Characterization and QTL Mapping of a Major Field Resistance Locus for Bacterial Blight in Rice
Abstract
:1. Introduction
2. Results
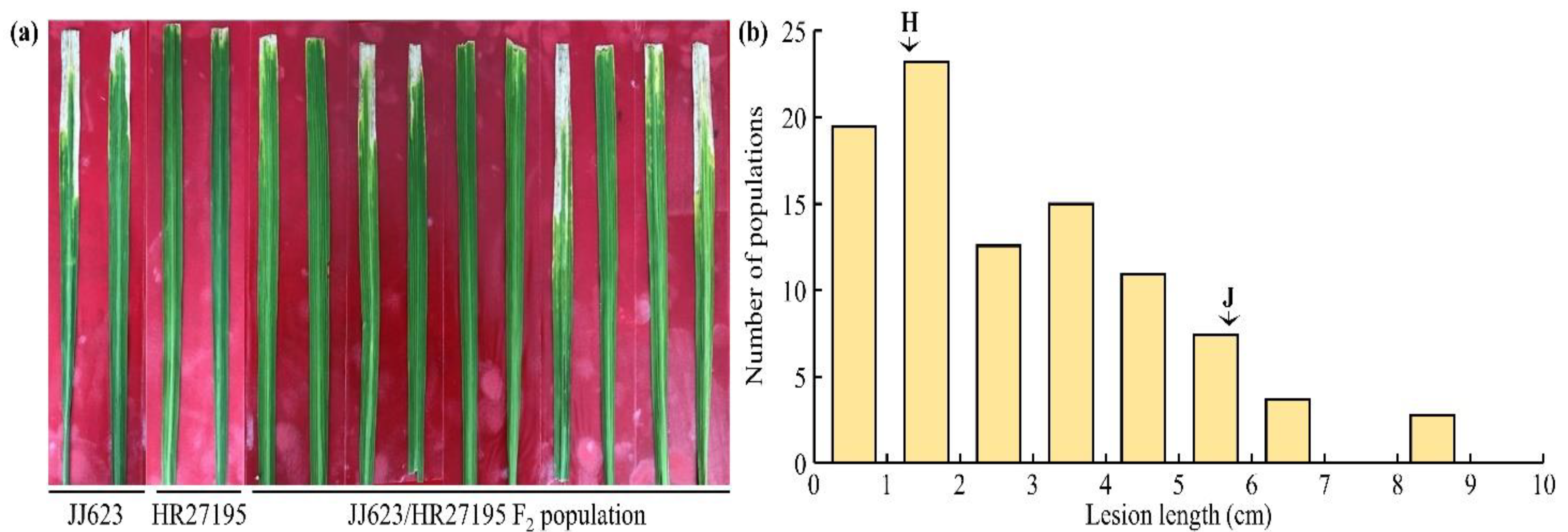
2.1. Evaluation of BB Resistance Using the RIL Population
2.2. Development of the Mapping Population and R-Gene Inheritance
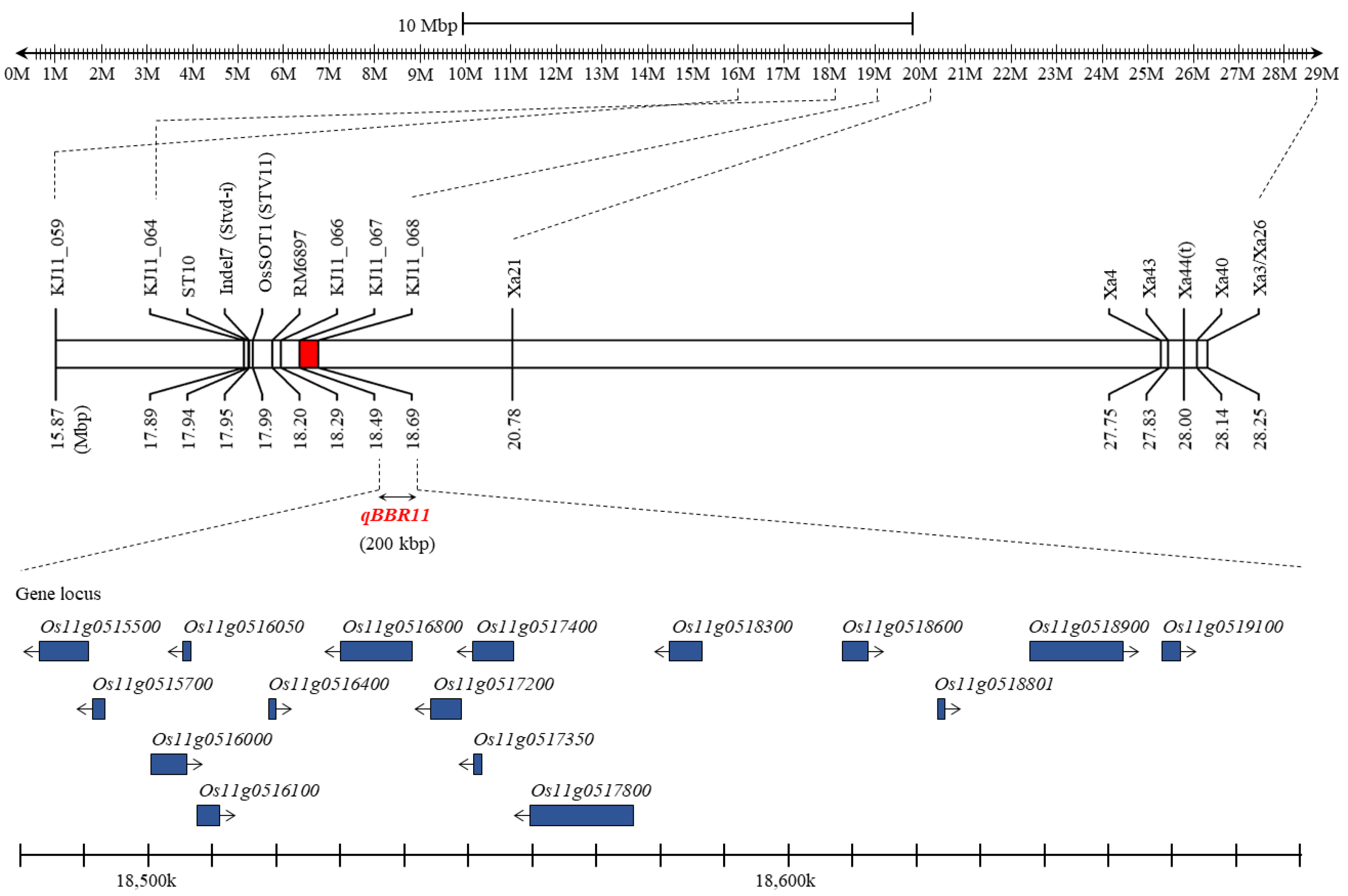
2.3. Analysis of QTLs for BB Resistance
2.4. Analysis of the Genetic Segregation Ratio of the Resistance Gene
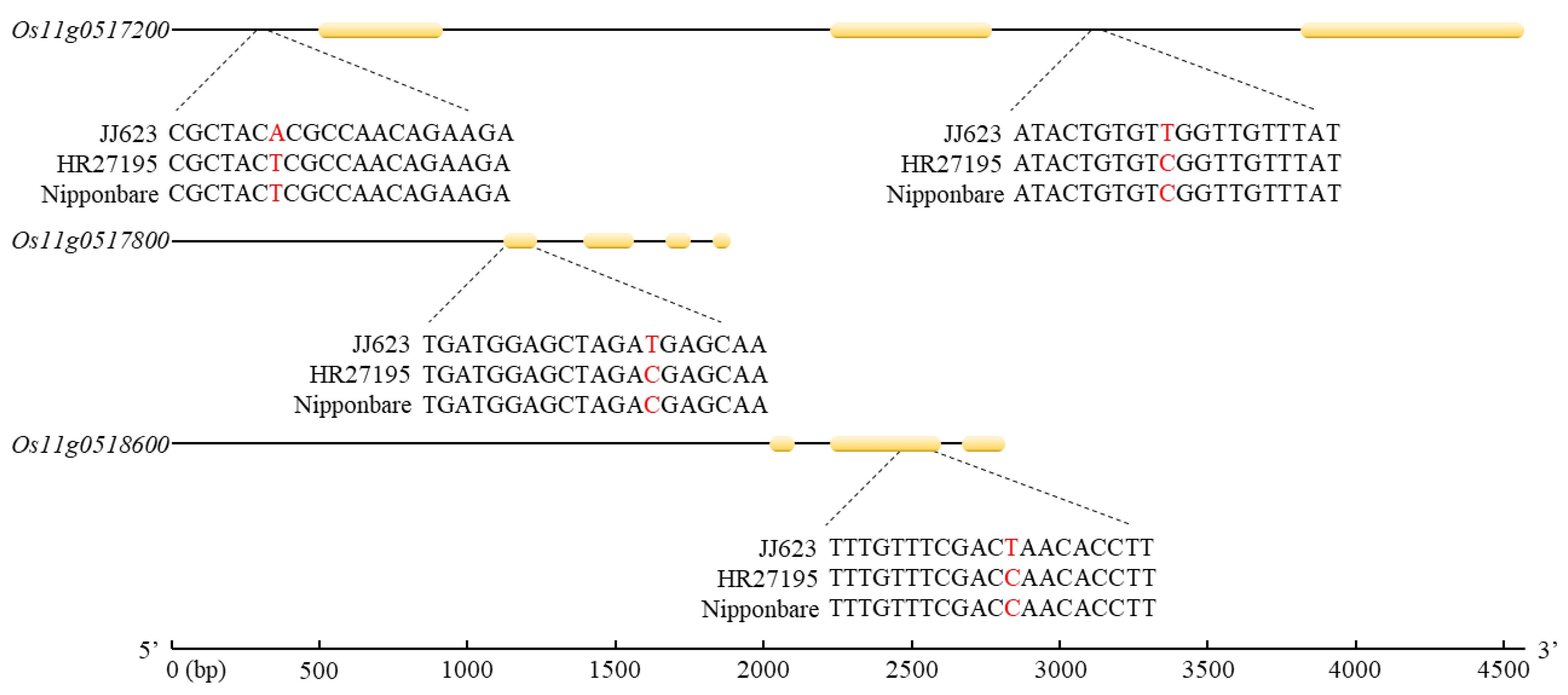
2.5. Candidate Genes and Sequencing Analysis
3. Discussion
4. Materials and Methods
4.1. Plant Material and Field Design
4.2. Bacterial Strains and Plant Inoculations
4.3. DNA Extraction and PCR Analysis
4.4. Genotyping and Linkage Mapping
4.5. Analysis of Putative Candidate Genes
4.6. Haplotype Sequence Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Khush, G.S. What it will take to feed 5.0 billion rice consumers in 2030. Plant Mol. Biol. 2005, 59, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Wing, R.A.; Purugganan, M.D.; Zhang, Q. The rice genome revolution: From an ancient grain to Green Super Rice. Nat. Rev. Genet. 2018, 19, 505–517. [Google Scholar] [CrossRef] [PubMed]
- Ritson, C. Population growth and global food supplies. In Food Education and Food Technology in School Curricula; Rutland, M., Turner, A., Eds.; Springer: Cham, Switzerland, 2020; pp. 261–271. [Google Scholar]
- Hunjan, M.S.; Lore, J.S. Climate change: Impact on plant pathogens, diseases, and their management. In Crop Protection under Changing Climate; Chauhan, B.S., Jabran, K., Florentine, S., Eds.; Springer: Cham, Switzerland, 2020; pp. 85–100. [Google Scholar]
- Watson, D. Adaption to climate change: Climate adaptive breeding of maize, wheat and rice. In Sustainable Solutions for Food Security; Sarkar, A., VanLoon, G.W., Sensarma, S.R., Eds.; Springer: Cham, Switzerland, 2019; pp. 67–89. [Google Scholar]
- Chukwu, S.; Rafii, M.; Ramlee, S.; Ismail, S.; Hasan, M.; Oladosu, Y.; Magaji, U.; Akos, I.; Olalekan, K. Bacterial leaf blight resistance in rice: A review of conventional breeding to molecular approach. Mol. Biol. Rep. 2019, 46, 1519–1532. [Google Scholar] [CrossRef] [PubMed]
- Fiyaz, R.A.; Shivani, D.; Chaithanya, K.; Mounika, K.; Chiranjeevi, M.; Laha, G.; Viraktamath, B.; Rao, L.S.; Sundaram, R. Genetic improvement of rice for bacterial blight resistance: Present status and future prospects. Rice Sci. 2022, 29, 118–132. [Google Scholar] [CrossRef]
- Niño-Liu, D.O.; Ronald, P.C.; Bogdanove, A.J. Xanthomonas oryzae pathovars: Model pathogens of a model crop. Mol. Plant Pathol. 2006, 7, 303–324. [Google Scholar] [CrossRef]
