Abstract
Blackcurrant reversion virus (BRV) is the most destructive currant-infecting and mite-transmitted pathogen from the genus Nepovirus. In this work, BRV transmission in the system Ribes ex vitro–Ribes in vitro was applied for the first time. Triple infection of BRV identified in blackcurrant cv. Gojai was used for phylogenetic analysis and inoculation assay. Transmission of BRV was successful due to its stability in the inoculum for up to 8 days at 4 °C; all BRV isolates were infectious. Our suggested inoculation method through roots was applied in six Ribes spp. genotypes with 100.0% reliability, and the expression levels of defence-related gene PR1 to biotic stress was observed. The prevalence of the virus in microshoots after 2–14 days post-inoculation (dpi) was established by PCR. In resistant genotypes, the BRV was identified up to 8 dpi; meanwhile, infection remained constant in susceptible genotypes. We established that BRV transmission under controlled conditions depends on the inoculum quality, post-inoculation cultivation temperature, and host-plant susceptibility to pathogen. This in vitro inoculation method opens possibilities to reveal the resistance mechanisms or response pathways to BRV and can be used for the selection of resistant Ribes spp. in breeding programs.
1. Introduction
Blackcurrant reversion virus (BRV) is transmitted by a specific biological vector—eriophyid mites from Cecidophyopsis spp. (Acari: Eriophyidae). The virus is the causative agent of blackcurrant reversion disease (BRD). The pathogen (BRV), vector (C. ribis), and disease (BRD) complex causes significant yield losses in blackcurrant plantations worldwide [1]. R. nigrum L. is the primary natural host of BRV, although natural infestations also occur in other species—R. pauciflorum, R. rubrum, R. alpinum, R. aureum, etc. One of the best ways to reduce BRV economical losses is the targeted selection of genetically resistant cultivars [2]. Species R. uva-crispa, R. cereum, R. dikuscha, R. nigrum var. sibiricum, etc., have resistance determined by genes to gall mite and consequently to BRV [3,4,5,6]. However, only several molecular markers of resistance genes are known and can be used for marker-assisted selection (MAS) in blackcurrant breeding programs [7,8,9].
The BRV belongs to the subgroup c of the genus Nepovirus, family Secoviridae [10,11]. Virus isometric particles are approximately 27 nm in diameter, and the viral genome consists of two polyadenylated, single-stranded positive-sense RNA1 and RNA2 molecules [12,13]. It is known that the 3′ nontranslated region (NTR) of RNAs are essential for virus lifecycles, promoting translation regulation and its efficiency [14], and consequently were used for BRV detection or phylogenetic analysis [6,15,16,17].
The BRD according to symptoms is divided into two forms—European (E) and Russian (R) [4,18]. The main difference between the forms is the severity and rate of symptoms expressed in the infected plants. Both forms cause a transformation in leaf shape, a decrease in marginal serration and in the number of main veins and hair numbers, and an increase in colour intensity on the flower buds. However, the flower sterility resulting in a complete loss of berry yield occurs only in the R form of reversion disease [4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19].
Atypical to other Nepovirus, BRV is not transmitted through seeds or pollen. The BRV, the biological vector, and the host plant have a specific relationship [20]. Mechanical inoculations using sap extracts are usually complicated and fail by the very low concentration of virus particles, their erratic distribution in plants, and the rapid inactivation of the virus by phenolic compounds in leaves of Ribes plants [10,19,21]. BRV transmission in vivo is possible by the graft inoculation method in R. nigrum [18,22,23]. In this case, reversion symptoms on plants occur after 2–4 vegetation seasons [20,23]. This inoculation technique in field or greenhouse conditions is not suitable for rapid and sensitive biotechnological and genetic research. A more reliable inoculation method of BRV for biotechnological purposes would be under in vitro conditions, but such research is limited. No scientific data about successful cases of BRV inoculation in the Ribes to Ribes system are available.
Inoculation of plants in vitro would offer the potential for great savings in time and for efficient experimentation under controlled conditions. It is an accurate tool to study the epidemiology of BRD and the interrelationships between the virus and the host at molecular and genetic levels. Thus, the aim of this study was to develop a rapid and efficient inoculation method for BRV under in vitro conditions, emphasizing genetic diversity, stability, and infectivity of BRV.
2. Results
2.1. Source of BRV for Inoculation In Vitro
Heterogeneous infection of BRV in the same plant of Lithuanian blackcurrant cv. Gojai was detected. This was proved by the multiple alignment analysis of 3′ NTR RNA2 sequences. Three different BRV isolates with various mutations were found: BRV_1-18_LT, BRV_7-18_LT, and BRV_3-18_LT. Virus isolates identified in this study were uploaded into the NCBI database with accession numbers MH891843, MH891844, and MH891845. Genetic identity among isolates ranges from 94.6 to 99.6% in the 3′ nontranslated region of RNA2 (Table 1).

Table 1.
Percent identity of BRV isolates at nucleotide level of RNA2 3′ NTR regions found in Lithuanian cv. Gojai.
