A Review of Integrative Omic Approaches for Understanding Rice Salt Response Mechanisms
Abstract
:1. Introduction
2. Effects of Salinity on Rice
2.1. Morphological Effects on Rice under Salinity
2.2. Physiology, Biochemistry and Molecular Response of Rice under Salinity Stress
3. Adaptive Mechanisms of Salinity Tolerance in Rice
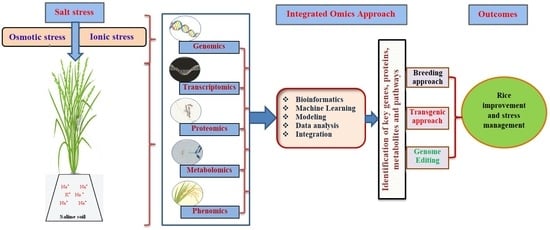
4. Omics Platforms Used for Rice Improvement
4.1. Genomics
| Parents | QTLs Number | Different Stage | References |
|---|---|---|---|
| Pokkali × IR29 | 23 | Seedlings | [84] |
| Nonabokra × Koshihikari | 11 | Seedlings | [85] |
| Ahlemi Tarom × Neda | 73 | Seedlings | [86] |
| Capsule × BRRI dhan29 | 27 | Seedlings | [87] |
| Pokkali × Bengal | 50 | Seedlings | [82] |
| Hasawi × IR29 | 34 | Seedlings | [88] |
| Kalarata × Azucena | 13 | Seedlings | [89] |
| Nonabokra × Jupiter | 33 | Seedlings | [90] |
| Dianjingyou × Sea Rice 86 | 1 | Seedlings | [91] |
| CSR27 × MI48 | 25 | Seedlings, vegetative and reproductive | [92] |
| OM7347 × OM5629 | 9 | Seedlings, vegetative and reproductive | [93] |
| Horkuch × IR29 | 14 | Seedlings and reproductive | [94] |
| Cheriviruppu8 × Pusa Bashmati 1 | 16 | Reproductive | [95] |
| Pokkali × IR36 | 6 | Maturity | [96] |
| CSR27 × MI48 | 8 | Maturity | [97] |
| Jiucaiqing × IR26 | 16 | Germination | [98] |
| Wujiaozhan × Nipponbare | 13 | Germination | [99] |
4.2. Transcriptomics
4.3. Proteomics
4.4. Metabolomics
4.5. Phenomics
| Omic Approach | Techniques | Description | References |
|---|---|---|---|
| Genomics | Map-based sequencing | Rice genome sequence. | [81] |
| Illumina-seq | 213 and 436 transcript tags of shoot and root were differentially expressed in response to salt. | [256] | |
| Genome-wide meta-analysis | 3449 DEGs were detected in rice tissues. Surprisingly, 23 possible-candidate salinity responsive genes for yield and ion homeostasis were discovered. | [257] | |
| Mutation breeding | Rice mutants improve salt tolerance. | [119,121,123,124] | |
| Illumina-seq | DMRs enhance salt tolerance. | [131] | |
| Genetic engineering | Developed salinity tolerant rice mutants through CRISPR-cas9. | [258,259] | |
| Transcriptomics | DNA microarray | 486 salt-responsive ESTs identified from rice shoot. | [148] |
| RNA-seq | Several salt-inducible genes have been identified | [152,153] | |
| RNA-seq | In hybrid rice LYP9 and from its two parents, salt- induced DEGs were found to be 8292, 8037 and 631, respectively. This research provided a new perspective on heterosis mechanisms in salinity tolerance. | [154] | |
| RNA-seq | More transporters, ion and sugar-related transports were also identified from Mulai roots to have a role in the control of salt tolerance. | [166] | |
| RNA-seq | Identify genetic SSR markers that will help in marker- assisted breeding to improve the agronomic traits under different stress conditions. | [163] | |
| RNA-seq | Identified important genes regulated during salt stress in rice, such as OsSOS1, OsHKT1;5, OsHKT2;1, OsNHX1, OsAKT1, OsNRT1;2, OsTPC1, OsCDPK7, OsARP, OsMAPK5, 44 and OsSERF1. | [76] | |
| Proteomics | 2-DE | Six salt responsive proteins identified | [181] |
| 2-DE and MALDI-TOF MS | During salt stress, 57 responsive proteins were regulated, among them several are novel salt responsive proteins. | [182] | |