- Noh, T.H.; Lee, D.K.; Park, J.C.; Shim, H.K.; Choi, M.Y.; Kang, M.H.; Kim, J.D. Effects of bacterial leaf blight occurrence on rice yield and grain quality in different rice growth stage. Res. Plant Dis. 2007, 13, 20–23. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Liu, J.; Triplett, L.; Leach, J.E.; Wang, G.L. Novel insights into rice innate immunity against bacterial and fungal pathogens. Annu. Rev. Phytopathol. 2014, 52, 213–241. [Google Scholar] [CrossRef] [Green Version]
- Pradhan, S.; Barik, S.; Nayak, D.; Pradhan, A.; Pandit, E.; Nayak, P.; Das, S.; Pathak, H. Genetics, molecular mechanisms and deployment of bacterial blight resistance genes in rice. Crit. Rev. Plant Sci. 2020, 39, 360–385. [Google Scholar] [CrossRef]
- Reddy, A. Bacterial blight: Crop loss assessment and disease management. In Proceedings of the international workshop on bacterial blight of rice, Manila, Philippines, 14–18 March 1988; pp. 79–88. [Google Scholar]
- Lee, S.W.; Han, M.; Park, C.J.; Seo, Y.S.; Bartley, L.E.; Jeon, J.S. The molecular mechanisms of rice resistance to the bacterial blight pathogen, Xanthomonas oryzae pathovar oryzae. In Advances in Botanical Research; Kader, J.C., Delseny, M., Eds.; Elsevier: Oxford, UK, 2011; Volume 60, pp. 51–87. [Google Scholar]
- Nasir, M.; Iqbal, B.; Hussain, M.; Mustafa, A.; Ayub, M. Chemical management of bacterial leaf blight disease in rice. J. Agric. Res. 2019, 57, 99–103. [Google Scholar]
- Laha, G.; Sailaja, B.; Srinivas Prasad, M.; Ladhalakshmi, D.; Krishnaveni, D.; Singh, R.; Prakasam, V.; Yugander, A.; Kannan, C.; Valarmathi, P. Changes in rice disease scenario in India: An analysis from production oriented survey. In Technical Bulletin; ICAR-Indian Institute of Rice Research: Hyderabad, India, 2016; Volume 91, p. 95. [Google Scholar]
- Liu, H.; Yang, W.; Hu, B.; Liu, F. Virulence analysis and race classification of Xanthomonas oryzae pv. oryzae in China. J. Phytopathol 2007, 155, 129–135. [Google Scholar] [CrossRef]
- Li, C.; Su, P.; Wang, D.; Peng, S.; Chen, Y.; Chen, J.; Tan, X.; Zhang, D.; Wang, G.L.; Liu, Y. Dissection of the genetic architecture of rice resistance to Xanthomonas oryzae pv. oryzae using a genomewide association study. J. Phytopathol. 2018, 166, 470–476. [Google Scholar] [CrossRef]
- Jiang, N.; Yan, J.; Liang, Y.; Shi, Y.; He, Z.; Wu, Y.; Zeng, Q.; Liu, X.; Peng, J. Resistance genes and their interactions with bacterial blight/leaf streak pathogens (Xanthomonas oryzae) in rice (Oryza sativa L.)—An updated review. Rice 2020, 13, 3. [Google Scholar] [CrossRef] [Green Version]