A phylogenetic dendrogram was constructed to determine the viral infection affinity using sequences of BRV identified in Lithuania and sequences submitted to the NCBI database (Figure 1). Worldwide isolates of BRV formed two reliably distinct branches at 90.0% bootstrap. The tree provides information on the geographic origin of the isolates and the implicit migration of infection. According to the symptoms of the disease caused in host plants, virus isolates in the phylogenetic tree were classified as the R (Russian) form of BRD in most cases, except for the isolate from Canada or plants with virus symptoms. All isolates from Lithuania were grouped into the phylogenetic dendrogram’s first branch. In this branch, BRV isolates were still reliably separated into several groups, and BRV_1_18_LT and BRV_7_18_LT formed a separate phylogenetic branch at 100.0% bootstrap. These sequences of RNA2 3′ NTR were genetically close to each other but significantly genetically diverse from other BRV isolates worldwide. Isolate BRV_3_18_LT showed diversity from BRV_1_18_LT and BRV_7_18_LT but was genetically close to an isolate from Poland (AF321570).
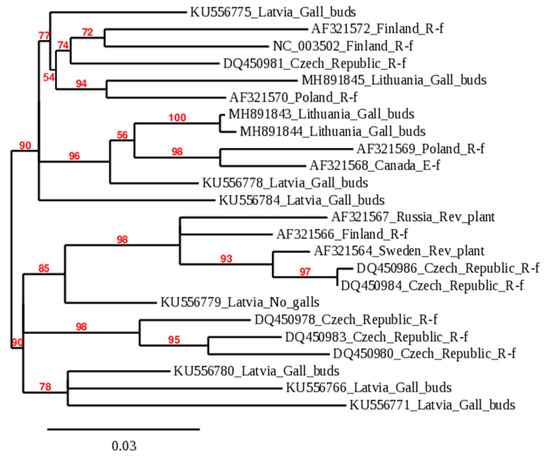
Figure 1.
Genetic relationship among worldwide isolates of BRV according to 3′ NTR RNA2 sequences. The label of members in the phylogenetic tree consists of accession number in NCBI_country of origin_blackcurrant reversion disease form or symptom. Accession numbers of BRV sequences obtained in this research are MH891843–MH891845; characteristics of isolates of other plants shown in Figure 1 are available in Table S2 in Supplementary Materials.
2.2. Reliability of the Inoculation In Vitro
Symptomatic leaves of blackcurrant cv. Gojai containing heterogeneous infection of BRV, obtained from the plant genetic resources collection of LAMMC, were used in the preparation of inoculum. Stability of RNA2 of BRV in inoculum was investigated. Prepared inoculum was maintained at 4 and 21 °C for up to 8 days. Specific primer pairs (Table 2) for RNA2 were used for BRV identification. Amplification results of BRV-specific fragments in inoculum during different storage temperatures are presented in Figure 2. Virus RNA2 was detected in inoculum at 4 °C during the entire period of the experiment (Figure 2A), while BRV-specific fragments were amplified and clearly shown in the inoculum only after the first day in storage at 21 °C (Figure 2B).
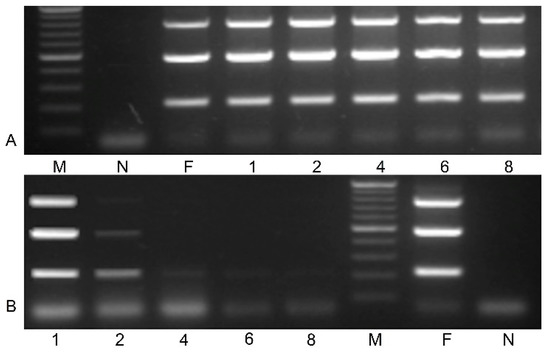
Figure 2.
Stability of virus RNA2 in inoculum at 4 °C (A) and 21 °C (B) storage temperature (M—gene ruler 100–1000 bp; N—negative control; F—fresh inoculum; inoculum after storage 1–8 days. PCR mix without cDNA was used as a negative control.
The standard curve for quantification of the virus concentration by RT-PCR with eight points was constructed (Figure 3). Plasmid vector with RNA2 3′ NTR of the BRV was used to determine the number of copies of the virus. Through the range from 1.98 × 103 to 1.98 × 1010 copies per reaction, the threshold cycle (Ct) and copy numbers displayed a linear relationship with an R2 of 0.9986 and the equation y = –3.6614x + 29.923 (Figure 3). The value of Ct obtained in the inoculum was 21.73 (±0.15).
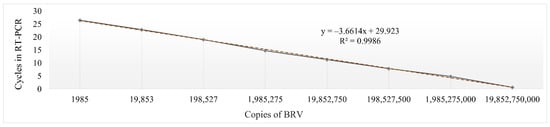
Figure 3.
The standard curve of RT-PCR detecting BRV. The plasmids were used to establish the standard curve and the software Microsoft Excel was used to analyse the relationship of Ct values and plasmid copy numbers.
To determine the efficiency of the inoculation method in vitro, microshoots with roots of R. aureum, R. dikuscha, and R. nigrum cvs. Aldoniai, Ben Gairn, Ben Tirran, and Vernisaz were inoculated by soaking in the same way as shown in Figure 4. The roots increase the contact area between the inoculum and the plant, and enhance the entry of the virus into the phloem. Based on our data in Figure 2, a post-inoculation period at 4 °C is necessary to ensure viability of the virus on the surface of the roots.

