. mors-panacis with defense-related TF expression and ginsenoside accumulation, indicating that ginsenosides act as defence compounds.
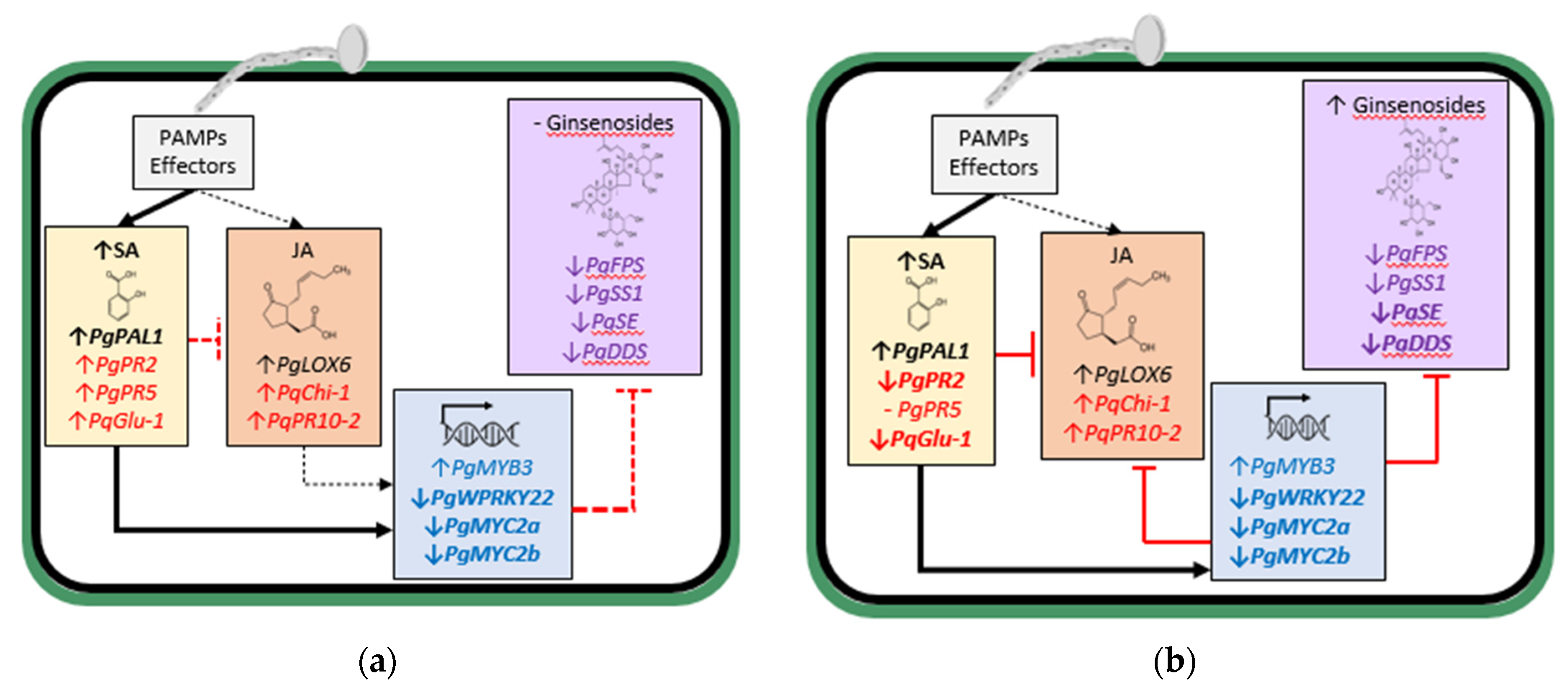
Based on the infection of ginseng roots by the more aggressive
I. mors-panacis resulting in decreased root ginsenoside content [
27], a follow-up study investigated SA levels and the expression of JA-, SA- and ginsenoside biosynthesis-regulated gene expression during infection by more or less aggressive
Ilyonectria spp. in
P. ginseng roots [
26]. SA concentration was significantly higher than the control for
I. mors-panacis at 4 hpi and was even higher at 16 dpi, but that did not occur with
I. robusta. Total ginsenoside root content did not change significantly until 16 dpi with
I. mors-panacis when it slightly greater than the control. By comparison,
I. robusta infection increased total ginsenoside root content first at 4 dpi with a much greater increase compared to the control by 16 dpi. At 4 dpi with
I. mors-panacis, there was reduced expression of the
P. ginseng ginsenoside biosynthesis-regulated genes of farnesyl pyrophosphate synthase (
PgFPS), squalene synthase 1 (
PgSS1), squalene epoxidase 1 (
PgSE1) and dammarenediol synthase (
PgDDS), while there was increased expression of the JA synthesis gene lipoxygenase 6 (
PgLOX6) and the SA synthesis gene phenylalanine ammonia-lyase 1 (
PgPAL1). At 4 dpi for
I. robusta, however, there was an increased expression of the ginsenoside biosynthesis-regulated genes and
PgPAL, but not
PgLOX6. At 16 dpi with
I. mors-panacis, there was reduced expression of all the ginsenoside biosynthesis-regulated genes, but increased expression of
PgPAL and
PgLOX6. In contrast at 16 dpi with
I. robusta, there was increased expression in the ginsenoside biosynthesis-regulated genes. There was also a much greater increase in
PgLOX6 expression but less of an increase in
PgPAL expression compared to