| 2-DE and LC-MS/MS | Four proteins were identified, among them 2 proteins, involved in salt stress response and the ubiquitin 26S proteasome system. | [183] | |
| 2-D and MALDI-TOF MS | 11 proteins were found to be differentially expressed. Most of them were new to being involved in rice salt response. | [185] | |
| 2-DE | 40 uniquely upregulated proteins were identified under ABA+salt stress. | [187] | |
| iTRAQ | Identified 5340 proteins, among them differentially expressed proteins involved in salt stress regulation and response to oxidation–reduction; photosynthesis and carbohydrate metabolisms. | [192] | |
| iTRAQ | Identified more than 2000 proteins in both root and shoot of salt-tolerant elite line FL478, during the early salinity stage. Among the identified proteins, some proteins are potential candidates, involved in the amino acid synthesis, antioxidant stress, and maintenance of mitochondrial activity, metabolism and Calvin cycle. | [193] | |
| iTRAQ | Identified 4598 proteins; among them, 279 were up- and downregulated and involved in oxidative phosphorylation, photosynthesis, phenylpropanoid biosynthesis, posttranslational modification and energy metabolism. | [194] | |
| Metabolomics | GC-MS | Metabolic profiling of ice seeds. | [215,216,217,218] |
| GC-MS | Rice metabolic profiling. | [219] | |
| H-NMR | Five conserved salts responsive metabolic markers were identified. | [220] | |
| H-NMR | Significant accumulation of sugar and amino acids under stress conditions. | [221] | |
| GC-MS | Characterised 92 primary metabolites in both shoots and roots in rice under stress and control conditions. Among them, 11 metabolites including amino acid and sugar significantly increased in tolerant varieties at the time of salt treatments. | [222] | |
| GC-MS | Two signalling molecules serotonin and gentisic acid are two significant biomarker compounds produced in tolerant varieties that contribute to NaCl tolerance | [224] | |
| GC-MS | A total of 84 metabolites were identified including amino acid, sugar, organic acid and other small molecular components. | [225] | |
| Phenomics | RGB and fluorescence images | A combined technique was applied for the screening of different salt tolerance traits of rice. | [246] |
| IR thermal images | Used to examine rice phenotyping under a salt stress environment. | [247] | |
| Automated imaging | Identify significant traits for subsequent QTL analysis, to deeper understand the genetic mechanisms driving RSA. | [248] | |
| X-ray tomography | Used to quantify the response of rice RSA to the soil environment. | [249] | |
| RGB and fluorescence images | Investigate the complex salinity tolerance in Australian wild rice species. | [251] |
5. Modernise Breeding Approaches for Rice Salinity Improvement
5.1. Marker-Assisted Selection (MAS)
5.2. Transgenic Approach
5.3. Genome Editing
5.4. Machine Learning (ML)
6. Integration of Omics and Role of Bioinformatics for Rice Improvement
7. Conclusions
8. Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pereira, A. Plant abiotic stress challenges from the changing environment. Front. Plant Sci. 2016, 7, 2013–2015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pareek, A.; Sopory, S.K.; Bohnert, H.J. (Eds.) Abiotic Stress Adaptation in Plants; Springer: Dordrecht, The Netherlands, 2010; ISBN 978-90-481-3111-2. [Google Scholar]
