Gene Expression, Histology and Histochemistry in the Interaction between Musa sp. and Pseudocercospora fijiensis
Abstract
1. Introduction
2. Results
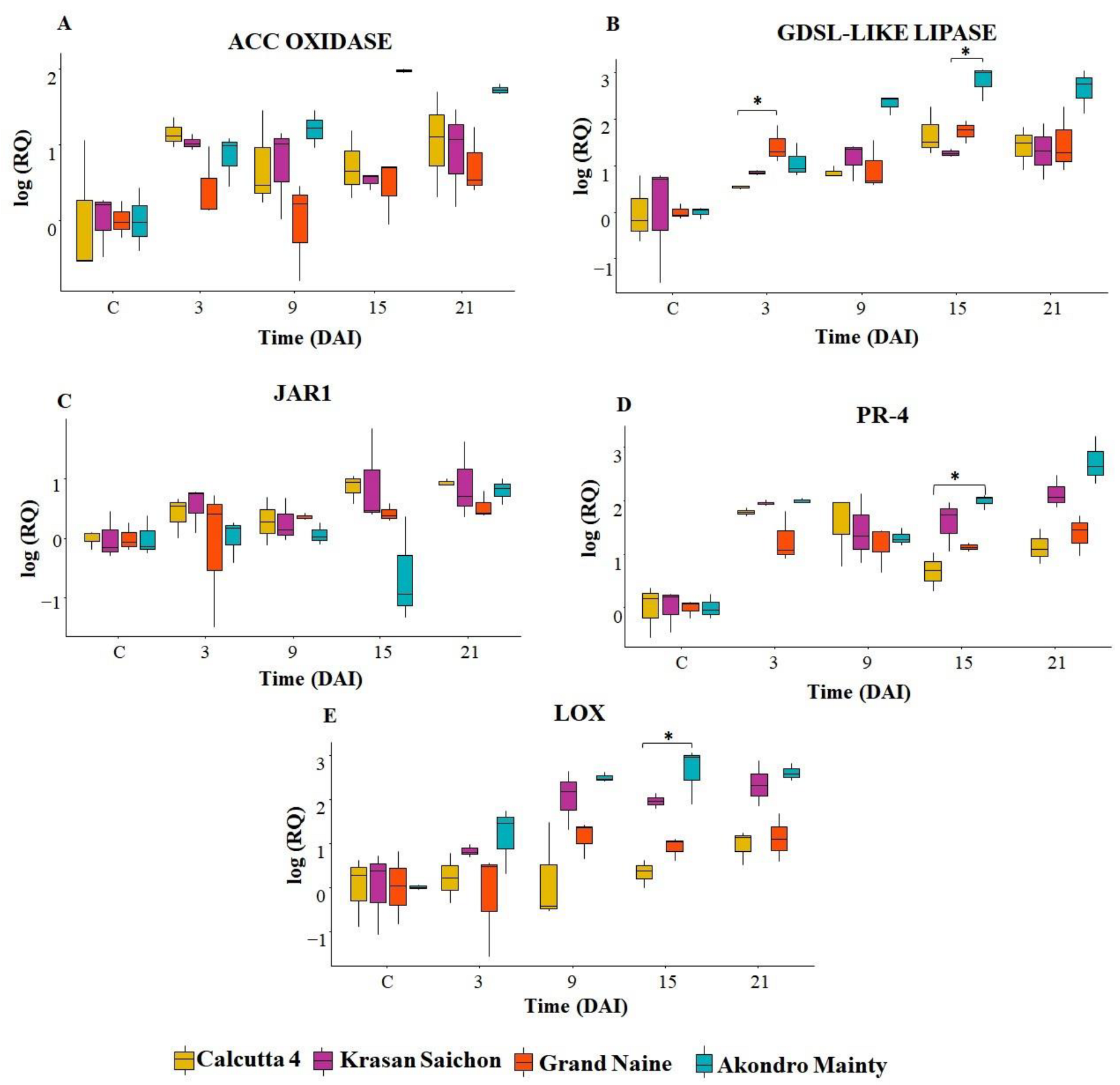
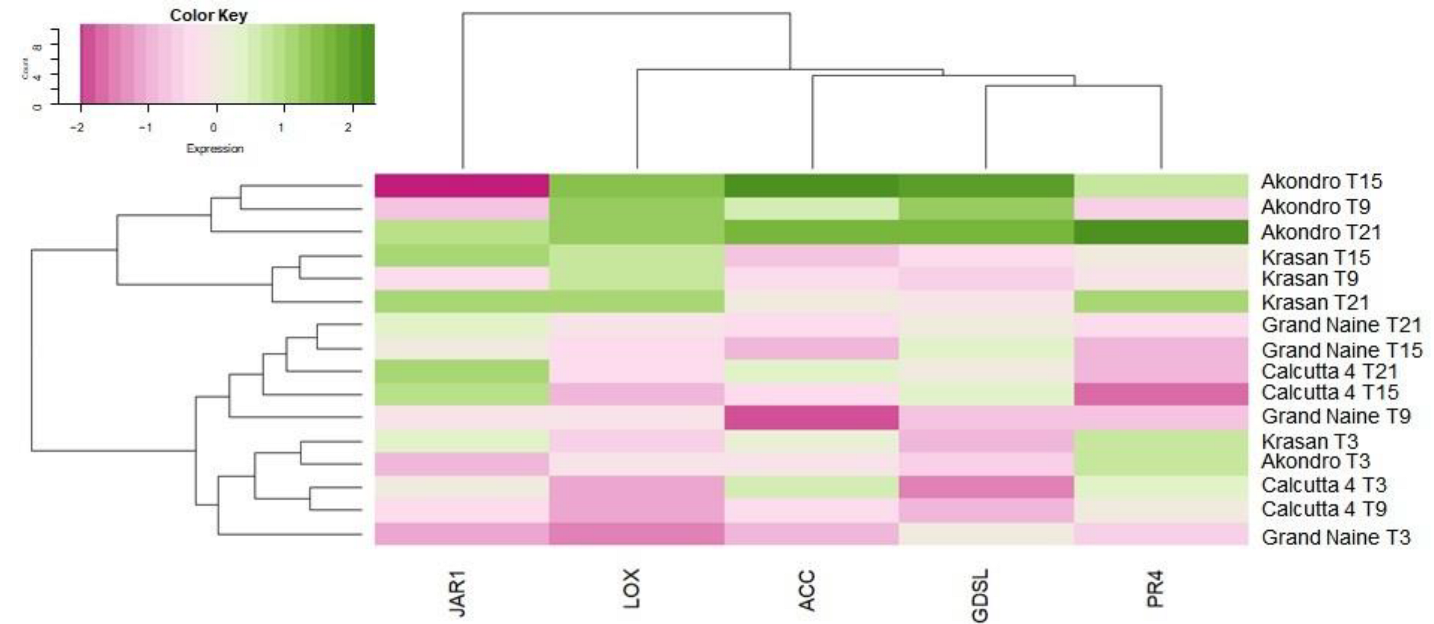
2.1. Analysis of Gene Expression
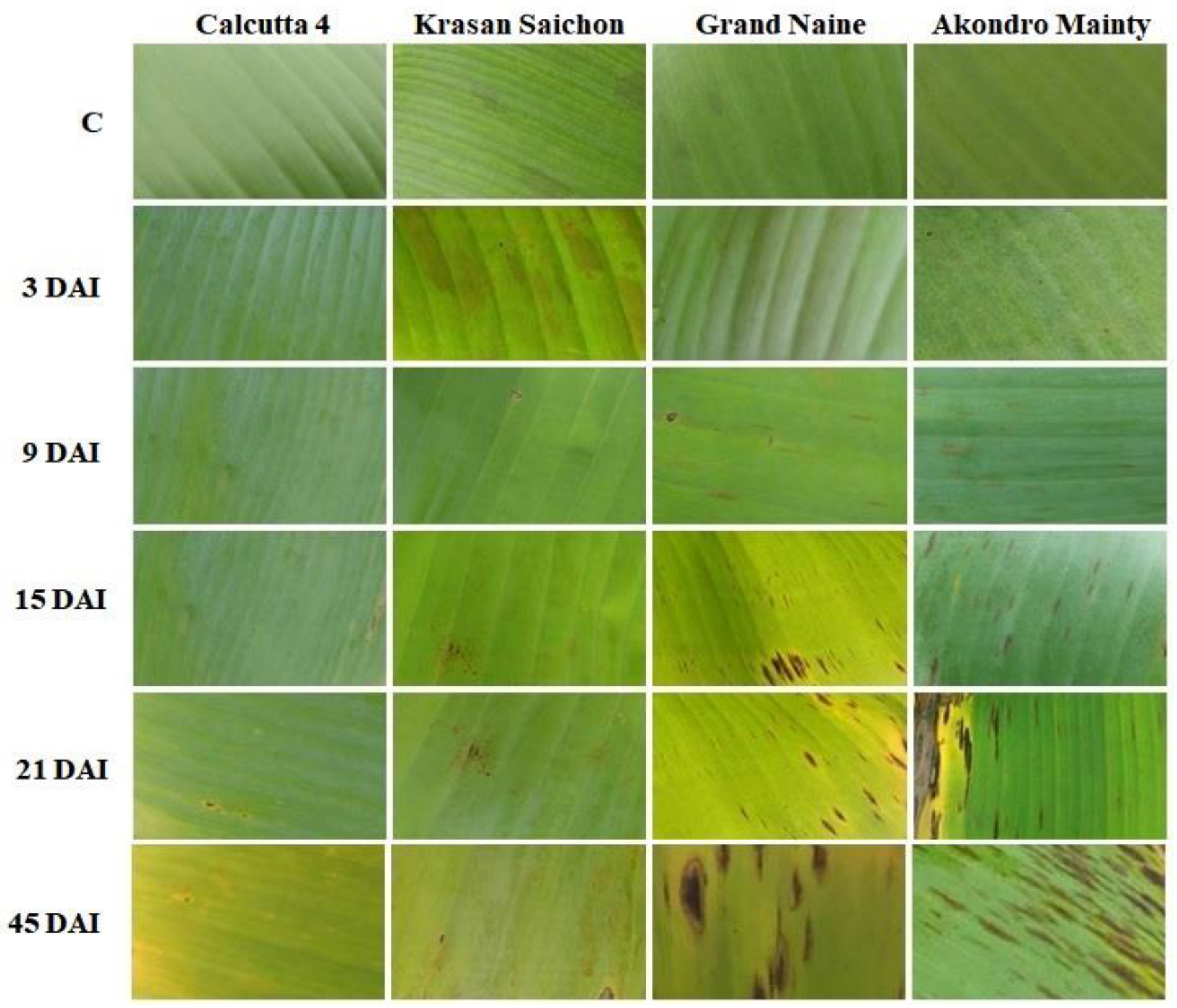
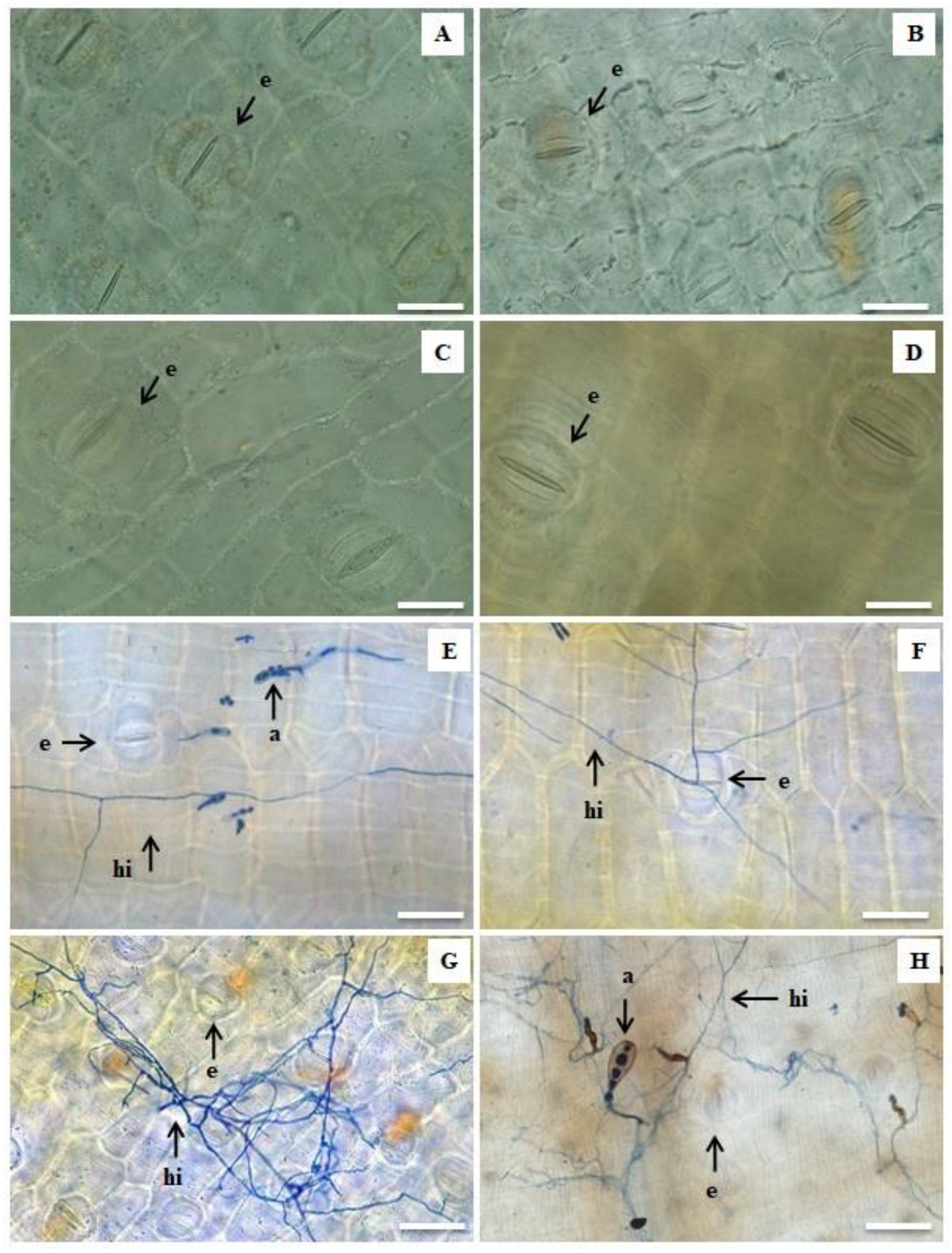
2.2. Histological Analyses
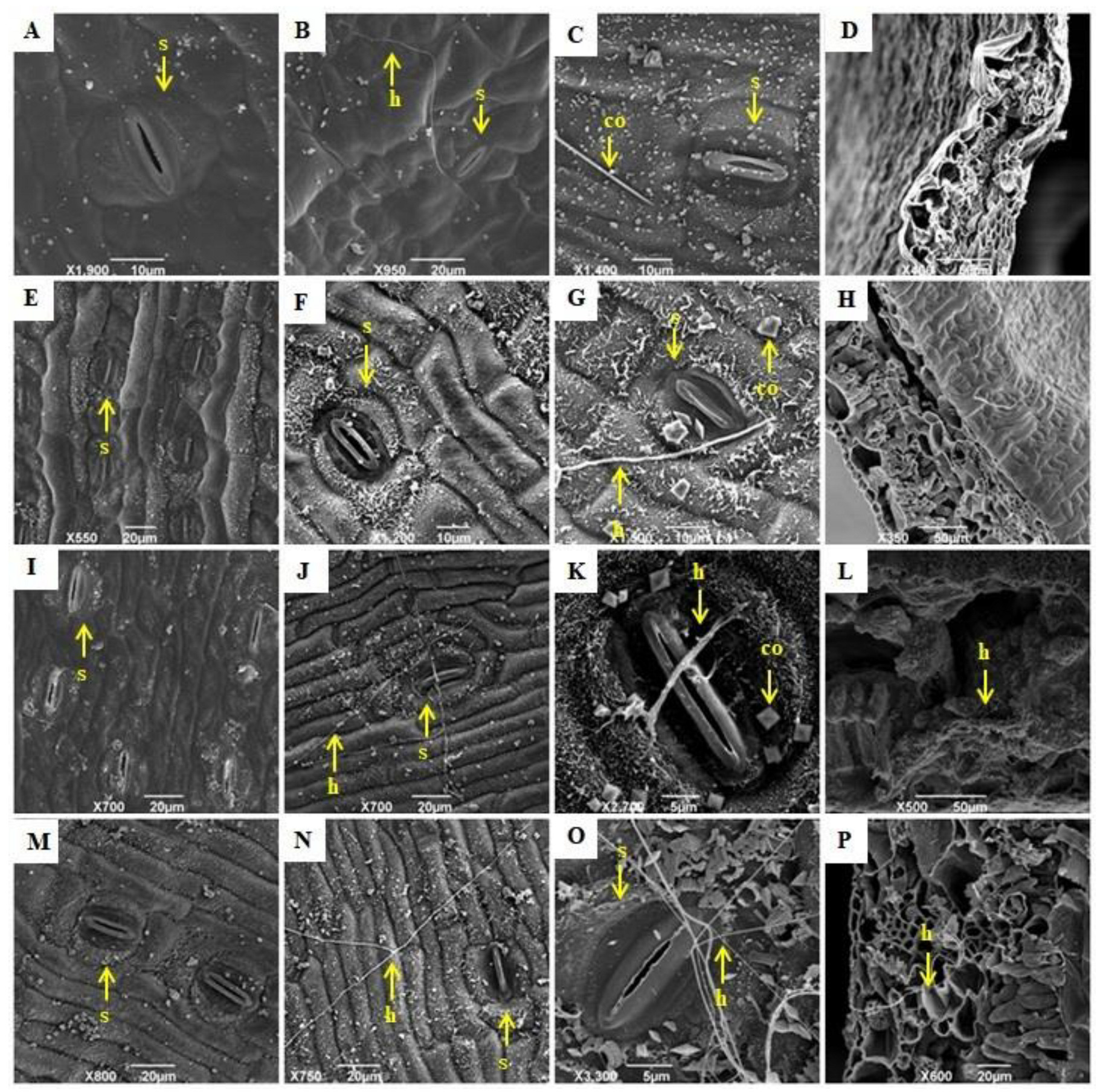
2.2.1. Leaf Clarification and Staining