I. mors-panacis. Thus, there was an inverse relationship between higher aggressiveness of
I. mors-panacis with ginsenoside and JA synthesis gene expression, whereas there was a direct relationship with SA and SA-synthesis gene expression. Down regulation of JA-regulated gene expression could be due to triggering of SA-regulated gene expression, as it is known that they are antagonistic to each other [
47]. However, it is possible that the down-regulation of the JA-regulated gene expression could be due to effectors from
I. mors-panacis that are suppressing it, as many fungal pathogen effectors are well known for suppressing PTI [
48].
From these studies, it appears that increased JA-regulated ginsenoside synthesis is an important element of the defense response of ginseng plants against
Ilyonectria spp. Early in the interaction, infection by the more aggressive
I. mors-panacis triggered both JA and SA-synthesis gene expression, likely due to PAMP activity, but much more for SA synthesis gene expression, which corresponded to increased SA content suppressing the JA response, including TFs and ginsenoside biosynthesis-regulated genes (
Figure 2a). Later in the interaction, this continued with even greater suppression of TFs and ginsenoside biosynthesis-regulated genes, and ginsenoside content only slightly increased (
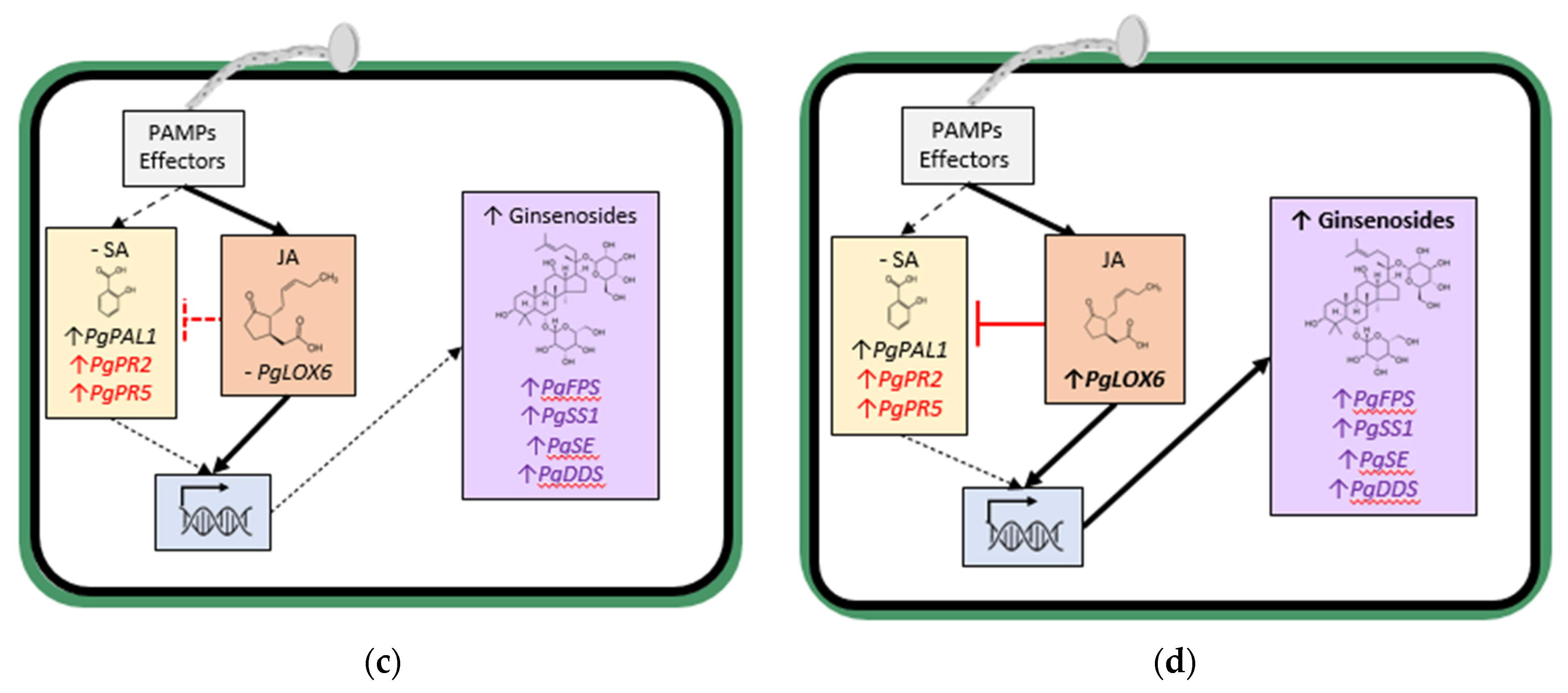
Figure 2b). In contrast, early in the interaction, infection by less aggressive
Ilyonectria spp. did not significantly increase SA content and only slightly induced SA synthesis gene expression but not JA synthesis gene expression, and ginsenoside synthesis genes were induced along with ginsenoside accumulation (
Figure 2c). Later in the interaction with the less aggressive
Ilyonectria sp., there was induced SA, JA and ginsenoside synthesis gene expression and much greater ginsenoside accumulation (
Figure 2d).
4. Pathogenesis-Related Proteins