- Mantri, N.; Patade, V.; Penna, S.; Ford, R.; Pang, E. Abiotic Stress Responses in Plants: Present and Future. In Abiotic Stress Responses in Plants; Springer: New York, NY, USA, 2012; pp. 1–19. [Google Scholar]
- Munns, R. Plant Adaptations to Salt and Water Stress. Adv. Bot. Res. 2011, 57, 1–32. [Google Scholar]
- Shahbaz, M.; Ashraf, M. Improving Salinity Tolerance in Cereals. CRC. Crit. Rev. Plant Sci. 2013, 32, 237–249. [Google Scholar] [CrossRef]
- Shabala, S.; Bose, J.; Hedrich, R. Salt bladders: Do they matter? Trends Plant Sci. 2014, 19, 687–691. [Google Scholar] [CrossRef]
- Liu, M.; Pan, T.; Allakhverdiev, S.I.; Yu, M.; Shabala, S. Crop Halophytism: An Environmentally Sustainable Solution for Global Food Security. Trends Plant Sci. 2020, 25, 630–634. [Google Scholar] [CrossRef]
- Mondal, M.M.A.; Puteh, A.B.; Malek, M.A.; Rafii, M.Y. Salinity induced morpho-physiological characters and yield attributes in rice genotypes. J. Food Agric. Environ. 2013, 11, 610–614. [Google Scholar]
- Jamil, A.; Riaz, S.; Ashraf, M.; Foolad, M.R. Gene Expression Profiling of Plants under Salt Stress. CRC. Crit. Rev. Plant Sci. 2011, 30, 435–458. [Google Scholar] [CrossRef]
- Shankar, R.; Bhattacharjee, A.; Jain, M. Transcriptome analysis in different rice cultivars provides novel insights into desiccation and salinity stress responses. Sci. Rep. 2016, 6, 23719. [Google Scholar] [CrossRef] [Green Version]
- Kaur, N.; Dhawan, M.; Sharma, I.; Pati, P.K. Interdependency of Reactive Oxygen Species generating and scavenging system in salt sensitive and salt tolerant cultivars of rice. BMC Plant Biol. 2016, 16, 131. [Google Scholar] [CrossRef] [Green Version]
- Munns, R.; Tester, M. Mechanisms of Salinity Tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [Green Version]
- Haque, M.A.; Rafii, M.Y.; Yusoff, M.M.; Ali, N.S.; Yusuff, O.; Datta, D.R.; Anisuzzaman, M.; Ikbal, M.F. Advanced Breeding Strategies and Future Perspectives of Salinity Tolerance in Rice. Agronomy 2021, 11, 1631. [Google Scholar] [CrossRef]
- Zhao, C.; Zhang, H.; Song, C.; Zhu, J.-K.; Shabala, S. Mechanisms of Plant Responses and Adaptation to Soil Salinity. Innovation 2020, 1, 100017. [Google Scholar] [CrossRef] [PubMed]
- Mehta, S.; James, D.; Reddy, M.K. Omics Technologies for Abiotic Stress Tolerance in Plants: Current Status and Prospects. In Recent Approaches in Omics for Plant Resilience to Climate Change; Springer International Publishing: Cham, Switzerland, 2019; pp. 1–34. [Google Scholar]
- Reddy, I.N.B.L.; Kim, B.K.; Yoon, I.S.; Kim, K.H.; Kwon, T.R. Salt Tolerance in Rice: Focus on Mechanisms and Approaches. Rice Sci. 2017, 24, 123–144. [Google Scholar] [CrossRef]
- Cortés, A.J.; López-Hernández, F. Harnessing Crop Wild Diversity for Climate Change Adaptation. Genes 2021, 12, 783. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Zhang, H.H.; Wang, X. Machine learning for Big Data analytics in plants. Trends Plant Sci. 2014, 19, 798–808. [Google Scholar] [CrossRef] [PubMed]
- Cortés, A.J.; Restrepo-Montoya, M.; Bedoya-Canas, L.E. Modern Strategies to Assess and Breed Forest Tree Adaptation to Changing Climate. Front. Plant Sci. 2020, 11, 1606. [Google Scholar] [CrossRef] [PubMed]