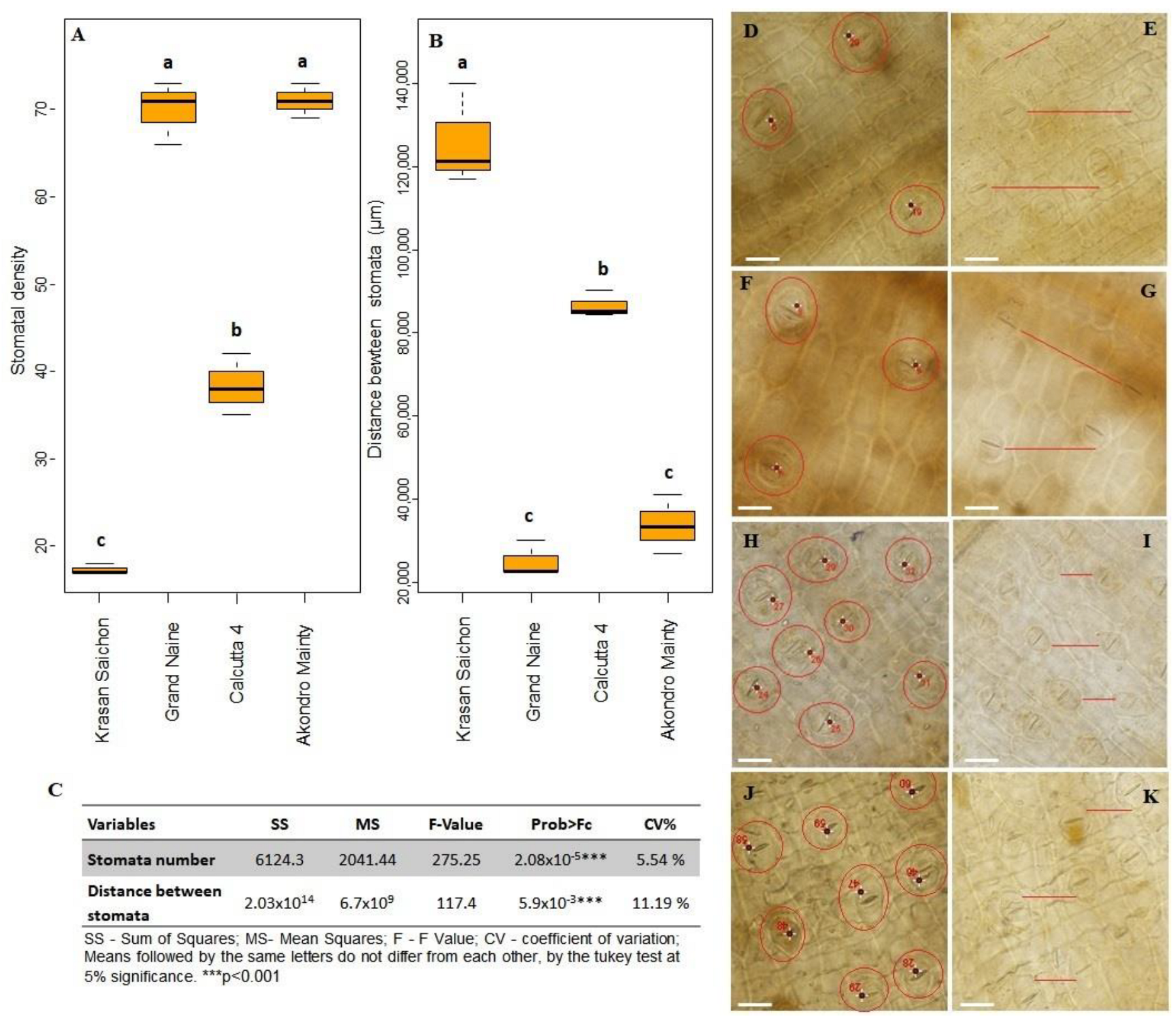
2.2.2. Stomatal Density
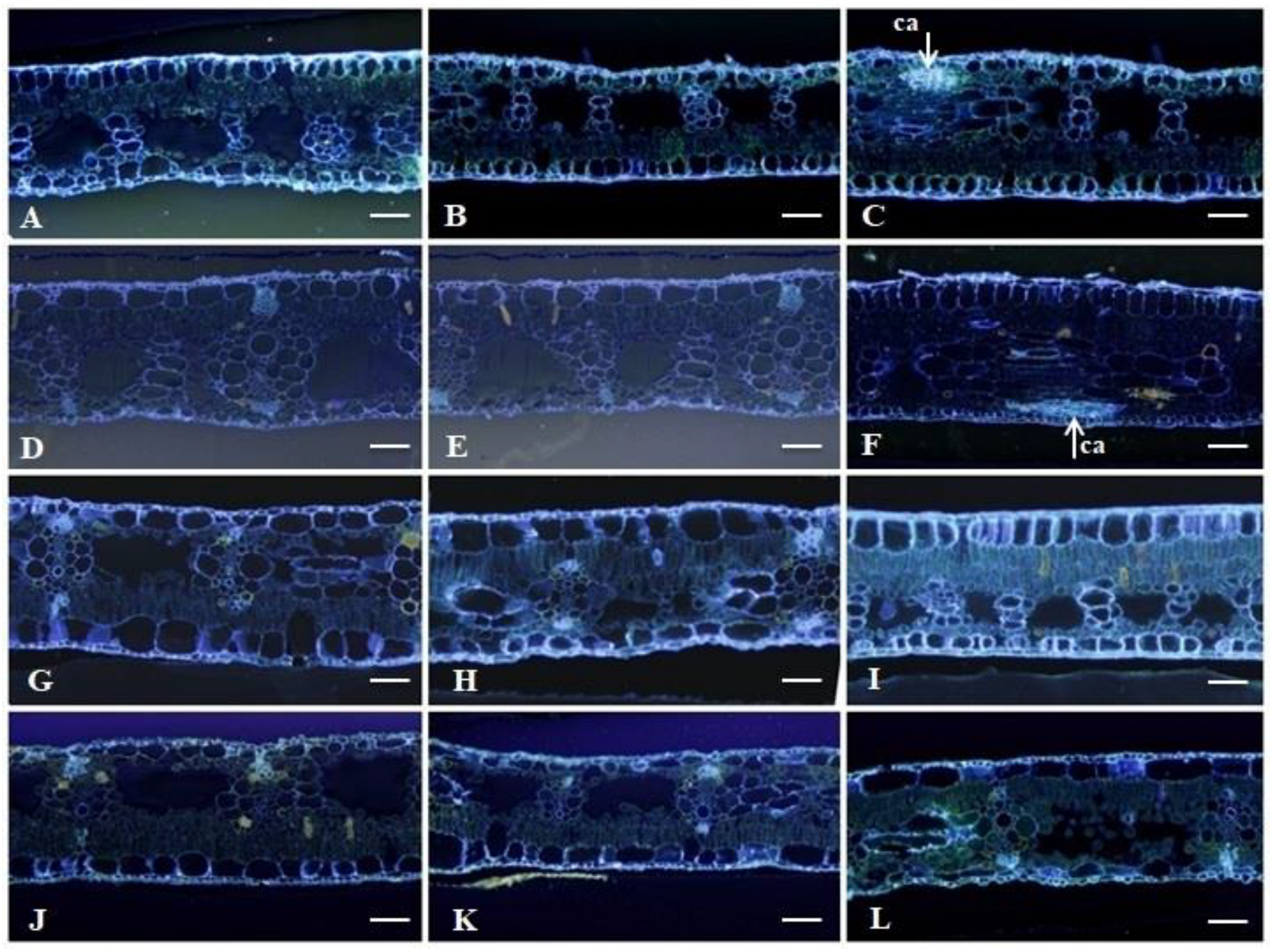
2.2.3. Histochemical Analysis
3. Discussion
3.1. Gene Expression
3.2. Histological Analyses
4. Material and Methods
4.1. Plant Material
4.2. Fungal Material
4.3. Plant Inoculation and Sample Collection
4.4. Molecular Analysis
4.4.1. Total RNA Extraction and cDNA Synthesis
4.4.2. Primers Used and RT–qPCR
4.4.3. Quantitative PCR Analysis
4.5. Histological Analysis
4.5.1. Clarification and Leaf Staining
4.5.2. Stomatal Density
4.5.3. Histochemistry
4.5.4. Scanning Electron Microscopy (SEM)
4.5.5. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Churchill, A.C.L. Mycosphaerella fijiensis, the black leaf streak pathogen of banana: Progress towards understanding pathogen biology and detection, disease development, and the challenges of control. Molec. Plant Pathol. 2011, 12, 307–328. [Google Scholar] [CrossRef] [PubMed]
- Weber, O.B.; Garruti, D.D.S.; Norões, N.P. Performance of banana genotypes with resistance to black leaf streak disease in Northeastern Brazil. Pesqui. Agropecu. Bras. 2017, 52, 161–169. [Google Scholar] [CrossRef]
- Faostat. Food and Agriculture Organization of the United Nations. Available online: http://www.fao.org/faostat/en/#home (accessed on 10 June 2022).