- Lu, J.; Wang, C.; Zeng, D.; Li, J.; Shi, X.; Shi, Y.; Zhou, Y. Genome-wide association study dissects resistance loci against bacterial blight in a diverse rice panel from the 3000 rice genomes project. Rice 2021, 14, 22. [Google Scholar] [CrossRef]
- He, Q.; Li, D.; Zhu, Y.; Tan, M.; Zhang, D.; Lin, X. Fine mapping of Xa2, a bacterial blight resistance gene in rice. Mol Breed 2006, 17, 1–6. [Google Scholar] [CrossRef]
- Bao, S.; Tan, M.; Lin, X. Genetic mapping of a bacterial blight resistance gene Xa14 in rice. Acta Agron. Sin. 2010, 36, 422–427. [Google Scholar] [CrossRef]
- Wang, C.; Wen, G.; Lin, X.; Liu, X.; Zhang, D. Identification and fine mapping of the new bacterial blight resistance gene, Xa31 (t), in rice. Eur. J. Plant Pathol. 2009, 123, 235–240. [Google Scholar] [CrossRef]
- Bhasin, H.; Bhatia, D.; Raghuvanshi, S.; Lore, J.S.; Sahi, G.K.; Kaur, B.; Vikal, Y.; Singh, K. New PCR-based sequence-tagged site marker for bacterial blight resistance gene Xa38 of rice. Mol. Breed. 2012, 30, 607–611. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, J.; Pan, J.; Gu, Z.; Chen, X.; Jin, Y.; Liu, F.; Zhang, H.; Ma, B. Identification and molecular mapping of the rice bacterial blight resistance gene allelic to Xa7 from an elite restorer line Zhenhui 084. Eur. J. Plant Pathol. 2009, 125, 235–244. [Google Scholar] [CrossRef]
- Wang, C.; Fan, Y.; Zheng, C.; Qin, T.; Zhang, X.; Zhao, K. High-resolution genetic mapping of rice bacterial blight resistance gene Xa23. Mol. Genet. Genom. 2014, 289, 745–753. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C.K.; Wang, C.L.; Yu, Y.J.; Liang, Y.T.; Zhao, K.J. Identification and molecular mapping of Xa32 (t), a novel resistance gene for bacterial blight (Xanthomonas oryzae pv. oryzae) in rice. Acta Agron. Sin. 2009, 35, 1173–1180. [Google Scholar] [CrossRef]
- Kim, S.M.; Suh, J.P.; Qin, Y.; Noh, T.H.; Reinke, R.F.; Jena, K.K. Identification and fine-mapping of a new resistance gene, Xa40, conferring resistance to bacterial blight races in rice (Oryza sativa L.). Theor. Appl. Genet. 2015, 128, 1933–1943. [Google Scholar] [CrossRef] [PubMed]
- Hur, Y.J.; Jeung, J.U.; Kim, S.Y.; Park, H.S.; Cho, J.H.; Lee, J.Y.; Sohn, Y.B.; Song, Y.C.; Park, D.S.; Lee, C.W. Functional markers for bacterial blight resistance gene Xa3 in rice. Mol. Breed. 2013, 31, 981–985. [Google Scholar] [CrossRef]