Figure 4.
In vitro inoculation of R. nigrum microshoots: (A) microshoot before rooting; (B) microshoot with roots; (C) microshoot soaked in the tube with inoculum.
In the post-inoculation period at 2, 4, 6, 8, 10, and 14 days, the virus infection in microshoots without roots was evaluated by PCR (Figure 5). According to the data for 481 bp-length BRV-specific fragment sequencing, all isolates of BRV were transmitted to infected plants by in vitro mechanical inoculation. The nucleotide sequences of these fragments completely coincided with the sequence provided to NCBI (MH891843, MH891844, and MH891845). This indicates that BRV strains obtained in the cultivar Gojai have the potential to contaminate and spread in an in vitro system.
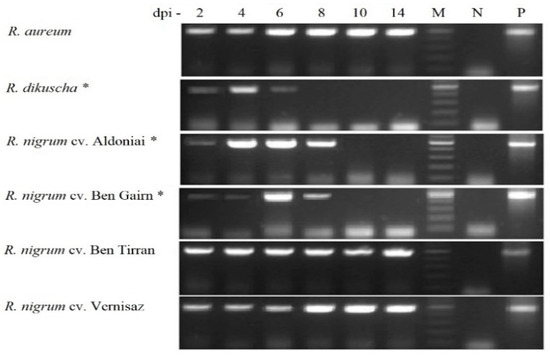
Figure 5.
PCR product of the BRV (481 bp) in Ribes microshoots after 2–14 dpi. M—gene ruler 100–500 bp; N—negative controls were virus free plants; P—positive control was inoculum; *—BRV-resistant genotypes.
In vitro plants of R. dikuscha and R. nigrum cvs. Aldoniai or Ben Gairn remained symptom-free in the post-inoculation period. Clearly, symptoms of response to biotic stress were visible on R. aureum microplants after 6 dpi (Figure 6). The leaves changed their colour, and the clear synthesis of the stress-characteristic phenolic compounds became visible.
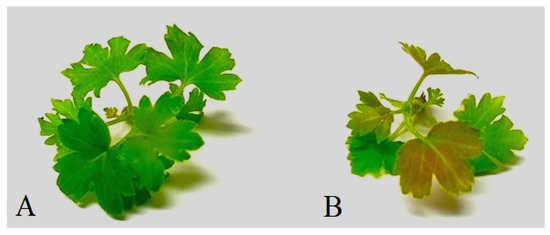
Figure 6.
Microshoots of R. aureum after 6 dpi: (A) BRV-free mock-inoculated microshoot; (B) BRV-infected microshoot.
The incidence rate of BRV in all Ribes cultivars and species was 100% for 20 microshoots in each treatment. However, the persistence of the infection in the plant was different. Until the end of the experiment, 2 weeks after inoculation, the infection remained constant in the BRV-susceptible genotypes: R. aureum and R. nigrum cvs. Ben Tirran and Vernisaz. The viral infection was detected in the resistant plants, R. dikuscha and R. nigrum cv. Ben Gairn, until 6 dpi, and in R. nigrum cv. Aldoniai until 8 days after infection (Figure 5).
2.3. Relative Gene Expression in Response to BRV Infection
The quantitative expression profiles of the pathogenesis-related 1 (PR1) gene after BRV infection were compared in both resistant and susceptible genotypes for treatment periods 2, 4, 6, and 8 dpi (Figure 7). We established that PR1 was rapidly induced after BRV infection in Ribes spp. The expression levels were significantly higher in all genotypes in comparison with the control plants during the entire period of inoculation. The highest expression values of PR1 were presented in virus-susceptible genotypes R. nigrum cvs. Ben Tirran and Vernisaz at 4- and 8-days post-inoculation. Relative PR1 expression confirms successful virus infection in all microshoots in vitro; gene response to biotic stress was found in all treatments after using our proposed in vitro inoculation method through roots.
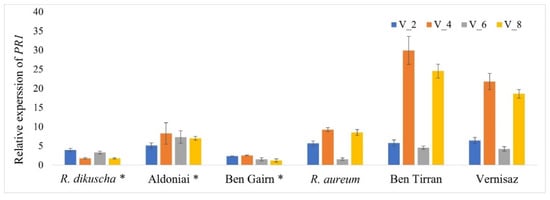
Figure 7.
Relative expression levels of PR1 in resistant and susceptible Ribes genotypes in response to BRV infection. The expression of each sample was calculated from the control mock-inoculated plant at points 2, 4, 6, and 8 dpi; *—BRV-resistant genotypes.
3. Discussion
A sensitive plant-testing system is an important tool for targeted and accelerated selection of resistant genotypes in Ribes breeding programs. The inoculation method under in vitro conditions is appropriate to screen genotypes for their responses to pathogens and could be a useful tool for better understanding the virus–host interaction and resistance mechanisms. Virus inoculation systems suitable for in vitro conditions have been developed in herbaceous plants such as potatoes, lettuce, and tomatoes [24,25,26]. However, inoculation systems for woody perennial plants under in vitro conditions are unexplored. In general, an in vitro system for orchard and berry cultures is usually used for virus elimination and micropropagation [27].