- Cordeiro, Z.J.M.; Matos, A.P.; Haddad, F. Doenças Fúngicas e Bacterianas. In O Agronegócio da Banana; Ferreira, C.F., Silva, S.O., Amorim, E.P., Santos-Serejo, J.A., Eds.; Embrapa: Brasília, Brazil, 2016; pp. 111–136. [Google Scholar]
- Studholme, D.J.; Wicker, E.; Abrare, S.M.; Aspin, A.; Bogdanove, A.; Broders, K.; Dubrow, Z.; Grant, M.; Jones, J.B.; Karamura, G.; et al. Transfer of Xanthomonas campestris pv. arecae and X. campestris pv. musacearum to X. vasicola (Vauterin) as X. vasicola pv. arecae comb. nov. and X. vasicola pv. musacearum comb. nov. and description of X. vasicola pv. vasculorum pv. nov. Phytopathology 2020, 110, 1153–1160. [Google Scholar] [CrossRef]
- Monteiro, J.D.; Santos, M.; Santos, J.R.P.; Cares, J.E.; Marchão, R.L.; Amorim, E.P.; Costa, D.D.C. Identification of plant parasitic nematodes in triploid and tetraploid bananas in brazil. Rev. Caatinga 2020, 33, 865–877. [Google Scholar] [CrossRef]
- Le Thi, L.; Mertens, A.; Vu, D.T.; Vu, T.D.; Minh, P.L.A.; Duc, H.N.; Backer, S.; Swennen, R.; Vandelook, F.; Panis, B.; et al. Diversity of Fusarium associated banana wilt in northern Viet Nam. MycoKeys 2022, 87, 53–76. [Google Scholar] [CrossRef] [PubMed]
- George, M.; Cherian, K.A.; Mathew, D. Symptomatology of Sigatoka leaf spot disease in banana landraces and identification of its pathogen as Mycosphaerella eumusae. J. Saudi Soc. Agric. Sci. 2021, 21, 278–287. [Google Scholar] [CrossRef]
- Nascimento, F.S.; Sousa, Y.M.; Rocha, A.J.; Ferreira, C.F.; Haddad, F.; Amorim, E.P. Sources of black Sigatoka resistance in wild banana diploids. Rev. Bras. Frutic. 2020, 42, e-038. [Google Scholar] [CrossRef]
- Noar, R.D.; Thomas, E.; Daub, M.E. Genetic Characteristics and Metabolic Interactions between Pseudocercospora fijiensis and Banana: Progress toward Controlling Black Sigatoka. Plants 2022, 11, 948. [Google Scholar] [CrossRef]
- Bebber, D.P. Climate change effects on Black Sigatoka disease of banana. Philos. Trans. R. Soc. 2019, 374, 2018026. [Google Scholar] [CrossRef]
- Arango-Isaza, R.E.; Diaz-Trujillo, C.; Dhillon, B.; Aerts, A.; Carlier, J.; Crane, C.F.; Jong, T.V.; Vries, I.; Dietrich, R.; Farmer, A.D.; et al. Combating a global threat to a clonal crop: Banana black sigatoka pathogen Pseudocercospora fijiensis (Synonym Mycosphaerella fijiensis) genomes reveal clues for disease control. PLoS Genet. 2016, 12, e1005876. [Google Scholar] [CrossRef] [PubMed]
- Marín, D.H.; Romero, R.A.; Guzman, M.; Sutton, T.B. Black Sigatoka: An increasing threat to banana cultivation. Plant Dis. 2003, 87, 208–222. [Google Scholar] [CrossRef] [PubMed]
- Chillet, M.; Castelan, F.P.; Abadie, C.; Hubert, O.; Chilin-Charles, Y.; de Bellaire, L.L. Effect of different levels of Sigatoka disease severity on banana pulp colour and early ripening. Can. J. Plant Pathol. 2014, 36, 48–53. [Google Scholar] [CrossRef]
- Pérez-Vicente, L.; Guzmán, M.; Pasberg-Gauhl, C.; Gauhl, F.; Jones, D.R. Fungal Diseases of the Foliage. In Handbook of Diseases of Banana, Abaca and Enset; Jones, D.R., Ed.; CABI: Boston, MA, USA, 2018; Volume 1, pp. 42–53. ISBN 9781780647203. [Google Scholar]
- Pelaez, V.; Silva, L.R.; Araujo, E.B. Regulation of pesticides: A comparative analysis. Sci. Public Policy 2013, 40, 644–656. [Google Scholar] [CrossRef]
- Buralli, R.J.; Ribeiro, H.; Mauad, T.; Amato-Lourenço, L.F.; Salge, J.M.; Diaz-Quijano, F.A.; Leão, R.S.; Marques, R.C.; Silva, D.S.; Guimarães, J.R.D. Respiratory Condition of Family Farmers Exposed to Pesticides in the State of Rio de Janeiro, Brazil. Int. J. Environ. Res. Public Health 2018, 15, 1203. [Google Scholar] [CrossRef]
- Alakonya, A.E.; Kimunye, J.; Mahuku, G.; Amah, D.; Uwimana, B.; Brown, A.; Swennenet, R. Progress in understanding Pseudocercospora banana pathogens and the development of resistant Musa germplasm. Plant Pathol. 2018, 67, 759–770. [Google Scholar] [CrossRef]
- Guzmán, M.; Romero, R.A.; Pérez-Vicente, L. Introduction of Resistant Cultivars. In Handbook of Diseases of Banana, Abaca and Enset; Jones, D.R., Ed.; CABI: Boston, MA, USA, 2018; Volume 1, pp. 112–113. ISBN 9781780647203. [Google Scholar]
- Jones, D.R. Sigatoka Leaf Spot. In Handbook of Diseases of Banana, Abaca and Enset; Jones, D.R., Ed.; CABI: Boston, MA, USA, 2018; Volume 1, pp. 115–127. ISBN 9781780647203. [Google Scholar]
- Soares, J.M.; Rocha, A.J.; Nascimento, F.S.; Santos, A.S.; Miller, R.N.; Ferreira, C.F.; Amorim, E.P. Genetic improvement for resistance to black Sigatoka in bananas: A systematic review. Front. Plant Sci. 2021, 12, 657916. [Google Scholar] [CrossRef]
- Mendoza-Rodríguez, M.; Sánchez, A.; Acosta-Suárez, M.; Roque, B.; Portal, O.; Jiménez, E. Construcción y secuenciación parcial de una biblioteca sustractiva en Calcutta 4 (Musa AA) en estadio temprano de infección con Mycosphaerella fijiensis Morelet. Biotecnol. Veg. 2006, 6, 213–217. [Google Scholar]
- Portal, O.; Izquierdo, Y.; Vleesschauwer, D.; Sánchez-Rodríguez, A.; Mendoza-Rodríguez, M.; Acosta-Suárez, M.; Ocaña, B.; Jiménez, E.; Höfte, M. Analysis of expressed sequence tags derived from a compatible Mycosphaerella fijiensis–banana interaction. Plant Cell Rep. 2011, 30, 913–928. [Google Scholar] [CrossRef]
- Rodriguez, H.A.; Hidalgo, W.F.; Sanchez, J.D.; Menezes, R.C.; Schneider, B.; Arango, R.E.; Morales, J.G. Differential regulation of jasmonic acid pathways in resistant (Calcutta 4 4) and susceptible (Williams) banana genotypes during the interaction with Pseudocercospora fijiensis. Plant Pathol. 2020, 69, 872–882. [Google Scholar] [CrossRef]
- Amorim, E.P.; Amorim, V.B.O.; Silva, M.S.; Haddad, F.; Ferreira, C.F.; Santos-Serejo, J.A. Developing Hybrid Banana Varieties with Improved Properties. In Achieving Sustainable Cultivation of Bananas: Germplasm and Genetic Improvement; Kema, G.H.J., Drenth, A., Eds.; Burleigh Dodds Science Publishing: Cambridge, UK, 2021; Volume 2, pp. 1–17. ISBN 9781786763440. [Google Scholar]
- De Vos, M.; Oosten, V.R.V.; Poecke, R.M.V.; Pelt, J.A.V.; Pozo, M.J.; Mueller, M.J.; Buchala, A.J.; Métraux, J.P.; Loon, L.C.V.; Dicke, M.; et al. Signal signature and transcriptome changes of Arabidopsis during pathogen and insect attack. Mol. Plant Microbe Interact. 2005, 18, 923–937. [Google Scholar] [CrossRef]