PAMPs or effectors of plant pathogens typically trigger the production of host pathogenesis-related proteins (PR proteins) [
49], which includes antioomycete and antifungal compounds, such as thaumatin-like proteins, chitinase and glucanase fungal cell wall degradation enzymes, proteinase inhibitors, and ribonucleases [
50]. Among the DEGs identified in
P. ginseng roots infected by
C. destructans [
29], there were 29 DEGs for defence response, which included genes for the PR proteins,
c55244_g1 described as part of the Bev v 1 family, and
c58299_g4 without a description. Expression of
c55244_g1 decreased from 0 to 0.5 dpi, increased at 4 dpi and decreased again at 7 dpi. Expression of
c58299_g4 increased from 0 to 0.25 dpi, decreased at 0.5 dpi, increased at 4 dpi and decreased again at 7 dpi. The authors concluded that 0.5 dpi was the critical time point for resistance given the peak in up-regulation of total DEGs and RGs, and thus the increase in expression of
c58299_g4 at 0.25 dpi could be part of the defence response. However, the fluctuating pattern of expression over time perhaps indicates periods of induction and suppression by PAMPs and effectors, respectively, or a relationship to other factors.
Polygalacturonase inhibiting proteins (PGIPs) are plant cell wall glycoproteins that are considered PR-like proteins, with leucine-rich repeats that bind and inhibit pathogen secreted pectinases and act in defence signalling [
51]. Infection by
C. destructans increased
PgPGIP expression by 1-fold at 6 h post-infection (hpi) and 10.5-fold at 1 dpi, followed by a decrease to basal levels at 2 dpi and 3 dpi [
32]. It is possible that
PgPGIP was part of PTI triggered early in the infection and then later suppressed by effectors from
C. destructans. The authors only referred to the pathogen as
C. destructans, and so it is not possible to know whether a more or less aggressive
Ilyonectria species was used.