- Song, W.Y.; Wang, G.L.; Chen, L.L.; Kim, H.S.; Pi, L.Y.; Holsten, T.; Gardner, J.; Wang, B.; Zhai, W.X.; Zhu, L.H. A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 1995, 270, 1804–1806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swamy, P.; Panchbhai, A.N.; Dodiya, P.; Naik, V.; Panchbhai, S.; Zehr, U.B.; Azhakanandam, K.; Char, B.R. Evaluation of bacterial blight resistance in rice lines carrying multiple resistance genes and Xa21 transgenic lines. Curr. Sci. 2006, 90, 6. [Google Scholar]
- Ke, Y.; Deng, H.; Wang, S. Advances in understanding broad-spectrum resistance to pathogens in rice. Plant J. 2017, 90, 738–748. [Google Scholar] [CrossRef]
- Pradhan, S.K.; Nayak, D.K.; Mohanty, S.; Behera, L.; Barik, S.R.; Pandit, E.; Lenka, S.; Anandan, A. Pyramiding of three bacterial blight resistance genes for broad-spectrum resistance in deepwater rice variety, Jalmagna. Rice 2015, 8, 51. [Google Scholar] [CrossRef]
- Suh, J.P.; Jeung, J.U.; Noh, T.H.; Cho, Y.C.; Park, S.H.; Park, H.S.; Shin, M.S.; Kim, C.K.; Jena, K.K. Development of breeding lines with three pyramided resistance genes that confer broad-spectrum bacterial blight resistance and their molecular analysis in rice. Rice 2013, 6, 5. [Google Scholar] [CrossRef] [Green Version]
- Pink, D.; Hand, P. Plant resistance and strategies for breeding resistant varieties. Plant Prot. Sci. 2003, 38, 9–14. [Google Scholar] [CrossRef] [Green Version]
- Liu, Q.; Yang, J.; Yan, S.; Zhang, S.; Zhao, J.; Wang, W.; Yang, T.; Wang, X.; Mao, X.; Dong, J. The germin-like protein OsGLP2-1 enhances resistance to fungal blast and bacterial blight in rice. Plant Mol. Biol. 2016, 92, 411–423. [Google Scholar] [CrossRef]
- Sundaram, R.M.; Chatterjee, S.; Oliva, R.; Laha, G.S.; Cruz, C.V.; Leach, J.E.; Sonti, R.V. Update on Bacterial Blight of Rice: Fourth International Conference on Bacterial Blight. Rice 2014, 7, 12. [Google Scholar] [CrossRef] [Green Version]
- Marli, G.K.M. Current challenges in plant breeding to achieve zero hunger and overcome biotic and abiotic stresses induced by the global climate changes: A review. J. Plant Sci. Phytopathol. 2021, 5, 053–057. [Google Scholar] [CrossRef]
- Liang, L.Q.; Wang, C.Y.; Zeng, L.X.; Wang, W.J.; Feng, J.Q.; Chen, B.; Su, J.; Chen, S.; Shang, F.D.; Zhu, X.Y. The rice cultivar Baixiangzhan harbours a recessive gene xa42 (t) determining resistance against Xanthomonas oryzae pv. oryzae. Plant Breed. 2017, 136, 603–609. [Google Scholar] [CrossRef]
- Zhao, K.; Zhang, Q. A climate-resilient R gene in rice traps two pathogen effectors for broad and durable resistance to bacterial blight. Mol. Plant 2021, 14, 366–368. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Hu, J.; Li, Z.; Liu, J.; Gao, G.; Zhang, Q.; Xiao, J.; He, Y. Evaluation and breeding application of six brown planthopper resistance genes in rice maintainer line Jin 23B. Rice 2018, 11, 22. [Google Scholar] [CrossRef]