BRV is one of the most serious pathogens in blackcurrant plantations; its control is complicated and the consequences are economically damaging [28]. Heterogenic infection of BRV in the same host plant of field-grown cv. Gojai was shown by the diversity of the sequences (Table 1) and the data of phylogenetic analysis (Figure 1). In this study, we found that multiple infections of BRV with different origins could be detected in one host plant. The natural vector transmitting BRV among bushes is the gall mite. Under controlled conditions, transmission assays showed that 3 h of mite feeding on blackcurrant seedlings was sufficient for virus transmission [21]. However, previous studies have shown that using the gall mite infection method in vitro had difficulties in controlling both the infection pressure and the influence of environmental conditions on the spread of mites and BRV [28,29].
The BRV genome consists of two molecules of positive-sense RNAs: RNA1 and RNA2, which are packed in viral particles. BRV particles are composed of two capsid proteins (54 and 55 kDa) and can replicate in a low range of hosts [22,30]. The 3′ NTRs of BRV are highly similar and the most stable parts of the genome, and are therefore used for detection by PCR [15,31]. Genetic identity among isolates, from 94.6 to 99.6% in the 3′ nontranslated region of RNA2, was also found in our study (Table 1). Only virus particles formed from the 54 kDa fragment can be used for indicator plant infection by mechanical inoculation, but the resulting progeny viruses contain both forms [32]. For the first time, blackcurrant reversion virus was successfully mechanically transmitted from symptomatic blackcurrant leaves to Chenopodium quinoa [22] and to other herbaceous plants, Nicotiana occidentalis and Nicotiana tabacum [17]. These cases of mechanical inoculation are the only two reported in the literature where the virus was transmitted from blackcurrants to herbaceous plants, despite the attempts of scientists to repeat it. Despite these difficulties, sap from C. quinoa symptom-bearing leaves was transmitted from 4 to 11 days using carborundum to other herbaceous plants: C. amaranticolor, C. murale, N. benthamiana, N. clevelandii, N. debnyi, and N. occidentalis [16,22]. The successful slash inoculation of young tissue-culture-propagated blackcurrants with the isolated virus from C. quinoa was performed by A. Lemmetty and K. Lehto [23]. This plant material is more sensitive due to its soft tissues and low content of phenolic compounds. The first symptoms on the leaves were observed after 5–7 months. Furthermore, the visual symptoms of the disease appeared only after 4–5 years when inoculating blackcurrant woody plants. Additional fulfilling of Koch’s postulates was achieved when mechanically inoculated plants developed typical BRD symptoms on leaves and flower buds [15,18].
It was observed that some plant viruses are difficult to inoculate mechanically because they are transmitted by arthropods during feeding and their localisation to vascular tissue (usually phloem) is limited [33]. The BRV can reach low titres at the beginning of the spread of the infection in the Ribes plants, and does not give a titre in genetically resistant plants under natural conditions. Specific antibodies for BRV proteins are not commercially available [10,20]; therefore, polymerase chain reaction is the most sensitive and reliable detection method at present. Using PCR in this study, BRV infection was approved until 8 dpi in BRV-resistant plants R. dikuscha, cv. Aldoniai (R. dikuscha in progeny) [34] and cv. Ben Gairn (R. uva-crispa in progeny) [5].
The amount of virus in the inoculum is an important factor for mechanical infection transmission. Too high or too low virus concentration in the inoculum leads to unsuccessful plant infection [35]. For example, wheat streak mosaic virus (WSMV) inoculum with 2.21 × 106 concentration was most suitable for mechanical inoculation in wheat [36]. In this study, for inoculum preparation we mixed the inoculation buffer with plant sap and triple BRV infection (approximately 2.24 × 104 copies in µL) (Figure 3). All three isolates of BRV (Figure 1) appeared infectious according to the homology of 481 bp fragment (Figure 5) sequencing in this study.
Pathogenesis-related 1 gene is the most abundant gene family of PRs, and the expression of PR1 is used as a molecular marker to indicate plant defence response to different types of insects and pathogens [37]. In our previous study, the biotic stress defence response PR1 homolog emphasizing the R. nigrum genome was found, and the primer pair identifying this gene was designed [38]. Therefore, in this work, RT-PCR was used to establish the host-plant defence response to viral infection, as biotic stress, for showing the success of the suggested inoculation method (Figure 7). We determined the expression profiles of PR1 in resistant and susceptible genotypes during the entire period of the experiment after mechanical BRV inoculation in vitro. The defence response pathway with the determined PR1 expression was characteristic for all genotypes, regardless of their resistance to BRV. The expression levels of PR1 in susceptible genotypes R. nigrum cvs. Ben Tirran and Vernisaz were higher compared to susceptible R. aureum and resistant genotypes. However, all susceptible genotypes had a similar tendency showing significant increase in PR1 at 4 and 8 dpi. It was found that in the Ribes spp., as in other plants, PRs are necessary for the establishment of systemic acquired resistance (SAR) in tissues distant from the primary infection site [39].