The expression of two SA-responsive PR proteins,
PgPR2 encoding an acidic β-1,3-glucanase and
PgPR5 encoding a neutral thaumatin-like protein was compared between the infection by more or less aggressive
Ilyonectria spp. in
P. ginseng roots [
28]. At 4 dpi with the more aggressive
I. mors-panacis,
PgPR2 expression was 1-fold higher and
PgPR5 expression was 0.2-fold higher than the control, but by 16 dpi,
PgPR2 expression was 0.2-fold and
PgPR5 expression was 0.8-fold lower than the control. With the less aggressive
I. robusta, however, expression was induced by 2.5-fold for
PgPR2 and 1-fold for
PgPR5 compared to the control at 4 dpi, while at 16 dpi, expression was induced 0.1-fold for
PgPR2 and 1-fold for
PgPR5 compared to the control. Although at 4 and 16 dpi, SA concentration was significantly greater than the control only for
I. mors-panacis, the pathogen less induced expression of
PgPR2 and
PgPR5 than
I. robusta, indicating that
I. mors-panacis prevented downstream defence signalling for SA-related PR protein production, possibly by PTI-suppressive effectors, whereas
I. robusta was unable to do that.
For
P. quinquefolius, infection by
I. mors-panacis caused expression of the JA-responsive
PqPR5, encoding a neutral thaumatin-like protein, to significantly increase 10-fold at 1 dpi, remaining similar at 6 dpi and then significantly decreasing at 12 dpi [
33]. Moreover, expression of the JA-responsive
PqChi-1 encoding a basic chitinase, significantly increased 4-fold at 1 dpi, a further 1.75-fold at 6 dpi and then remained unchanged at 12 dpi, and expression of the JA-responsive
PqPR10-2, encoding a neutral ribonuclease, significantly increased 1.5-fold at 1 dpi and remained similar at 6 and 12 dpi. Expression of the SA-responsive PR protein,
PqGlu-1 encoding an acidic β-1,3-glucanase, significantly increased 3-fold at 1 dpi and then a further 1.6-fold at 12 dpi. Thus, both JA and SA-regulated PR protein expression were triggered early in the infection with SA-regulated PR protein gene expression further increasing late in the infection.
Based on these studies, the more aggressive
I. mors-panacis induced SA accumulation early in the interaction with increased SA- and JA-related PR gene expression (
Figure 2a). Later in the interaction, there were greater SA levels but with reduced expression of most SA-related PR gene expression indicating downstream suppression, possibly by effectors (
Figure 2b). Less aggressive
Ilyonectria spp. also induced SA-related PR protein gene expression early in the interaction, even though SA levels were not significantly increased (
Figure 2c). Although SA levels were still not significantly increased later in the interaction with less aggressive
Ilyonectria species, increased SA-related PR protein gene expression persisted (
Figure 2d). As some of the previous studies only refer to the pathogen as
C. destructans [
29,
32], a comparison between more and less aggressive species in their studies is not possible.