- Tester, M.; Langridge, P. Breeding technologies to increase crop production in a changing world. Science 2010, 327, 818–822. [Google Scholar] [CrossRef]
- Kim, K.Y.; Ko, J.C.; Shin, W.C.; Park, H.S.; Baek, M.K.; Nam, J.K.; Kim, B.K.; Lee, J.H. Effect of low radiation during grain filling stage on rice yield and grain quality. Korean J. Crop Sci. 2014, 59, 174–180. [Google Scholar] [CrossRef] [Green Version]
- Kim, K.Y.; Shin, M.S.; Kim, W.J.; Park, H.S.; Ko, J.C.; Nam, J.K.; Shin, W.C.; Mo, Y.J.; Jeung, J.U.; Kim, B.K. Development of near-isogenic lines (NILs) conferring Xa4, xa5 and Xa21 genes resistant to bacterial blight (Xanthomonas oryzae pv. oryzae) in japonica rice genetic background. Korean J. Breed. Sci. 2011, 43, 383–390. [Google Scholar]
- Ferreira, A.; Da Silva, M.F.; Cruz, C.D. Estimating the effects of population size and type on the accuracy of genetic maps. Genet. Mol. Biol. 2006, 29, 187–192. [Google Scholar] [CrossRef] [Green Version]
- Yang, G.; Chen, S.; Chen, L.; Sun, K.; Huang, C.; Zhou, D.; Huang, Y.; Wang, J.; Liu, Y.; Wang, H. Development of a core SNP arrays based on the KASP method for molecular breeding of rice. Rice 2019, 12, 21. [Google Scholar] [CrossRef]
- Steele, K.A.; Quinton-Tulloch, M.J.; Amgai, R.B.; Dhakal, R.; Khatiwada, S.P.; Vyas, D.; Heine, M.; Witcombe, J.R. Accelerating public sector rice breeding with high-density KASP markers derived from whole genome sequencing of Indica rice. Mol. Breed. 2018, 38, 38. [Google Scholar] [CrossRef] [Green Version]
- Nagasaki, H.; Ebana, K.; Shibaya, T.; Yonemaru, J.I.; Yano, M. Core single-nucleotide polymorphisms—A tool for genetic analysis of the Japanese rice population. Breed. Sci. 2010, 60, 648–655. [Google Scholar] [CrossRef] [Green Version]
- Bao, J.; Corke, H.; Sun, M. Microsatellites, single nucleotide polymorphisms and a sequence tagged site in starch-synthesizing genes in relation to starch physicochemical properties in nonwaxy rice (Oryza sativa L.). Theor. Appl. Genet. 2006, 113, 1185–1196. [Google Scholar] [CrossRef] [PubMed]
- Varshney, R.; Thiel, T.; Sretenovic-Rajicic, T.; Baum, M.; Valkoun, J.; Guo, P.; Grando, S.; Ceccarelli, S.; Graner, A. Identification and validation of a core set of informative genic SSR and SNP markers for assaying functional diversity in barley. Mol. Breed. 2008, 22, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Würschum, T.; Langer, S.M.; Longin, C.F.H.; Korzun, V.; Akhunov, E.; Ebmeyer, E.; Schachschneider, R.; Schacht, J.; Kazman, E.; Reif, J.C. Population structure, genetic diversity and linkage disequilibrium in elite winter wheat assessed with SNP and SSR markers. Theor. Appl. Genet. 2013, 126, 1477–1486. [Google Scholar] [CrossRef]