The in vitro inoculation technique described here is the first reported in the literature where the BRV was successfully transmitted from blackcurrants to blackcurrants with maximum reliability. Several virus transmission methods (leaf damage with carborundum powder; stem soaking in the inoculum; inoculum dripping with syringe into the stem) have been tested on microshoots during our research work, but all of them have failed. Possibilities to successful viral infection through inoculation of Ribes plants grown in vitro by using a mechanical approach are possible when the appropriate conditions are used (Figure 4). Rooted microshoots were the most suitable plant material for easy virus entry and movement in the plant because of its soft tissues and low content of phenolic compounds. Based on studies by P. Susi [10] and our data (Figure 2), cold storage (4 °C) in the first period after inoculation is necessary to ensure virus viability and for causing additional abiotic stress in the plant.
It is suggested that the procedure described in this study may be widely applicable for blackcurrant reversion virus mechanical inoculation using sap from R. nigrum. The principle for the inoculation method depends on soaking the roots of microshoots in the PBS buffer containing an infectious BRV isolate. The method was used successfully to inoculate susceptible Ribes spp. plants with BRV and to study the genetic response for BRV in resistance plants.
4. Materials and Methods
4.1. Plant Material
Leaves of R. nigrum cv. Gojai with symptoms of BRD presented in field conditions were used for preparation of inoculum. Inoculum containing BRV was tested for virus stability, infectivity, and genetic analysis. Rooted microshoots of the species R. aureum, R. dikuscha and R. nigrum cvs. Aldoniai, Ben Gairn, Ben Tirran, Vernisaz were used for inoculation treatments under in vitro conditions.
4.2. In Vitro Culture of Ribes spp.
All manipulations with plants in the in vitro system were performed using modified MS medium (Table S1 in Supplementary Materials) [40]. Microshoots of Ribes species and cultivars were propagated on medium I (3 weeks), grown on medium II up to 3–4 cm (2 weeks), and rooted on nutrition medium III (4 weeks). Plants grown in this way were used in the inoculation assay in vitro. Micropropagation and rooting of Ribes were carried out under controlled conditions in a growth chamber at 21 ± 2 °C, 50–150 µmol m–2 s–1 light intensity, and 16/8 h photoperiod.
4.3. Preparation of Inoculum
The inoculum was prepared from 3.0 g of ground symptomatic blackcurrant cv. Gojai leaves and 10.0 mL of 0.1 M phosphate buffered saline (PBS) buffer (pH 7.5) enriched with 2.0% polyvinylpyrrolidone (PVP), 0.2% sodium sulphite (Na2SO3), 0.08% 2-mercaptoethanol, and 0.2% Tween 20. The inoculum was filtered through 0.22 µm pore-size sterile filters for microorganism contamination.
4.4. Evaluation of BRV Stability in the Inoculum
Aliquots of the prepared inoculum were stored at 4 and 21 °C for 8 days to assess virus stability. RNA was extracted from each treatment of inoculums (fresh and after 1, 2, 4, 6, and 8 days). Virus RNA was evaluated by the PCR method using 3 primer pairs from different sites of the RNA2 3′ NTR regions in the BRV genome (Table 2).

Table 2.
Primer pairs used for BRV detection and sequencing.
Table 2.
Primer pairs used for BRV detection and sequencing.
| Primer | Sequence 5′ to 3′ | Annealing T, °C | Length, bp | Purpose in Research | Reference |
|---|---|---|---|---|---|
| P1/P2 | GTAATACGCTGGTGTCTC/ GAAAGGACATTTCAGCTC | 49 | 215 | For detection of RNA2 plants | [15] |
| P5/P6 | AAACCAGACCCAGGTGAGTG/GGACACTTCCATATAAGTCGGC | 60 | 481 | For detection of RNA2 in plants and inoculum | [31] |
| BCP11/P6 | ATTTCGAGCTGTATGGTCG/CTCGGAAGCAGTAGACCT | 51 | 787 | For detection of RNA2 in inoculum | [15] |
| BCP11/P2 | ATTTCGAGCTGTATGGTCG/GAAAGGACATTTCAGACTC | 51 | ~1449 | For sequencing and genetic analysis of RNA2 | [15] |
4.5. Total RNA Isolation and cDNA Synthesis
Total RNA was isolated from 100.0 µL of inoculum or 0.1 g fresh tissue of microshoots. Plant samples were homogenized in liquid nitrogen. For extraction of total RNA, we used a GeneJET Plant RNA Purification Mini Kit (Thermo Scientific, Vilnius, Lithuania) according to manufacturer’s protocol. Total RNA was used directly as a template for cDNA synthesis with a Maxima H Minus First Strand cDNA Synthesis Kit (Thermo Scientific, Vilnius, Lithuania), and oligo d(T)20 primer was used in the reaction.
4.6. BRV Detection by PCR
The detection of BRV was carried out by polymerase chain reaction (PCR) in the inoculum and in the Ribes spp. microshoots after mechanical inoculation in vitro. BRV-specific oligonucleotide primers and their characteristics are presented in Table 2. PCR reaction was performed in 20.0 µL reaction volume consisting of 11.1 µL H2O, 2.5 µL 10× Taq buffer + (NH4)2SO4 + MgCl2, 2.0 µL 25 mM MgCl2, 2.0 µL 2 mM dNTP mix, 0.1 µL of each 0.1 µM forward and reverse primers, 0.2 µL Taq DNA polymerase (Thermo Scientific, Vilnius, Lithuania), and 2.0 µL cDNA (about 100 ng). The amplification reaction was performed in a Mastercycler X50a (Eppendorf, Stevenage, UK) under the following conditions: 95 °C for 3 min, 35 cycles at 95 °C for 30 s, 40 s at temperature suitable for primers (Table 2), 72 °C for 40 s, and the final elongation step at 72 °C for 10 min. The amplification products were analysed in 1.5% (w/v) agarose gel using electrophoresis and visualized by ethidium bromide staining and UV illumination. The size of the PCR products was determined with O’GeneRuler 1kb DNA Ladder (Thermo Scientific, Vilnius, Lithuania).