5. Altered Root Morphology
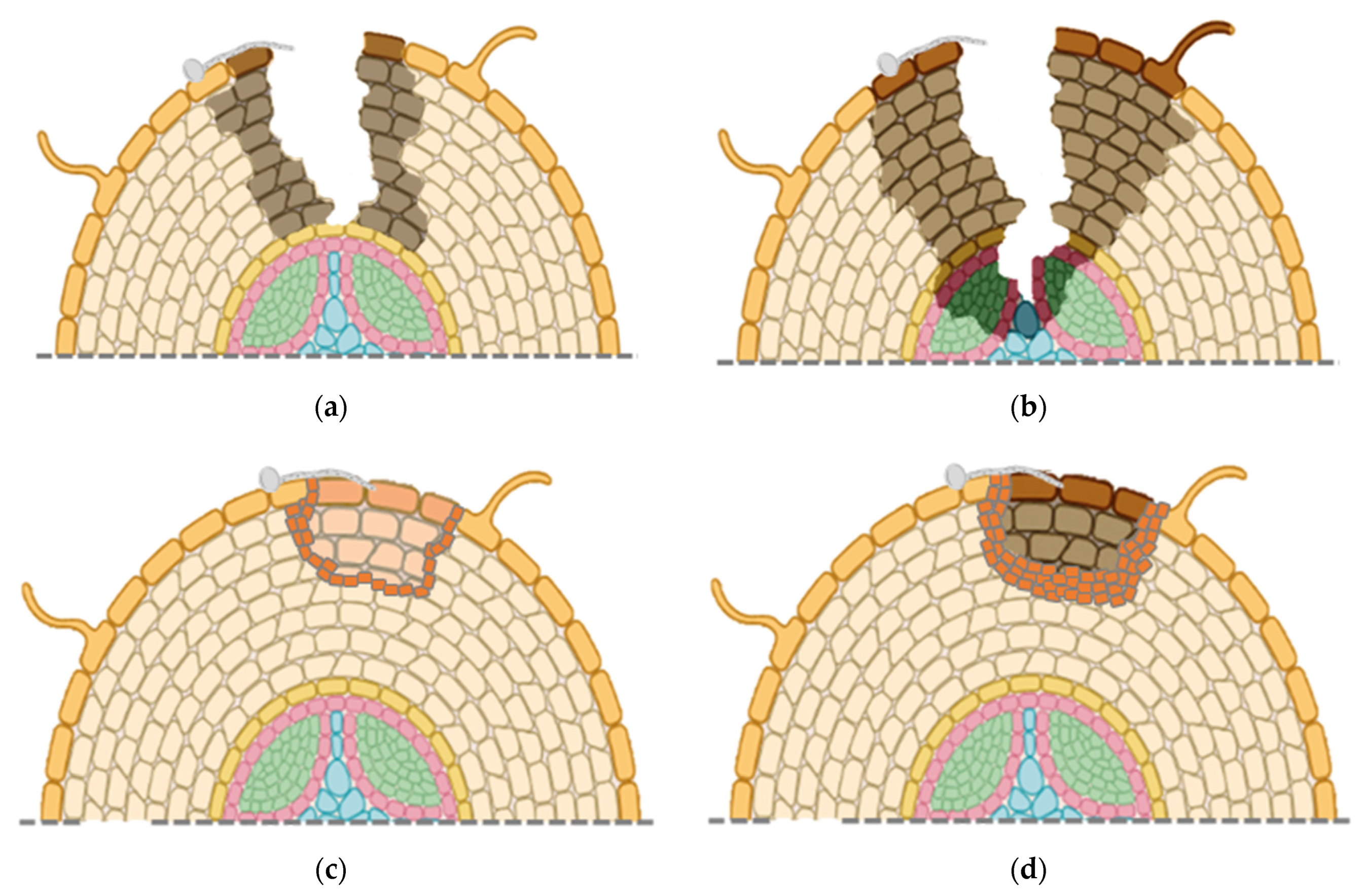
Invasion by plant pathogenic fungi can trigger morphological changes in the host, such as plant cell wall strengthening related to lignin deposition or wound-periderm formation [
52]. An examination of structural changes in the cell wall of
P. ginseng roots infected with a high and weakly virulent
C. destructans isolate showed that only the highly virulent isolate produced black and soft necrotic lesions reaching the cortex of the roots by 7 dpi [
31]. The weakly virulent isolate only caused yellow and dry lesions that were superficial with necrotic tissue restricted to the infection site. Infection by the weakly virulent isolate was associated with new layers of cell wall at 6 dpi with the number of layers expanding up to 12 dpi. At 20 dpi, there was complete separation of the infection site due to the layers of cell wall thickening, with an eventual detachment of the infected area. Based on this, it was concluded that the cell wall thickening layer was a wound periderm or an abscission layer, preventing further invasion by the weakly virulent
C. destructans or damage from its enzymes and other secretion products [
31]. The authors proposed that black root rot symptoms are caused by highly virulent
C. destructans isolates, whereas rusty root symptoms are caused by weakly virulent
C. destructans isolates. It is possible that highly virulent isolates are able to suppress cell wall strengthening with effectors enabling them to spread past the wound-periderm, but weakly virulent isolates lack those effectors, thus being contained by the wound-periderm. However, it is unknown if the highly and weakly virulent isolates were different species of
Ilyonectria as the authors only referred to the pathogen as
C. destructans.Another study found that
P. ginseng roots infected by
C. destructans showed yellowish and dry superficial lesions at 1–4 dpi, but at 7 dpi, lesions became necrotic, with thickened cell walls partially disintegrated, damaged organelles, and intercellular hyphae reaching the xylem [
30]. It was concluded that
P. ginseng might have triggered defence responses at 4 dpi that became ineffective at 7 dpi. Necrotic tissue and the eventual death of the plant were attributed to the blockage of the xylem, restraining the adsorption of water and inorganic minerals. As the authors only referred to the pathogen as
C. destructans, it is not possible to know which
Ilyonectria species from this complex was used.