- Liu, H.; Carvalhais, L.C.; Kazan, K.; Schenk, P.M. Development of marker genes for jasmonic acid signaling in shoots and roots of wheat. Plant Signal. Behav. 2016, 11, e1176654. [Google Scholar] [CrossRef] [PubMed]
- Deuner, C.; Borges, C.T.; Almeida, A.S.; Meneghello, G.E.; Tunes, L.V.M. Ácido jasmônico como promotor de resistência em plantas. Rev. Ciências Agrárias 2015, 38, 275–281. [Google Scholar] [CrossRef]
- Ballaré, C.L. Light regulation of plant defense. Annu. Rev. Plant Biol. 2014, 65, 335–363. [Google Scholar] [CrossRef]
- Weber, H. Fatty acid-derived signals in plants. Trends Plant Sci. 2002, 7, 217–224. [Google Scholar] [CrossRef]
- Wasternack, C.; Hause, B. Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 2013, 111, 1021–1058. [Google Scholar] [CrossRef] [PubMed]
- Ruan, J.; Zhou, Y.; Zhou, M.; Yan, J.; Khurshid, M.; Weng, W.; Cheng, J.; Zhang, K. Jasmonic acid signaling pathway in plants. Int. J. Mol. Sci. 2019, 20, 2479. [Google Scholar] [CrossRef]
- Glazebrook, J. Contrasting Mechanisms of Defense Against Biotrophic and Necrotrophic Pathogens. Annu. Rev. Phytopathol. 2005, 43, 205–227. [Google Scholar] [CrossRef]
- Yang, J.; Duan, G.; Li, C.; Liu, L.; Han, G.; Zhang, Y.; Wang, C. The crosstalks between jasmonic acid and other plant hormone signaling highlight the involvement of jasmonic acid as a core component in plant response to biotic and abiotic stresses. Front. Plant Sci. 2019, 10, 1349. [Google Scholar] [CrossRef]
- Wang, K.L.C.; Li, H.; Ecker, J.R. Ethylene biosynthesis and signaling networks. Plant Cell 2002, 14, 31–51. [Google Scholar] [CrossRef]
- Han, L.; Li, G.J.; Yang, K.Y.; Mao, G.; Wang, R.; Liu, Y.; Zhang, S. Mitogen-activated protein kinase 3 and 6 regulate Botrytis cinerea-induced ethylene production in Arabidopsis. Plant J. 2010, 64, 114–127. [Google Scholar] [CrossRef]
- Wu, J.; Baldwin, I.T. New insights into plant responses to the attack from insect herbivores. Annu. Rev. Genet. 2010, 44, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Langin, G.; Gouguet, P.; Üstün, S. Microbial effector proteins–a journey through the proteolytic landscape. Trends Microbiol. 2020, 28, 523–535. [Google Scholar] [CrossRef] [PubMed]
- De Wit, P.J.; Mehrabi, R.; van den Burg, H.A.; Stergiopoulos, I. Fungal effector proteins: Past, present and future. Mol. Plant Pathol. 2009, 10, 735–747. [Google Scholar] [CrossRef] [PubMed]
- DeYoung, B.J.; Innes, R.W. Plant NBS-LRR proteins in pathogen sensing and host defense. Nat. Immunol. 2006, 7, 1243–1249. [Google Scholar] [CrossRef] [PubMed]
- Selin, C.; de Kievit, T.R.; Belmonte, M.F.; Fernando, W. Elucidating the role of effectors in plant-fungal interactions: Progress and challenges. Front. Microbiol. 2016, 7, 600. [Google Scholar] [CrossRef]
- Rodríguez-García, C.M.; Canché-Gómez, A.D.; Sáenz-Carbonell, L.; Peraza-Echeverría, L.; Canto-Canché, B.; Islas-Flores, I.; Peraza-Echeverría, S. Expression of MfAvr4 in banana leaf sections with black leaf streak disease caused by Mycosphaerella fijiensis: A technical validation. Australas. Plant Pathol. 2016, 45, 481–488. [Google Scholar] [CrossRef]
- Bormann, C.; Baier, D.; Horr, I.; Raps, C.; Berger, J.; Jung, G.; Schwarz, H. Characterization of a novel, antifungal, chitin-binding protein from Streptomyces tendae Tu901 that interferes with growth polarity. J. Bacteriol. 1999, 181, 7421–7429. [Google Scholar] [CrossRef]
- Sels, J.; Mathys, J.; DeConinck, B.M.; Cammue, B.P.; DeBolle, M.F. Plant pathogenesis-related (PR) proteins: A focus on PR peptides. Plant Physiol. Biochem. 2008, 46, 941–950. [Google Scholar] [CrossRef]
- Wang, N.; Xiao, B.; Xiong, L. Identification of a cluster of PR4-like genes involved in stress responses in rice. J. Plant Physiol. 2011, 168, 2212–2224. [Google Scholar] [CrossRef]
- Dai, L.; Wang, D.; Xie, X.; Zhang, C.; Wang, X.; Xu, Y.; Wang, Y.; Zhang, J. The novel gene VpPR4-1 from Vitis pseudoreticulata increases powdery mildew resistance in transgenic Vitis vinifera L. Front. Plant Sci. 2016, 7, 695. [Google Scholar] [CrossRef]
- Ali, S.; Mir, Z.A.; Tyagi, A.; Bhat, J.A.; Chandrashekar, N.; Papolu, P.K.; Rawat, S.; Grover, A. Identification and comparative analysis of Brassica juncea pathogenesis-related genes in response to hormonal, biotic and abiotic stresses. Acta Physiol. Plant. 2017, 39, 268. [Google Scholar] [CrossRef]
- Concepción-Hernández, M.; Acosta-Suárez, M.; Mendoza-Rodríguez, M.; Portal, O. Perfil de expresión de una lipasa de tipo GDSL durante interacciones compatible e incompatible entre Musa spp. y Pseudocercospora fijiensis. Biotecnol. Veg. 2018, 18, 57–60. [Google Scholar]
- Ding, L.N.; Li, M.; Wang, W.J.; Cao, J.; Wang, Z.; Zhu, K.M.; Yang, Y.H.; Li, H.L.; Tan, X.L. Advances in plant GDSL lipases: From sequences to functional mechanisms. Acta Physiol. Plant. 2019, 41, 151. [Google Scholar] [CrossRef]
- Kim, H.G.; Kwon, S.J.; Jang, Y.J.; Chung, J.H.; Nam, M.H.; Park, O.K. GDSL lipase 1 regulates ethylene signaling and ethylene-associated systemic immunity in Arabidopsis. FEBS Lett. 2014, 588, 1652–1658. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.S.; Kim, B.K.; Kwon, S.J.; Jin, H.C.; Park, O.K. Arabidopsis GDSL lipase 2 plays a role in pathogen defense via negative regulation of auxin signaling. Biochem. Biophys. Res. Commun. 2009, 379, 1038–1042. [Google Scholar] [CrossRef] [PubMed]
- Timm, E.S.; Pardo, L.H.; Coello, R.P.; Navarrete, T.C.; Villegas, O.N.; Ordóñez, E.S. Identification of differentially-expressed genes in response to Mycosphaerella fijiensis in the resistant Musa accession ‘Calcutta 4-4’ using suppression subtractive hybridization. PLoS ONE 2016, 11, e0160083. [Google Scholar] [CrossRef][Green Version]
- Vásquez, A.; Tapia, A.; Galindo, J. Ultrastructural studies of the infection process of Mycosphaerella fijiensis of Musa cultivars. Euphytica 1990, 88, 25–34. [Google Scholar]