4.7. Purification, Cloning, Sequencing, and Digestion of RNA2 3′ NTR of BRV
Amplified fragments for sequencing with primer pair BCP11/P2 (Table 2) were purified using a GeneJET Gel Extraction Kit (Thermo Scientific, Vilnius, Lithuania). Fragments were ligated into pJET 1.2 blunt vector using a CloneJET™ PCR Cloning Kit (Thermo Scientific, Vilnius, Lithuania). The strain E. coli JM107 was transformed using TransformAid Bacterial Transformation Kit (Thermo Scientific, Vilnius, Lithuania). Ten samples of plasmids with BRV insertions were sequenced using a Big Dye Terminator v 3.1 Cycle Sequencing Kit and performed on a 3130 Genetic Analyzer (Applied Biosystem, Waltham, MA, USA).
4.8. Plant Inoculation In Vitro
Before inoculation, virus-free rooted microshoots were placed in the dark for 48 h at 21 ± 2 °C. Under sterile conditions, roots of 4-week-old microshoots were shortened with a scalpel to a length of 4 mm and soaked for 2 min in sterile tubes with fresh inoculum containing BRV (Figure 4). Inoculated plants were replanted on nutrition medium II (Table S1 in Supplementary Materials) and cultivated for 2 weeks at 4 °C, 16/8 h photoperiod. Mock-inoculated microshoots in PBS buffer were used as the negative control. For RNA isolation, 20 microshoots of each genotype were frozen in liquid nitrogen after cutting their roots and immediately used for RNA isolation.
4.9. Gene Expression Analysis in Inoculated Ribes Microplants
Changes in the putative PR1 gene in mRNA of inoculated blackcurrant genotypes in vitro were analysed using quantitative real-time RT-PCR (qPCR). Reaction was performed in a 11.0 µL reaction volume containing 5.95 μL H2O, 1.25 μL 10× Taq buffer + (NH4)2SO4 + MgCl2, 1.0 μL 2 mM dNTP mix, 1.0 μL 25 mM MgCl2, 1.0 µL cDNA, 0.1 μL Taq DNA polymerase (Thermo Scientific, Vilnius, Lithuania), 1.0 μL 20× EvaGreen dye (Biotium, Inc., Fremont, CA, USA), and 0.1 μL of each forward and reverse primer [38]. The analysis was carried out on three biological replicates using a Realplex RT-PCR cycler (Eppendorf, Stevenage, UK) under following conditions: 95 °C for 3 min, 35 cycles at 95 °C for 30 s, 55 °C for 40 s, and 72 °C for 40 s; melting point of PCR products: 95 °C for 15 s, 60 °C for 15 s, 60–95 °C for 20 min, 95 °C for 15 s, and hold at 4 °C.
4.10. Statistical Analysis
Sequences of the three Lithuanian BRV isolates were uploaded into the NCBI database (accession numbers MH891843, MH891844, and MH891845). A percent identity matrix among the 3′ NTR of RNA2 sequences of BRV was created by the Clustal 2.1 program. The phylogenetic tree was constructed comparing 3 Lithuanian BRV sequences with 21 sequences of homologous region of BRV with reference to the NCBI database. Phylogenetic analysis was performed using the maximum likelihood method implemented in the PhyML program; a bootstrap analysis with 100 replications was performed [41]. Relative expression was assessed by the 2(-Delta Delta C (T)) method [42]. An actin gene, having constant expression levels (data not shown), was used to normalize raw data and calculate relative transcript levels. Means and SEM (standard error of the mean) from independent experiments were subjected to STAT-ENG.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/plants11131635/s1, Table S1: Composition of modified MS [35] nutrition medium for propagation, growth, and rooting of blackcurrant microshoots, Table S2: Accession numbers and details of blackcurrant BRV sequences obtained from the NCBI database and included in the phylogenetic tree of this study.
