1. Introduction
Crop diseases pose a serious threat to global food security [
1]. Disease identification methods that function well outside of the lab are needed to correctly identify diseases and prevent instances of incorrect chemical control [
1]. Crop disease monitoring by image analysis using a hand-held device such as a mobile phone is a goal of precision agriculture [
2]. Such a tool could be provided free to resource-limited smallholder farmers [
3]. It could also aid in high throughput phenotyping for rapid breeding of resistant crop varieties [
4].
Gray leaf spot (GLS) is caused by the foliar fungal pathogens
Cercospora zeina or
Cercospora zeae-maydis that can be responsible for significant yield losses [
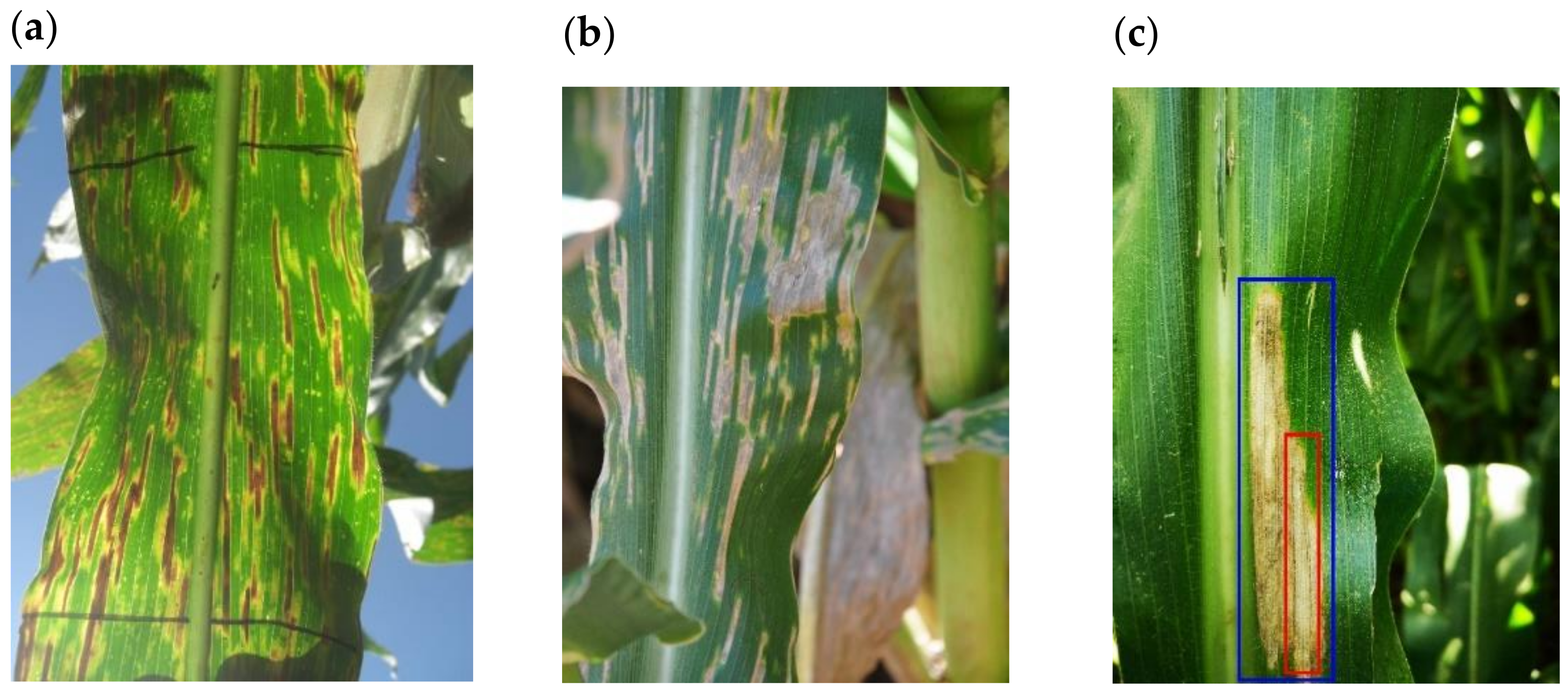
2]. It presents as small, rectangular, matchstick-like lesions that expand parallel to the leaf vein and rarely, if ever, cross it [
3]. These lesions start off as small yellowish discolorations on the leaf surface and gradually shift to a grayish-brown hue as the disease progresses.