- Homa, K.; Barney, W.P.; Ward, D.L.; Wyenandt, C.A.; Simon, J.E. Morphological characteristics and susceptibility of basil species and cultivars to Peronospora belbahrii. HortScience 2016, 51, 1389–1396. [Google Scholar] [CrossRef]
- Hernández, Y.; Portillo, F.; Portillo, M.; Navarro, C.; Rodriguez, M.; Velazco, J. Stomatal density in plantains materials (Musa AAB, AAAB and ABB) susceptible and resistant to Black Sigatoka (Mycosphaerella fijiensis, Morelet). Ver. Fac. Agron. 2006, 23, 294–300. [Google Scholar]
- Cruz Arévalo, S.D.; Romero Meza, R.F.; Cedeño Moreira, Á.V.; Verdosoto Valencia, Á.V.; Peñafiel Jaramillo, M.F.; Canchignia Martínez, H.F. Densidad estomática, contenido de clorofila y relación filogenética en 17 cultivares de Musa spp. Sci. Agropecu. 2019, 10, 47–54. [Google Scholar] [CrossRef]
- Sharma, R.C.; Sharma, S.; Gupta, A.K. Stomatal characters of Populus ciliata in relation to leaf rust and growth parameters. Phytomorphology 2001, 51, 199–205. [Google Scholar]
- Alonso-Villaverde Iglesias, V.; Boso Alonso, S.; Santiago Blanco, J.L.; Gago Montaña, P.; Martínez Rodríguez, M.D.C. Variability of the stomata among ’Albariño’ (Vitis vinifera L.) clones and its relationship with susceptibility to downy mildew. Vitis 2011, 50, 45–46. [Google Scholar]
- Stenglein, S.A.; Arambarri, A.M.; Menendez Sevillano, M.C.; Balatti, P.A. Leaf epidermal characters related with plant’s passive resistance to pathogens vary among accessions of wild beans Phaseolus vulgaris var. aborigineus (Leguminosae-Phaseoleae). Flora 2005, 200, 285–295. [Google Scholar] [CrossRef]
- Ekanayake, I.J.; Ortiz, R.; Vuylsteke, D.R. Leaf stomatal conductance and stomatal morphology of Musa germplasm. Euphytica 1998, 99, 221–229. [Google Scholar] [CrossRef]
- Vermerris, W.; Nicholson, R. Families of Phenolic Compounds and Means of Classification. In Phenolic Compound Biochemistry; Springer: Berlin/Heidelberg, Germany, 2008; pp. 1–34. ISBN 978-1-4020-5164-7. [Google Scholar]
- Agrios, G.N. Parasitism and Disease Development. In Plant Pathology; Elsevier: Amsterdam, The Netherlands; Academic Press: Cambridge, MA, USA, 2004; pp. 77–104. [Google Scholar]
- Torres, J.M.; Calderón, H.; Rodríguez-Arango, E.; Morales, J.G.; Arango, R. Differential induction of pathogenesis-related proteins in banana in response to Mycosphaerella fijiensis infection. Eur. J. Plant Pathol. 2012, 133, 887–898. [Google Scholar] [CrossRef]
- Hidalgo, W.; Chandran, J.N.; Menezes, R.C.; Otálvaro, F.; Schneider, B. Phenylphenalenones protect banana plants from infection by Mycosphaerella fijiensis and are deactivated by metabolic conversion. Plant Cell Environ. 2016, 39, 492–513. [Google Scholar] [CrossRef]
- Zavaliev, R.; Ueki, S.; Epel, B.L.; Citovsky, V. Biology of callose (β-1,3- glucan) turnover at plasmodesmata. Protoplasma 2011, 248, 117–130. [Google Scholar] [CrossRef] [PubMed]
- Ueki, S.; Citovsky, V. The systemic movement of a tobamovirus is inhibited by a cadmium-ion-induced glycine-rich protein. Nat. Cell Biol. 2002, 4, 478–486. [Google Scholar] [CrossRef]
- Jacobs, A.K.; Lipka, V.; Burton, R.A.; Panstruga, R.; Strizhov, N.; Schulze-Lefert, P.; Fincher, G.B. An Arabidopsis callose synthase, GSL5, is required for wound and papillary callose formation. Plant Cell 2003, 15, 2503–2513. [Google Scholar] [CrossRef]
- Hofmann, J.; Youssef-Banora, M.; de Almeida-Engler, J.; Grundler, F.M. The role of callose deposition along plasmodesmata in nematode feeding sites. Mol. Plant-Microbe Interact. 2010, 23, 549–557. [Google Scholar] [CrossRef]
- Weber, J.; Schwark, L. Epicuticular wax lipid composition of endemic European betula species in a simulated ontogenetic/diagenetic continuum and its application to chemotaxonomy and paleobotany. Sci. Total Environ. 2020, 730, 138324. [Google Scholar] [CrossRef]
- Wan, H.; Liu, H.; Zhang, J.; Lyu, Y.; Li, Z.; He, Y.; Zhang, X.; Deng, X.; Brotman, Y.; Fernie, A.R.; et al. Lipidomic and transcriptomic analysis reveals reallocation of carbon flux from cuticular wax into plastid membrane lipids in a glossy “Newhall” navel orange mutant. Hortic. Res. 2020, 7, 41. [Google Scholar] [CrossRef] [PubMed]
- Aragón, W.; Reina-Pinto, J.J.; Serrano, M. The intimate talk between plants and microorganisms at the leaf surface. J. Exp. Bot. 2017, 68, 5339–5350. [Google Scholar] [CrossRef] [PubMed]
- McNair, J.B. The interrelation between substances in plants: Essential oils and resins, cyanogen and oxalate. Am. J. Bot. 1932, 19, 255–272. [Google Scholar] [CrossRef]
- Franceschi, V.R.; Nakata, P.A. Calcium oxalate in plants: Formation and function. Annu. Rev. Plant Biol. 2005, 56, 41–71. [Google Scholar] [CrossRef]
- Horner, H.T.; Wagner, B.L. Calcium Oxalate Formation in Higher Plants. In Calcium Oxalate in Biological Systems; CRC Press: Boca Raton, FL, USA, 1995; pp. 53–72. [Google Scholar]
- Oliveira Ceita, G.; Macêdo, J.N.A.; Santos, T.B.; Alemanno, L.; da Silva Gesteira, A.; Micheli, F.; Mariano, A.C.; Gramacho, K.P.; Silva, D.C.; Meinhardt, L.; et al. Involvement of calcium oxalate degradation during programmed cell death in Theobroma cacao tissues triggered by the hemibiotrophic fungus Moniliophthora perniciosa. Plant Sci. 2007, 173, 106–117. [Google Scholar] [CrossRef]
- Ruiz, N.; Ward, D.; Saltz, D. Calcium oxalate crystals in leaves of Pancratium sickenbergeri: Constitutive or induced defence? Funct. Ecol. 2002, 16, 99–105. [Google Scholar] [CrossRef]
- Tillman-Sutela, E.; Kauppi, A. Calcium oxalate crystals in the mature seeds of Norway spruce, Picea abies (L.) Karst. Trees 1999, 13, 131–137. [Google Scholar] [CrossRef]
- Molano-Flores, B. Herbivory and calcium concentrations affect calcium oxalate crystal formation in leaves of Sida (Malvaceae). Ann. Bot. 2001, 88, 387–391. [Google Scholar] [CrossRef]
- Nakata, P.A. Advances in our understanding of calcium oxalate crystal formation and function in plants. Plant Sci. 2003, 164, 901–909. [Google Scholar] [CrossRef]
- Talbot, N.J. On the trail of a cereal killer: Exploring the biology of Magnaporthe grisea. Annu. Rev. Microbiol. 2003, 57, 177–202. [Google Scholar] [CrossRef]