Author Contributions
Conceptualization, I.M. and A.D.J.; methodology, A.D.J. and I.M.; software, I.M. and A.D.J.; formal analysis, V.S.; investigation, A.D.J. and I.M.; resources, V.S. and A.D.J.; data curation, A.D.J.; writing—original draft preparation, A.D.J.; writing—review and editing, I.M. and V.S.; visualization, A.D.J. and I.M.; supervision, I.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Šutic, D.D.; Ford, R.E.; Tošic, M.T. Virus diseases of small fruits. In Handbook of Plant Virus Diseases; Šutic, D.D., Ford, R.E., Tošic, M.T., Eds.; CRC Press: Boca Raton, FL, USA, 1999; pp. 433–475. [Google Scholar]
- Šikšnianas, T.; Stanys, V.; Stanienė, G.; Sasnauskas, A.; Rugienius, R. American black currant as donor of leaf disease resistance in black currant breeding. Biologija 2005, 3, 65–68. [Google Scholar]
- Anderson, M.M. Resistance to gall mite (Phytoptus ribis Nal.) in the Eucoreosma section of Ribes. Euphytica 1971, 20, 422–426. [Google Scholar] [CrossRef]
- Adams, A.N.; Thresh, J.M. Reversion of black currant. In Virus Diseases of Small Fruits; Converse, R.H., Ed.; USDA Agriculture: Washington, DC, USA, 1987; pp. 133–136. [Google Scholar]
- Brennan, R.M. Currants and gooseberries. In Temperate Fruit Crop Breeding; Hancock, J.F., Ed.; Springer: Dordrecht, The Netherlands, 2008; pp. 177–196. [Google Scholar] [CrossRef]
- Zulģe, N.; Gospodaryka, A.; Moročko-Bičevska, I. Occurrence and genetic diversity of Blackcurrant reversion virus found on various cultivated and wild Ribes in Latvia. Plant Pathol. 2018, 67, 210–220. [Google Scholar] [CrossRef]
- Brennan, R.M.; Jorgensen, L.; Gordon, S.; Loades, K.; Hackett, C.; Russell, J. The development of a PCR-based marker linked to resistance to the blackcurrant gall mite (Cecidophyopsis ribis Acari: Eriophyidae). Theor. Appl. Genet. 2009, 118, 205–211. [Google Scholar] [CrossRef]
- Mazeikiene, I.; Bendokas, V.; Stanys, V.; Siksnianas, T. Molecular markers linked to resistance to the gall mite in blackcurrant. Plant Breed. 2012, 131, 762–766. [Google Scholar] [CrossRef]
- Mazeikiene, I.; Juskyte, A.D.; Stanys, V. Application of marker-assisted selection for resistance to gall mite and Blackcurrant reversion virus in Ribes genus. Zemdirbyste 2019, 106, 359–366. [Google Scholar] [CrossRef]
- Susi, P. Black currant reversion virus, a mite-transmitted nepovirus. Mol. Plant Pathol. 2004, 5, 167–173. [Google Scholar] [CrossRef]
- Thompson, J.R.; Dasgupta, I.; Fuchs, M.; Iwanami, T.; Karasev, A.V.; Petrzik, K.; Sanfaçon, H.; Tzanetakis, I.; van Der Vlugt, R.; Wetzel, T.; et al. ICTV virus taxonomy profile: Secoviridae. J. Gen. Virol. 2017, 98, 529–531. [Google Scholar] [CrossRef]
- Pacot-Hiriart, C.; Latvala-Kilby, S.; Lehto, K. Nucleotide sequence of black currant reversion associated nepovirus RNA1. Virus Res. 2001, 79, 145–152. [Google Scholar] [CrossRef]
- Latvala-Kilby, S.; Lehto, K. The complete nucleotide sequence of RNA2 of blackcurrant reversion nepovirus. Virus Res. 1999, 65, 87–92. [Google Scholar] [CrossRef]
- Karetnikov, A.; Keränen, M.; Lehto, K. Role of the RNA2 3′ non-translated region of Blackcurrant reversion nepovirus in translational regulation. Virology 2006, 354, 178–191. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lemmetty, A.; Latvala, K.S.; Lehto, K. Comparison of different isolates of black currant reversion virus. Acta Hortic. 2001, 551, 45–49. [Google Scholar] [CrossRef]
- Lehto, K.; Lemmetty, A.; Keränen, M. The long 3’ non-translated regions of Blackcurrant reversion virus RNAs are highly conserved between virus isolates representing different phenotypes and geographic origins. Arch. Virol. 2004, 149, 1867–1875. [Google Scholar] [CrossRef]
- Přibylová, J.; Špak, J.; Petrzik, K.; Kubelková, D.; Špaková, V. Sequence comparison and transmission of Blackcurrant reversion virus isolates in black, red and white currants with black currant reversion disease and full blossom disease symptoms. Eur. J. Plant Pathol. 2008, 121, 67–75. [Google Scholar] [CrossRef]
- Jones, A.T.; McGavin, W.J. Improved PCR detection of Blackcurrant reversion virus in Ribes and further evidence that it is the causal agent of reversion disease. Plant Dis. 2002, 86, 1333–1338. [Google Scholar] [CrossRef]
- Jones, A.T. Black currant reversion disease – the probable causal agent, eriophyid mite vectors, epidemiology and prospects for control. Virus Res. 2000, 71, 71–84. [Google Scholar] [CrossRef]
- Dolan, A.; MacFarlane, S.A.; McGavin, W.J.; Brennan, R.M.; McNicol, J.W. Blackcurrant reversion virus: Validation of an improved diagnostic test, accelerating testing in breeding and certification of blackcurrants. J. Berry Res. 2011, 1, 201–208. [Google Scholar] [CrossRef]