Deep Learning (DL) is a technology that began gaining popularity in the late 1990s [
4,
5,
6], which enables the identification of features inside dynamic environments. The applications of DL are wide and varied. They have seen use in audio denoising [
7], land classification from satellite images [
8], self-driving cars [
9], drone detection [
10], and more. Over the past decade, DL has also been used for plant stress phenotyping, primarily using image data [
11,
12].
Most attempts to use DL to identify plant disease (
Table 1) do so using datasets generated under highly controlled lab conditions such as PlantVillage [
13]. These datasets typically lack the complications endemic to the field and omit confounding features such as insect damage, multiple diseases per leaf, coalescing lesions of the same or different diseases, varied backgrounds, heterogenous lighting conditions, foreign objects in a frame such as hands and feet, and so on. When models trained on these controlled datasets are asked to perform outside of ideal conditions, they tend to perform poorly [
13,
14].
There are a few papers that investigate DL and related approaches for disease identification in maize leaves [
15,
16,
17,
18,
19,
20]. However, most of these papers make use of PlantVillage images, apart from a series of studies on northern corn leaf blight (NCLB) detection in maize field trial images where only this disease was prevalent due to artificial inoculation [
16,
20,
21].
The PlantVillage dataset [
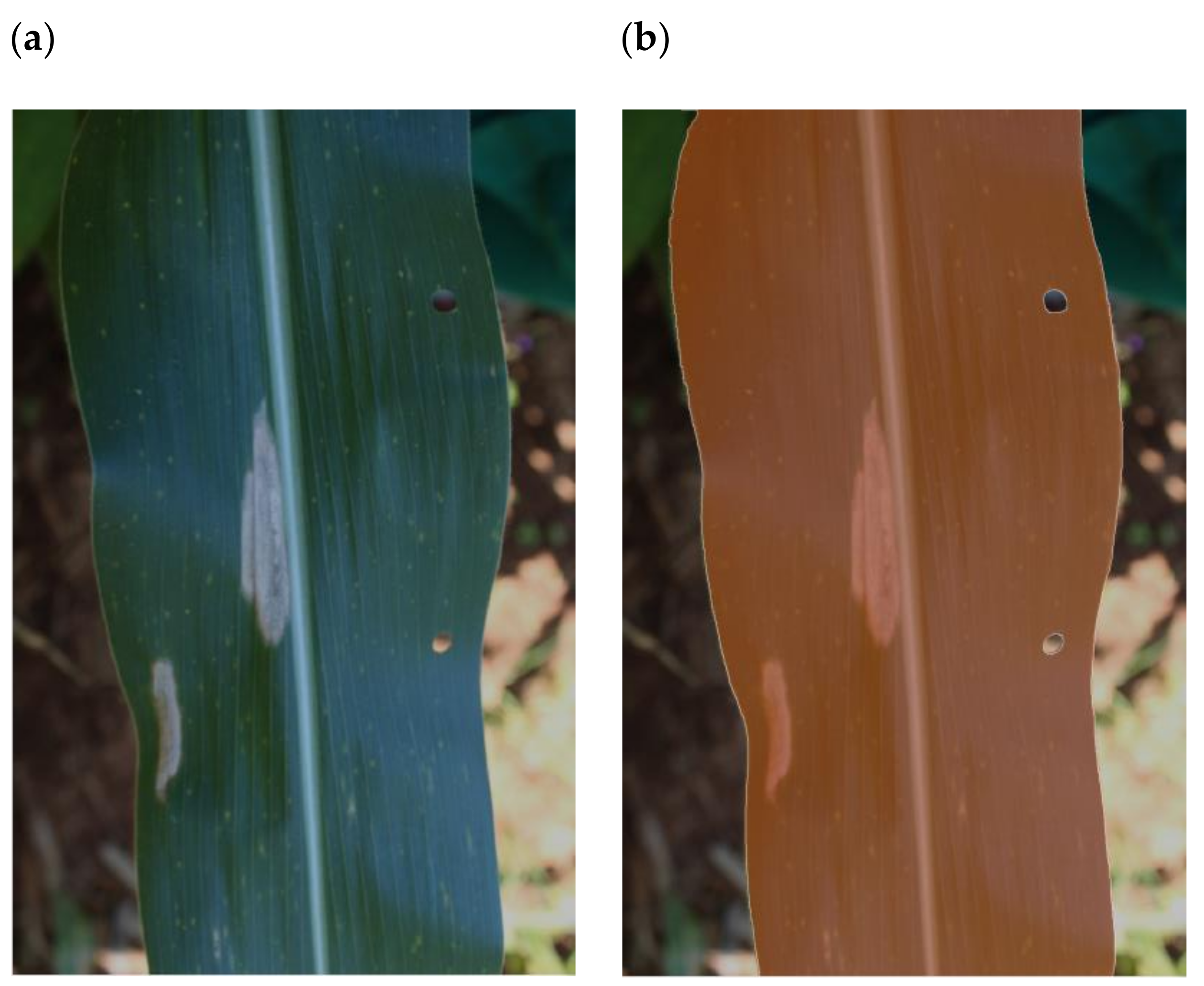
13] also contains several confounding features that impact the generalizability of models trained using this data. Namely, both GLS and NCLB had grey backgrounds (
Figure 1a,c), all images with Common Rust (CR) had a black background (
Figure 1b), and all healthy images had no background (
Figure 1d). In effect, this means that models trained using these images could make predictions based on the presence or absence of background pixels alone.