- Andaya, V.; Mackill, D. QTLs conferring cold tolerance at the booting stage of rice using recombinant inbred lines from a japonica× indica cross. Theor. Appl. Genet. 2003, 106, 1084–1090. [Google Scholar] [CrossRef]
- Li, H.; Wang, J.; Liu, A.; Liu, K.; Zhang, Q.; Zou, J. Genetic basis of low-temperature-sensitive sterility in indica-japonica hybrids of rice as determined by RFLP analysis. Theor. Appl. Genet. 1997, 95, 1092–1097. [Google Scholar] [CrossRef]
- Zheng, Y.; Zhang, G.; Lin, F.; Wang, Z.; Jin, G.; Yang, L.; Wang, Y.; Chen, X.; Xu, Z.; Zhao, X. Development of microsatellite markers and construction of genetic map in rice blast pathogen Magnaporthe grisea. Fungal Genet. Biol. 2008, 45, 1340–1347. [Google Scholar] [CrossRef]
- Hori, K.; Yamamoto, T.; Yano, M. Genetic dissection of agronomically important traits in closely related temperate japonica rice cultivars. Breed. Sci. 2017, 67, 427–434. [Google Scholar] [CrossRef] [Green Version]
- Cheon, K.S.; Jeong, Y.M.; Lee, Y.Y.; Oh, J.; Kang, D.Y.; Oh, H.; Kim, S.L.; Kim, N.; Lee, E.; Baek, J. Kompetitive allele-specific PCR marker development and quantitative trait locus mapping for bakanae disease resistance in Korean japonica rice varieties. Plant Breed. Biotechnol. 2019, 7, 208–219. [Google Scholar] [CrossRef]
- Kim, S.M. Identification of novel recessive gene xa44 (t) conferring resistance to bacterial blight races in rice by QTL linkage analysis using an SNP chip. Theor. Appl. Genet. 2018, 131, 2733–2743. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.B.; Kim, N.; Hur, Y.J.; Cho, S.M.; Kim, T.H.; Lee, J.Y.; Cho, J.H.; Lee, J.H.; Song, Y.C.; Seo, Y.S. Fine mapping of qBK1, a major QTL for bakanae disease resistance in rice. Rice 2019, 12, 36. [Google Scholar] [CrossRef] [Green Version]
- Kumar, J.; Hussain, A.; Singh, P.; Baksh, S.; Kar, M.; Mukherjee, A.; Singh, N.; Sinha, B.; Pramesh, K.; Singh, N. Genotypic and phenotypic dissection of Xanthomonas oryzae pv. oryzae on Indica rice cultivars for bacterial blight resistance. J. Environ. Biol. 2021, 42, 1578–1590. [Google Scholar] [CrossRef]
- Zhang, F.; Zhuo, D.L.; Zhang, F.; Huang, L.Y.; Wang, W.S.; Xu, J.L.; Vera Cruz, C.; Li, Z.K.; Zhou, Y.L. Xa39, a novel dominant gene conferring broad-spectrum resistance to Xanthomonas oryzae pv. oryzae in rice. Plant Pathol. 2015, 64, 568–575. [Google Scholar] [CrossRef]