- Dean, R.A.; Talbot, N.J.; Ebbole, D.J.; Farman, M.L.; Mitchell, T.K.; Orbach, M.J.; Thon, M.; Kulkarni, R.; Xu, J.R.; Huaqin, P.; et al. The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 2005, 434, 980–986. [Google Scholar] [CrossRef] [PubMed]
- Goos, R.D.; Tschirch, M. Greenhouse studies on the Cercospora leaf spot of banana. Trans. Br. Mycol. Soc. 1963, 46, 321–330. [Google Scholar] [CrossRef]
- Passos, M.A.; Cruz, V.O.; Emediato, F.L.; Teixeira, C.C.; Azevedo, V.C.R.; Brasileiro, A.C.M.; Amorim, E.P.; Ferreira, C.F.; Martins, N.F.; Togawa, R.C.; et al. Analysis of the leaf transcriptome of Musa acuminata during interaction with Mycosphaerella musicola: Gene assembly, annotation and marker development. BMC Genom. 2013, 14, 78. [Google Scholar] [CrossRef] [PubMed]
- Mendoza-Rodríguez, M.F. Avances en los estudios sobre la interacción Musa spp.-Mycosphaerella fijiensis morelet. Biot. Veg. 2014, 14, 131–144. [Google Scholar]
- Jesus, O.N.; Silva, S.O.; Amorim, E.P.; Ferreira, C.F.; Campos, J.M.S.; Silva, G.G.; Figueira, A. Genetic diversity and population structure of Musa accessions in ex situ conservation. BMC Plant Biol. 2013, 13, 22. [Google Scholar] [CrossRef] [PubMed]
- Christelová, P.; de Langhe, E.; Hribová, E.; Cížková, J.; Sardos, J.; Hušáková, M.; Houve, I.V.D.; Sutanto, A.; Kepler, A.K.; Swennen, R.; et al. Molecular and cytological characterization of the global Musa germplasm collection provides insights into the treasure of banana diversity. Biodivers. Conserv. 2016, 26, 801–824. [Google Scholar] [CrossRef]
- Arzanlou, M.; Abeln, E.C.; Kema, G.H.; Waalwijk, C.; Carlier, J.; Vries, I.D.; Guzmán, M.; Crous, P.W. Molecular diagnostics for the Sigatoka disease complex of banana. Phytopathology 2007, 97, 1112–1118. [Google Scholar] [CrossRef]
- Cordeiro, Z.J.M.; Rocha, H.S.; Araújo, A.G. Metodologias Para Manuseio de Mycosphaerella musicola em Laboratório. In Documentos da Embrapa Mandioca e Fruticultura, 1st ed.; Embrapa Mandioca e Fruticultura: Cruz das Almas, Brazil, 2011; p. 32. ISSN 1516-5728. [Google Scholar]
- Leiva-Mora, M.; Alvarado-Capó, Y.; Acosta-Suárez, M.; Cruz-Martín, M.; Sánchez-García, C.; Roque, B. Protocolo para la inoculación artificial de plantas de Musa spp. con Mycosphaerella fijiensis y evaluación de su respuesta mediante variables epifitiológicas y componentes de la resistencia. Biot. Veg. 2010, 10, 79–88. [Google Scholar]
- Mendoza-Rodríguez, M.F.; Portal, O.; Oloriz, M.I.; Ocaña, B.; Rojas, L.E.; Acosta-Suárez, M.; Roque, B.; Canales, E.; Borrás-Hidalgo, O.; Jimenéz, E. Early regulation of primary metabolism, antioxidant, methyl cycle and phenylpropanoid pathways during the Mycosphaerella fijiensis-Musa spp. interaction. Trop. Plant Pathol. 2018, 43, 1–9. [Google Scholar] [CrossRef]
- Zhao, L.; Ding, Q.; Zeng, J.; Wang, F.R.; Zhang, J.; Fan, S.J.; He, X.Q. An improved CTAB–ammonium acetate method for total RNA isolation from cotton. Phytochem. Anal. 2012, 23, 647–650. [Google Scholar] [CrossRef] [PubMed]
- Podevin, N.; Krauss, A.; Henry, I.; Swennen, R.; Remy, S. Selection and validation of reference genes for quantitative RT-PCR expression studies of the non-model crop Musa. Mol. Breed. 2012, 30, 1237–1252. [Google Scholar] [CrossRef] [PubMed]
- Phillips, J.M.; Haymann, A.S. Improved procedures for clearing roots and staining parasitic and vesicular-arbuscular mycorrhizal fungi for assessment of infection. Trans. Br. Mycol. Soc. 1970, 55, 158–161. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Johansen, D.A. Plant Microtechnique; McGraw-Hill Book Company: New York, NY, USA; London, UK, 1940; p. 530. ISBN 19410700679. [Google Scholar]
- Foster, J.W. Chemical Activities of Fungi. In Chemical Activities of Fungi; Academic Press: Cambridge, MA, USA, 1949; p. 648. ISBN 19512202464. [Google Scholar]
- Pfaffl, M.W.; Horgan, G.W.; Dempfle, L. Relative expression software tool (REST) for groupwise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002, 30, e36. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2016; Available online: https://www.R-project.org (accessed on 10 June 2022).

| Gene | Sequence (5′-3′) | Amplicon (bp) | Reference |
|---|---|---|---|
| Acc oxidase | F: CATAAACACCGGAGACCAGA | 114 | [23] |
| R: TTGTAGAAGGAAGCGATGGA | |||
| Jar1 | F: TCAACATCGACAAGAACACC | 154 | [23] |
| R: CACAACTCCCAGAAGATCAC | |||
| Pr-4 | F: CAGGTACTTCTTCCTCCACT | 100 | [23] |
| R: CTACTACCCGGCTCAGAATAA | |||
| Gdsl-like lipase | F: GCGCATCACAAAAGAAGAAG | 120 | [23] |
| R: CCCCGTCAACATTCATCAG | |||
| LOX | F: ACTACTTGCTCTTCCCCAGCGG | 120 | [24] |
| R: CGTCCCTCACACCACACGC | |||
| 25S | F: ACATTGTCAGGTGGGGAGTT | 106 | [92] |
| R: CCTTTTGTTCCACACGAGATT | |||
| TUB | F: TGTTGCATCCTGGTACTGCT | 112 | [92] |
| R: GGCTTTCTTGCACTGGTACAC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soares, J.M.d.S.; Rocha, A.d.J.; Nascimento, F.d.S.; Amorim, V.B.O.d.; Ramos, A.P.d.S.; Ferreira, C.F.; Haddad, F.; Amorim, E.P. Gene Expression, Histology and Histochemistry in the Interaction between Musa sp. and Pseudocercospora fijiensis. Plants 2022, 11, 1953. https://doi.org/10.3390/plants11151953
Soares JMdS, Rocha AdJ, Nascimento FdS, Amorim VBOd, Ramos APdS, Ferreira CF, Haddad F, Amorim EP. Gene Expression, Histology and Histochemistry in the Interaction between Musa sp. and Pseudocercospora fijiensis. Plants. 2022; 11(15):1953. https://doi.org/10.3390/plants11151953
Chicago/Turabian StyleSoares, Julianna Matos da Silva, Anelita de Jesus Rocha, Fernanda dos Santos Nascimento, Vanusia Batista Oliveira de Amorim, Andresa Priscila de Souza Ramos, Cláudia Fortes Ferreira, Fernando Haddad, and Edson Perito Amorim. 2022. "Gene Expression, Histology and Histochemistry in the Interaction between Musa sp. and Pseudocercospora fijiensis" Plants 11, no. 15: 1953. https://doi.org/10.3390/plants11151953
APA StyleSoares, J. M. d. S., Rocha, A. d. J., Nascimento, F. d. S., Amorim, V. B. O. d., Ramos, A. P. d. S., Ferreira, C. F., Haddad, F., & Amorim, E. P. (2022). Gene Expression, Histology and Histochemistry in the Interaction between Musa sp. and Pseudocercospora fijiensis. Plants, 11(15), 1953. https://doi.org/10.3390/plants11151953