No approach could be found that accounts for the range of complexity in crop fields. There are three reasons thought to contribute to this: (i) the difficulty of generating a dataset of sufficient size and complexity; (ii) the need for plant pathology experts to label sufficient numbers of images for training the DL models; and (iii) datasets that produce high accuracies are more likely to be popularized than those that produce less useful results.
There is limited use in digital plant pathology of validation algorithms to confirm that DL models do indeed detect disease symptoms in images, for example, tools such as Grad-CAM [
22] and Grad-CAM++ [
23]. These algorithms produce heatmaps that overlay images that correlate with regions associated with class activation by a DL model. The heatmaps must be manually inspected and thus this approach is impractical at scale. However, they are vital in aiding explainability.
Image backgrounds are known to impact model performance [
24]. Some researchers may opt to remove the background from their images via segmentation tools such as GrabCut [
25,
26]. These interventions require human input which, as with Grad-CAM, limits their applications at scale. Some practitioners have used DL models in a pre-processing step to remove background from images, using tools such as MaskRCNN [
27,
28,
29].
The main shortcomings we identified in the field of artificial intelligence-based crop disease identification were the lack of research that makes use of in-field data in realistic conditions and a lack of methodologies that address the poor generalization exhibited by models trained on lab-based image datasets. There is limited research that accounts for multiple disease symptoms on one leaf. There are few papers that make use of explainability tools that report which pixels of the image were detected as a positive identification by the DL model [
38]. We aimed to address this by demonstrating how models perform under realistic (i.e., uncontrolled) conditions versus idealized conditions. In addition to this, we also propose a method for segmentation using a MaskRCNN network and investigate the effect of background removal on model performance.
To our knowledge, this paper is the first contribution to the DL-driven identification of GLS in maize with mixed diseases under field conditions. In this work, it is shown that DL is capable of scaling outside of lab conditions, provided that sufficient data can be made available.
4. Discussion
The main finding from this study was the development of a CNN named GLS_net, which could identify GLS disease symptoms on maize leaf disease images at an accuracy of 73.4%. Importantly, this accuracy was achieved from field images with symptoms of mixed diseases common in sub-Saharan Africa [
45]. The main diseases in addition to GLS, which has thin matchstick-like lesions, were NCLB which has larger cigar-shaped lesions with pointed ends, CR which is characterized by reddish-brown pustules, and PLS with white spots [
3,
45,
46]. The GLS_net CNN was developed using a relatively small dataset of 2332 images, but augmentation was used to increase the dataset 8-fold prior to training.