- Jacob, H. Investigations on symptomatology, transmission, etiology and host specificity of blackcurrant reversion virus. Acta Hortic. 1976, 66, 99–104. [Google Scholar] [CrossRef]
- Lemmetty, A.; Latvala, S.; Jones, A.T.; Susi, P.; McGavin, W.J.; Lehto, K. Purification and properties of a new virus from black currant, its affinities with nepoviruses, and its close association with black currant reversion disease. Phytopathology 1997, 87, 404–413. [Google Scholar] [CrossRef]
- Lemmetty, A.; Lehto, K. Successful back-inoculation confirms the role of black currant reversion associated virus as the causal agent of reversion disease. Eur. J. Plant Pathol. 1999, 105, 297–301. [Google Scholar] [CrossRef]
- Russo, P.; Slack, S.A. Tissue culture methods for the screening and analysis of putative virus-resistant transgenic potato plants. Phytopathology 1998, 88, 437–441. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mazier, M.; German-Retana, S.; Flamain, F.; Dubois, V.; Botton, E.; Sarnette, V.; Le Gall, O.; Candresse, T.; Maisonneuve, B. A simple and efficient method for testing Lettuce mosaic virus resistance in in vitro cultivated lettuce. J. Virol. Methods 2004, 116, 123–131. [Google Scholar] [CrossRef]
- Al Abdallat, A.M.; Al Debei, H.S.; Asmar, H.; Misbeh, S.; Quraan, A.; Kvarnheden, A. An efficient in vitro-inoculation method for Tomato yellow leaf curl virus. Virol. J. 2010, 7, 84. [Google Scholar] [CrossRef] [PubMed]
- Magyar-Tábori, K.; Mendler-Drienyovszki, N.; Hanász, A.; Zsombik, L.; Dobránszki, J. Phytotoxicity and other adverse effects on the in vitro shoot cultures caused by virus elimination treatments: Reasons and solutions. Plants 2021, 10, 670. [Google Scholar] [CrossRef] [PubMed]
- Moročko-Bičevska, I.; Stalažs, A.; Lācis, G.; Laugale, V.; Baļķe, I.; Zuļģe, N.; Strautiņa, S. Cecidophyopsis mites and blackcurrant reversion virus on Ribes hosts: Current scientific progress and knowledge gaps. Ann. Appl. Biol. 2021, 180, 26–43. [Google Scholar] [CrossRef]
- Gelvonauskienė, D.; Šikšnianas, T.; Rugienius, R.; Stankienė, J.; Bendokas, V.; Stanys, V.; Sasnauskas, A. Distribution of gall mite and black currant reversion virus vectors in black currants. Sodinink. Daržinink. 2007, 26, 135–143. [Google Scholar]
- Latvala, S.; Susi, P.; Kalkkinen, N.; Lehto, K. Characterization of the coat protein gene of mite-transmitted blackcurrant reversion associated nepovirus. Virus Res. 1998, 53, 1–11. [Google Scholar] [CrossRef]
- Lemmetty, A.; Susi, P.; Latvala, S.; Lehto, K. Detection of the putative causal agent of Blackcurrant reversion disease. Acta Hortic. 1998, 471, 93–98. [Google Scholar] [CrossRef]
- Seitsonen, J.J.T.; Susi, P.; Lemmetty, A.; Butcher, S.J. Structure of the mite-transmitted Blackcurrant reversion nepovirus using electron cryo-microscopy. Virology 2008, 378, 162–168. [Google Scholar] [CrossRef]
- Hull, R. Mechanical inoculation of plant viruses. Curr Protoc Microbiol 2009, 13, 16B.6.1–16B.6.4. [Google Scholar] [CrossRef]
- Mažeikienė, I.; Stanys, V.; Juškytė, A.D.; Sasnauskas, A.; Šikšnianas, T. Black currant varieties ‘Aldoniai’ ir ‘Didikai’. Sodinink. Daržinink. 2017, 36, 3–14. [Google Scholar]
- Sundaresha, S.; Sreevathsa, R.; Balol, G.B.; Keshavareddy, G.; Rangaswamy, K.T.; Udayakumar, M. A simple, novel and high efficiency sap inoculation method to screen for tobacco streak virus. Physiol. Mol. Biol. Plants 2012, 18, 365–369. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ranabhat, N.B.; Bruce, M.A.; Fellers, J.P.; Shoup Rupp, J.L. A reproducible methodology for absolute viral quantification and viability determination in mechanical inoculations of wheat streak mosaic virus. Trop. Plant Pathol. 2022, 1–9. [Google Scholar] [CrossRef]
- Sudisha, J.; Sharathchandra, R.G.; Amruthesh, K.N.; Kumar, A.; Shetty, H.S. Pathogenesis Related Proteins in Plant Defense Response. In Plant Defence: Biological Control; Merillon, J.M., Ramawat, K.G., Eds.; Springer: Dordrecht, The Netherlands, 2011; Volume 12, pp. 379–403. [Google Scholar]
- Juškytė, A.D.; Mažeikienė, I.; Stanys, V. Putative genes of pathogenesis-related proteins and coronatine-insensitive protein 1 in Ribes spp. Plants 2022, 11, 355. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Sonbol, F.M.; Huot, B.; Gu, Y.; Withers, J.; Mwimba, M.; Yao, J.; He, S.Y.; Dong, X. Salicylic acid receptors activate jasmonic acid signalling through a non-canonical pathway to promote efector-triggered immunity. Nat. Commun. 2016, 7, 13099. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F.A. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Livak, K.; Schmittgen, T. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C (T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).