- Xiao, J.; Li, J.; Yuan, L.; Tanksley, S. Identification of QTLs affecting traits of agronomic importance in a recombinant inbred population derived from a subspecific rice cross. Theor. Appl. Genet. 1996, 92, 230–244. [Google Scholar] [CrossRef] [PubMed]
- Faus, I.; Niñoles, R.; Kesari, V.; Gadea, J. The ABCF3 gene of Arabidopsis is functionally linked with GCN1 but not with GCN2 during stress and development. Plant Mol. Biol. Rep. 2021, 39, 663–672. [Google Scholar] [CrossRef]
- Liu, X.; Afrin, T.; Pajerowska-Mukhtar, K.M. Arabidopsis GCN2 kinase contributes to ABA homeostasis and stomatal immunity. Commun. Biol. 2019, 2, 302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, N.; Zhang, S.J.; Zhao, Q.; Long, Y.; Guo, H.; Jia, H.F.; Yang, Y.X.; Zhang, H.Y.; Ye, X.F.; Zhang, S.T. Overexpression of tobacco GCN2 stimulates multiple physiological changes associated with stress tolerance. Front. Plant. Sci. 2018, 9, 725. [Google Scholar] [CrossRef] [PubMed]
- Tsuge, S.; Furutani, A.; Fukunaka, R.; Oku, T.; Tsuno, K.; Ochiai, H.; Inoue, Y.; Kaku, H.; Kubo, Y. Expression of Xanthomonas oryzae pv. oryzae hrp genes in XOM2, a novel synthetic medium. J. Gen. Plant Pathol. 2002, 68, 363–371. [Google Scholar] [CrossRef]
- Kauffman, H.; Reddy, A.; Hsieh, S.; Merca, S.; Kauffman, E. An improved technique for evaluating resistance of rice varieties to Xanthomonas oryzae. Plant Dis. Rep. 1973, 57, 537–541. [Google Scholar]
- RDA. Manual for Standard Evaluation Method in Agricultural Experiment and Research; RDA Press: Suwon, Korea, 2012. [Google Scholar]
- Meng, L.; Li, H.; Zhang, L.; Wang, J. QTL IciMapping: Integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J. 2015, 3, 269–283. [Google Scholar] [CrossRef] [Green Version]
- Kumagai, M.; Nishikawa, D.; Kawahara, Y.; Wakimoto, H.; Itoh, R.; Tabei, N.; Tanaka, T.; Itoh, T. TASUKE+: A web-based platform for exploring GWAS results and large-scale resequencing data. DNA Res. 2019, 26, 445–452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruden, D.M.; Cingolani, P.; Patel, V.M.; Coon, M.; Nguyen, T.; Land, S.J.; Lu, X. Using Drosophila melanogaster as a model for genotoxic chemical mutational studies with a new program, SnpSift. Front. Genet. 2012, 3, 35. [Google Scholar] [CrossRef] [Green Version]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef] [PubMed]

| Cultivar | K1 | K2 | K3 | K3a |
|---|---|---|---|---|
| Sindongjin | R | R | R | S |
| JJ623 | R | R | R | MR |
| HR27195 | R | R | R | R |
| Cross | Number of Resistance | Number of Susceptible | Total Number | Segregation Ratio | X2 | p-Value |
| JJ623/HR27195 | 29 | 61 | 90 | 1:3 | 2.50 | 0.113 |