In this study, a second CNN named GLS_net_pv was trained using the PlantVillage maize disease dataset. This is a standardized dataset photographed in the lab against a homogenous background with single leaf images labelled as GLS, NCLB, CR, or no disease. GLS_net_pv achieved an accuracy of 94.1% on the PV testing set, which is similar to accuracies in the 90th percentile from previous deep learning models trained on the PV dataset [
13,
17,
19,
34,
35]. However, GLS_net_pv performed poorly at identifying GLS in the field-derived mixed disease dataset with an accuracy of 55.1%, which illustrates the problem of applying a lab-image trained model to more complex field images. In contrast, the mixed-disease field image trained model GLS_net performed well in GLS identification in the PV dataset (78.6% accuracy) and the field disease dataset (73.4% accuracy). We conclude that (i) models can be trained using data obtained under realistic conditions and still provide reliable disease predictions; and (ii) these models are more robust and consistent across datasets. The accuracy of GLS_net is likely to increase as more images are added to the dataset. Hyper-parameter tuning is an additional approach that could be used to improve the GLS_net model [
47].
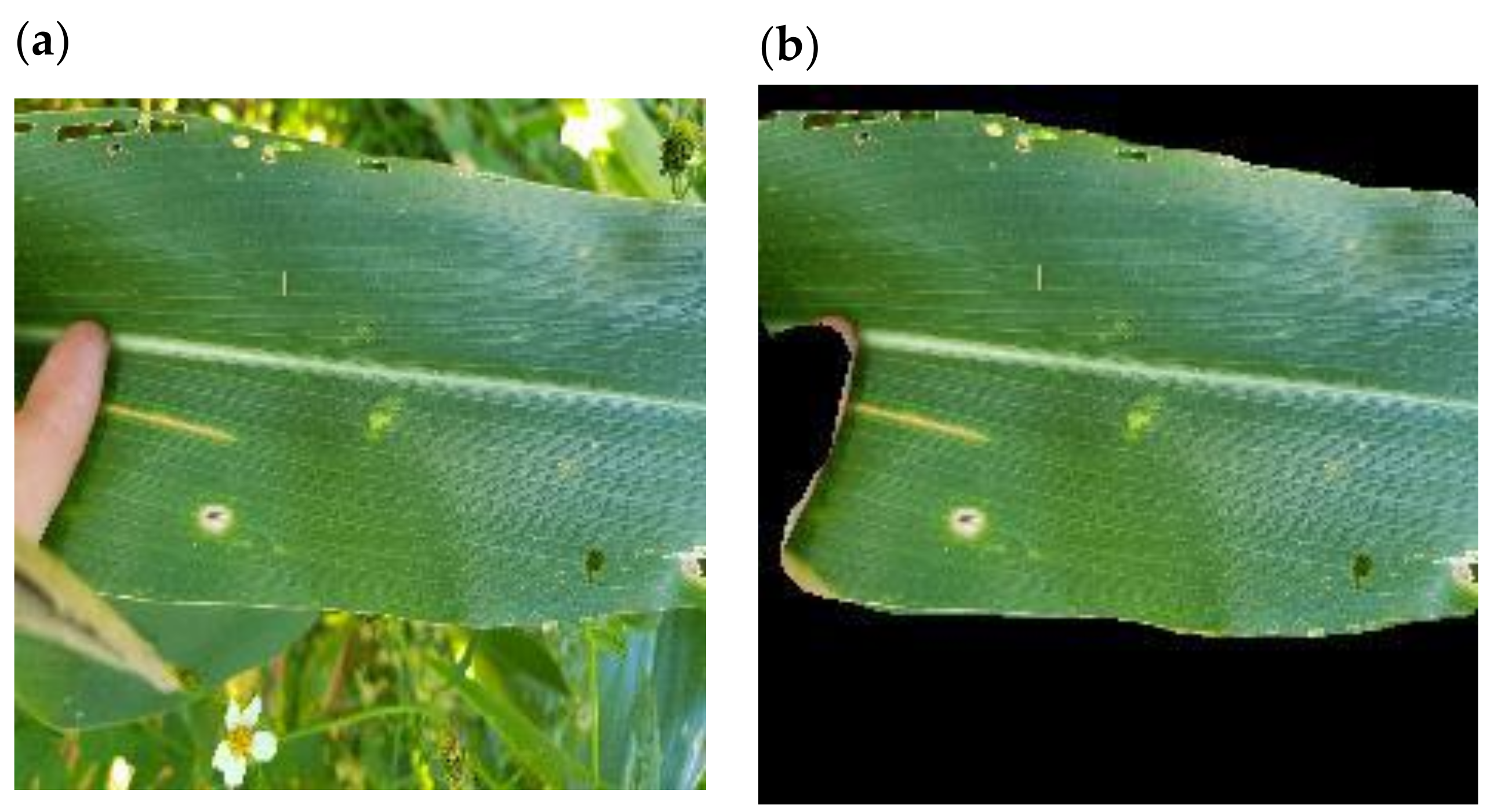
Background removal has been considered a method to improve the performance of CNNs by removing confounding objects from images [
27]. Field images of maize leaves with disease symptoms were thought to be good candidates for background removal since most images were made up of the main leaf in focus with different backgrounds. MaskRCNN has proven to be a useful tool for image segmentation [
28,
48]. In this study, it was adapted to produce a model called LeafRCNN which extracted maize leaves from their backgrounds. Importantly this was achieved with a relatively small subset of training images from the IF dataset, which were manually labelled using segments.ai [
40] for ground-truthing. LeafRCNN was then used to automatically remove the background of the complete IF dataset. Alternative methods of background removal such as GrabCut are potentially more time-consuming since GrabCut requires more manual intervention to be performed effectively [
49]. For this approach to work best, datasets should be comparatively homogenous, as is the case with the IF dataset where most images had a single leaf in focus.