- Tong, H.; Nikoloski, Z. Machine learning approaches for crop improvement: Leveraging phenotypic and genotypic big data. J. Plant Physiol. 2021, 257, 153354. [Google Scholar] [CrossRef]
- George, E.B., Jr. Research Databases. In Bibliography on Salt Tolerance; USDA-ARS—U.S. Department of Agriculture, Agricultural Research Service: Riverside, CA, USA, 2008. [Google Scholar]
- Lawlor, D.W.; Mengel, K.; Kirkby, E.A. Principles of plant nutrition. Ann. Bot. 2004, 93, 479–480. [Google Scholar] [CrossRef] [Green Version]
- Isayenkov, S.V.; Maathuis, F.J.M. Plant Salinity Stress: Many Unanswered Questions Remain. Front. Plant Sci. 2019, 10, 80. [Google Scholar] [CrossRef] [Green Version]
- Sahi, C.; Singh, A.; Kumar, K.; Blumwald, E.; Grover, A. Salt stress response in rice: Genetics, molecular biology, and comparative genomics. Funct. Integr. Genom. 2006, 6, 263–284. [Google Scholar] [CrossRef]
- Waziri, A.; Kumar, P.; Purty, R.S. Saltol QTL and Their Role in Salinity Tolerance in Rice. Austin J. Biotechnol. Bioeng. 2016, 3, 1063–1067. [Google Scholar]
- Jamil, M.; Lee, D.B.A.E.; Jung, K.Y.; Ashraf, M.; Chun, S.; Rha, E.U.I.S.; Jamil, M.; Lee, D.B.A.E.; Jung, K.Y.; Ashraf, M.; et al. Effect of Salt (Nacl) Stress on Germination and Early Seedling Growth of Four Vegetables Species. J. Cent. Eur. Agric. 2006, 7, 273–282. [Google Scholar]
- Zhang, H.; Han, B.; Wang, T.; Chen, S.; Li, H.; Zhang, Y.; Dai, S. Mechanisms of plant salt response: Insights from proteomics. J. Proteome Res. 2012, 11, 49–67. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Ansari, M.W.; Pareek, A.; Singla-Pareek, S.L. Raising salinity tolerant rice: Recent progress and future perspectives. Physiol. Mol. Biol. Plants 2008, 14, 137–154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Zelm, E.; Zhang, Y.; Testerink, C. Salt Tolerance Mechanisms of Plants. Annu. Rev. Plant Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef] [Green Version]
- Rahman, A.; Mahmud, S.H.J. Manganese-induced salt stress tolerance in rice seedlings: Regulation of ion homeostasis, antioxidant defense and glyoxalase systems. Physiol. Mol. Biol. Plants 2016, 22, 291–306. [Google Scholar] [CrossRef]
- Rahman, A.; Nahar, K.; Hasanuzzaman, M.; Fujita, M. Calcium Supplementation Improves Na+/K+ Ratio, Antioxidant Defense and Glyoxalase Systems in Salt-stressed Rice Seedlings. Front. Plant Sci. 2016, 7, 609. [Google Scholar] [CrossRef] [Green Version]
- Flowers, T.J.; Yeo, A.R. Variability in the resistance of sodium chloride salinity within rice (Oryza sativa l.) varieties. New Phytol. 1981, 88, 363–373. [Google Scholar] [CrossRef]
- Dolferus, R.; Ji, X.; Richards, R.A. Abiotic stress and control of grain number in cereals. Plant Sci. 2011, 181, 331–341. [Google Scholar] [CrossRef]
- Khatun, S.; Rizzo, C.A.; Flowers, T.J. Genotypic variation in the effect of salinity on fertility in rice. Plant Soil 1995, 173, 239–250. [Google Scholar] [CrossRef]
- Abdullah, Z.; Khan, M.A.; Flowers, T.J. Causes of Sterility in Seed Set of Rice under Salinity Stress. J. Agron. Crop Sci. 2001, 187, 25–32. [Google Scholar] [CrossRef]
- Ghosh, B.; Ali, N.; Gantait, S. Response of Rice under Salinity Stress: A Review Update. Rice Res. Open Access 2016, 4, 2–9. [Google Scholar] [CrossRef] [Green Version]