Overall, changes in root morphology indicate that more aggressive isolates of
C. destructans, possibly
I. mors-panacis, first cause superficial infections (
Figure 3a), but then can rapidly penetrate rotting the root cortex and xylem (
Figure 3b). Weakly aggressive isolates of
C. destructans, possibly other
Ilyonectria spp., only colonize near the root surface with host cell wall strengthening early in the interaction (
Figure 3c). Later in the interaction, there is separation of infected host tissue, possibly due to an abscission layer (
Figure 3d).
6. Virulence Factors
Fungal plant pathogens can suppress PTI through secretion of effectors, which has been described as effector-triggered susceptibility (ETS) [
53]. ETS by
I. mors-panacis is supported by a decline in expression of the JA-regulated
PgPR5 gene at 12 dpi from a peak at 6 dpi [
33], 2 SA-responsive PR protein genes at 16 dpi from a peak at 4 dpi [
28], 13 ET-regulated DEGs and 7 JA-regulated at 12 dpi from a peak at 0.5 dpi [
29], and 7 PRRs DEGs, 35 ET-regulated DEGs and 202 components of PTI/ETI DEGs at 12 dpi from a peak at 1.5 dpi [
42]. Thus, results from several studies indicate that induction of many different types of ginseng defense genes early in the interaction (0.5–6 dpi) is followed by a later decrease in expression (12–16 dpi) implying early triggering of PTI followed by ETS.
There is some evidence for effectors produced by
I. mors-panacis. Fungal small-secreted proteins (SSPs) have been showed to play a role as effectors suppressing the defense response and altering the physiology of their hosts [
54]. Analysis of the secretome of
I. mors-panacis found that 14.9% was composed of small-secreted proteins non-cysteine rich (SSNPs), and 7.9% was composed of small-secreted cysteine-rich proteins (SSCPs) [
33]. Among the SSCPs, one showed homology to the
CRX1 effector for virulence by
Fusarium oxysporum f. sp.
cepae and another to the xylem 6 effector (
SIX6) for virulence by
F. oxysporum f. sp.
lycopersici. There were 121 potential effectors genes in the secretome of
I. robusta based on matches to the PHI database [
42]. Among 20 genes examined, three were significantly up-regulated at 1.5, 3 and 6 dpi, another three were significantly up-regulated just at 3 and 6 dpi, and another four were significantly up-regulated just at 6 dpi. Thus, not all the potential effectors in the fungal genome were expressed during infection, and those that were expressed had different timings of induction indicating that they could have different targets in the host.
Ginsenoside saponins appear to be part of the defense response against
Cylindrocarpon/
Ilyonectria spp. [
27,
45]. However, the fungus can respond to ginsenoside exposure as 93
I. mors-panacis genes were found to be highly expressed when the fungus was cultured with
P. notoginseng ginsenosides [
34]. These included transcription factors, transporters, glycoside hydrolases and oxidoreductases, which might be responsible for regulation, transportation and detoxification of ginseng saponins. However, the role of the particular genes in virulence was not determined.