Surprisingly, the removal of backgrounds from the maize mixed disease image set did not produce a CNN (GLS_net_noBackground) with better GLS identification than the CNN trained on the original images with backgrounds (GLS_net). GLS_net_noBackground had a 72.6% accuracy compared to GLS_net accuracy of 73.4% (
Table 5 and
Table 10). A possible reason may be that some networks employ contextual cues to perform classification. In this regard, Xiao et al. [
24] noted that some models in their study were able to achieve “non-trivial accuracy by relying on the background alone”. However, background removal has proven useful in some cases in improving CNN performance [
27,
38]. Further research is required to determine why the removal of backgrounds around maize leaf disease field images did not result in significant improvements to the DL-based mixed disease identification.
In this study, versions of GradCAM [
22,
23] were employed to attempt to identify which regions of images were activated by the GLS_net CNN. It was found that the GradCAM heatmaps were activated in the correct areas of GLS lesions in some images, however, GradCAM++ did not perform well since it activated non-disease regions of high contrast on the images. GradCAM has been used previously to interrogate CNNs developed for plant disease images [
38], however, performance was better for images where backgrounds had been removed. This indicates that further optimisation of validation tools is required to deal with complex subjects such as mixed disease images. Improved validation tools are required since it has been noted that implementing DL models in practise with a lack of explainability may hold ethical and legal implications [
50].
Plant disease image datasets that have been used for training DL models for disease identification have to date been focused on single diseases on a single leaf, for example, PlantVillage (54,306 images for 14 plant species) [
13] and the maize image database with NCLB images (18,222 images) [
51]. Such public datasets are commendable and have been used by others for the development of single disease/single leaf DL models [
15,
20,
30].
The goal of our study was to address the challenge of identifying GLS disease in field images where symptoms of more than one disease were present on one leaf, and thus we developed a custom dataset of 2332 images, which was increased to 18,656 by augmentation. We initially attempted to develop a GLS disease identification CNN (GLSnet_pv) using the PlantVillage dataset for training, however, the accuracy was not sufficient compared to the GLS_net trained on the more complex multi-disease dataset. This highlighted some of the limitations of lab image datasets such as PlantVillage. First, images are only labelled with a single disease, however, some leaves had additional disease symptoms (see
Figure 1b,c for examples). Second, the maize no-disease images were zoomed in so that the leaf filled the image with no background, whereas most maize disease images showed leaf pieces with either a grey or black homogenous background. A CNN trained on this dataset to distinguish between maize disease and no-disease could achieve an inappropriately high level of accuracy based on the presence or absence of background pixels.
There is a need in the discipline of plant disease diagnosis to expand the current image datasets that are available for developing artificial intelligence solutions with deep learning. In this study, maize disease images were labelled for the presence or absence of different diseases by experienced field plant pathologists, a low throughput process. In addition, leaf areas were extracted using an online tool [
40]. The bottleneck in generating useful datasets is labelling each image to indicate either (i) the presence/absence of a disease symptom; or (ii) segmenting each image to define the positions of disease symptoms. Segmentation is important for applications where disease quantification is required, such as in crop breeding for disease resistance [
28,
47]. Current image datasets have the limitation that they are static, and not updated. There is a need for a collaborative image database platform that is (i) open access, (ii) actively maintained and curated, and (iii) searchable.