| QTL | Chr. | Position (cM) | Physical Position (Mbp) | Left Marker | Right Marker | LOD z | PVE y(%) | Add x | Dom w |
|---|---|---|---|---|---|---|---|---|---|
| qBBR11 | 11 | 242.7 | 18.49–18.69 | KJ11_067 | KJ11_068 | 5.47 | 24.5 | 1.26 | −0.09 |
| Gene ID | Physical Position (bp) | Putative Function |
|---|---|---|
| Os11g0515500 | 18,485,308–18,490,843 | Similar to transport inhibitor response 1 protein |
| Os11g0515700 | 18,493,370–18,494,393 | Nonprotein coding transcript |
| Os11g0516000 | 18,503,539–18,507,701 | Similar to Serine palmitoyltransferase (Fragment) |
| Os11g0516050 | 18,506,918–18,507,468 | Nonprotein coding transcript |
| Os11g0516100 | 18,509,086–18,512,628 | Ribosomal protein L31 domain containing protein. |
| Os11g0516400 | 18,520,372–18,521,065 | Hypothetical conserved gene |
| Os11g0516800 | 18,531,653–18,541,501 | Hypothetical protein |
| Os11g0517200 | 18,544,614–18,548,898 | Hypothetical conserved gene |
| Os11g0517350 | 18,550,900–18,551,191 | Similar to Maturase K (Fragment) |
| Os11g0517400 | 18,552,567–18,556,300 | Conserved hypothetical protein |
| Os11g0517800 | 18,560,178–18,575,490 | eIF-2-alpha kinase GCN2 |
| Os11g0518300 | 18,581,847–18,585,958 | Hypothetical protein |
| Os11g0518600 | 18,608,152–18,611,496 | Conserved hypothetical protein |
| Os11g0518801 | 18,622,623–18,622,684 | Nonprotein coding transcript |
| Os11g0518900 | 18,636,476–18,650,782 | Conserved hypothetical protein |
| Os11g0519100 | 18,655,353–18,658,368 | Similar to OSIGBa0132O24.2 protein |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, J.-R.; Lee, C.-M.; Ji, H.; Baek, M.-K.; Seo, J.; Jeong, O.-Y.; Park, H.-S. Characterization and QTL Mapping of a Major Field Resistance Locus for Bacterial Blight in Rice. Plants 2022, 11, 1404. https://doi.org/10.3390/plants11111404
Park J-R, Lee C-M, Ji H, Baek M-K, Seo J, Jeong O-Y, Park H-S. Characterization and QTL Mapping of a Major Field Resistance Locus for Bacterial Blight in Rice. Plants. 2022; 11(11):1404. https://doi.org/10.3390/plants11111404
Chicago/Turabian StylePark, Jae-Ryoung, Chang-Min Lee, Hyeonso Ji, Man-Kee Baek, Jeonghwan Seo, O-Young Jeong, and Hyun-Su Park. 2022. "Characterization and QTL Mapping of a Major Field Resistance Locus for Bacterial Blight in Rice" Plants 11, no. 11: 1404. https://doi.org/10.3390/plants11111404
APA StylePark, J.-R., Lee, C.-M., Ji, H., Baek, M.-K., Seo, J., Jeong, O.-Y., & Park, H.-S. (2022). Characterization and QTL Mapping of a Major Field Resistance Locus for Bacterial Blight in Rice. Plants, 11(11), 1404. https://doi.org/10.3390/plants11111404