An examination of 22 isolates of several
Ilyonectria species showed that four species that were avirulent to ginseng had similar metabolic profiles with each other, but they differed from those of
I. mors-panacis and
I. robusta, which had similar profiles and were virulent to ginseng [
55]. The virulent species to ginseng produced the resorcylic acid lactone (RAL), radicicol. There were eight RALs isolated from an
I. mors-panacis strain, which had antimicrobial activity and caused phytotoxicity to duckweed [
56]. They noted that similar polyketides have been identified as virulence factors in several fungal plant pathogens suppressing basal plant defenses with RALs acting as strong inhibitors of plant Hsp90 chaperones negatively affecting defense responses, including reducing PTI. The authors proposed that RALs of
I. mors-panacis may promote its virulence by suppressing ginseng defense responses.
Pathogens and their host compete for iron during infection with iron being involved in plant defense responses, and plants can withhold iron to limit the aggressiveness of pathogens [
57]. However, plant pathogens can act against those defenses by secreting iron-scavenging siderophores to increase iron uptake and decrease iron-regulated host defense responses. Examples of the importance of siderophores for plant pathogens are the compromised virulence in siderophore deficient
Alternaria alternata on citrus [
58] and
Erwinia amylovora on apple [
59]. A siderophore N, N′, N″-triacetylfusarinine C (TAFC) was found to be only present in
Ilyonectria species that were pathogenic to ginseng [
60]. Based on the same siderophore in
Aspergillus fumigatus, it was believed that iron was released from the siderophore by an esterase once taken up inside the cell. To test this, an
A. fumigatus esterase that acts on TAFC was applied to ginseng roots inoculated with
I. mors-panacis, and the esterase protected the roots from infection, supporting a role of this siderophore in
I. mors-panacis virulence to ginseng [
61].
As root rot is associated with pectolytic enzymes [
62] and lesion browning is associated with phenol oxidation [
63], an examination was undertaken into the production of plant cell wall degrading and phenol oxidizing enzymes as virulence factors of
C. destructans against
P. quinquefolius [
20]. Highly virulent isolates differed from weakly virulent isolates by being able to directly penetrate the root epidermis, growing mostly intercellularly, with apparent degradation of the plant cell wall. At 14 dpi in wounded roots, highly virulent isolates created larger sunken dark brown lesions, whereas weakly virulent isolates created smaller sunken brown lesions. In culture, highly virulent isolates produced 1.8 units mg
−1 dry mycelial weight
−1 of pectinase, while weakly virulent isolates produced no detectable levels. Moreover, in culture, highly virulent isolates produced 61 units mg
−1 dry mycelial weight
−1 of polyphenoloxidase, while weakly virulent isolates produced just 30 units mg
−1 dry mycelial weight
−1. It was concluded that sunken brown lesions correlated with the ability of highly virulent isolates to produce higher levels of pectinase and polyphenoloxidase in culture, thus indicating that pectinase and polyphenoloxidase are key factors for
C. destructans virulence. However, there were no measurements of the enzymes in roots, and so it is unknown if the correlation with virulence in culture also occurred during infection. As the authors only referred to the pathogen as
C. destructans, it is not possible to know whether the highly and weakly virulent isolates were different
Ilyonectria species.
In summary, virulence of
Ilyonectria species to ginseng appears at least to be related to the production of potential effectors that may down-regulate SA and JA-regulated PTI, production of cell-wall degrading enzymes to damage host cells, proteins to transport and detoxify saponins, polyketides to suppress defense response, and siderophores to scavenge host iron. In addition, highly aggressive
I. mors-panacis, but not the weakly aggressive
I. robusta, significantly induced production of SA and ROS after infection, although the mechanism was not investigated [
28]. It possibly could be similar to certain necrotrophic plant pathogens that can produce SA analogues, such as 5-formylsalicylic acid, that can be perceived by plants similar to SA to trigger defence responses [
64]. Whatever the mechanism, higher virulence may result from
I. mors-panacis triggering SA thus suppressing JA-related defenses.