- Yeo, A.R.; Lee, À.S.; Izard, P.; Boursier, P.J.; Flowers, T.J. Short- and long-term effects of salinity on leaf growth in rice (Oryza sativa L.). J. Exp. Bot. 1991, 42, 881–889. [Google Scholar] [CrossRef]
- Senguttuvel, P.; Vijayalakshmi, C.; Thiyagarajan, K.; Kannanbapu, J.R.; Kota, S.; Padmavathi, G.; Geetha, S.; Sritharan, N.; Viraktamath, B.C. Changes in photosynthesis, chlorophyll fluorescence, gas exchange parameters and osmotic potential to salt stress during early seedling stage in rice (Oryza sativa L.). SABRAO J. Breed. Genet. 2014, 46, 120–135. [Google Scholar]
- Kibria, M.G.; Hossain, M.; Murata, Y.; Hoque, M.A. Antioxidant Defense Mechanisms of Salinity Tolerance in Rice Genotypes. Rice Sci. 2017, 24, 155–162. [Google Scholar] [CrossRef]
- Morales, S.G.; Trejo-Téllez, L.I.; Merino, F.C.G.; Caldana, C.; Espinosa-Victoria, D.; Cabrera, B.E.H. Crescimento, atividade fotossintética, concentração de K+ e Na+ em plantas de arroz em condições de estresse salino. Acta Sci.—Agron. 2012, 34, 317–324. [Google Scholar]
- Baker, N.R. Chlorophyll Fluorescence: A Probe of Photosynthesis In Vivo. Annu. Rev. Plant Biol. 2008, 59, 89–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suo, J.; Zhao, Q.; David, L.; Chen, S.; Dai, S. Salinity response in chloroplasts: Insights from gene characterization. Int. J. Mol. Sci. 2017, 18, 1011. [Google Scholar] [CrossRef]
- Yamane, K.; Kawasaki, M.; Taniguchi, M.; Miyake, H. Differential effect of NaCl and polyethylene glycol on the ultrastructure of chloroplasts in rice seedlings. J. Plant Physiol. 2003, 160, 573–575. [Google Scholar] [CrossRef]
- Jung, J.-Y.; Shin, R.; Schachtman, D.P. Ethylene Mediates Response and Tolerance to Potassium Deprivation in Arabidopsis. Plant Cell 2009, 21, 607–621. [Google Scholar] [CrossRef] [Green Version]
- Razzaque, M.A.; Talukder, N.M.; Islam, M.T.; Dutta, R.K. Salinity effect on mineral nutrient distribution along roots and shoots of rice (Oryza sativa L.) genotypes differing in salt tolerance. Arch. Agron. Soil Sci. 2011, 57, 33–45. [Google Scholar] [CrossRef]
- Lodeyro, A.F.; Carrillo, N. Salt Stress in Higher Plants: Mechanisms of Toxicity and Defensive Responses. In Stress Responses in Plants; Springer International Publishing: Cham, Switzerland, 2015; pp. 1–33. [Google Scholar]
- Martínez-Atienza, J.; Jiang, X.; Garciadeblas, B.; Mendoza, I.; Zhu, J.-K.; Pardo, J.M.; Quintero, F.J. Conservation of the Salt Overly Sensitive Pathway in Rice. Plant Physiol. 2007, 143, 1001–1012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanwar, P.; Sanyal, S.K.; Tokas, I.; Yadav, A.K.; Pandey, A.; Kapoor, S.; Pandey, G.K. Comprehensive structural, interaction and expression analysis of CBL and CIPK complement during abiotic stresses and development in rice. Cell Calcium 2014, 56, 81–95. [Google Scholar] [CrossRef] [PubMed]
- Saijo, Y.; Hata, S.; Kyozuka, J.; Shimamoto, K.; Izui, K. Over-expression of a single Ca2+ -dependent protein kinase confers both cold and salt/drought tolerance on rice plants. Plant J. 2000, 23, 319–327. [Google Scholar] [CrossRef]
- Campo, S.; Baldrich, P.; Messeguer, J.; Lalanne, E.; Coca, M. Overexpression of a Calcium-Dependent Protein Kinase Confers Salt and Drought Tolerance in Rice by Preventing Membrane Lipid Peroxidation. Plant Physiol. 2014, 165, 688–704. [Google Scholar] [CrossRef] [Green Version]
- Asano, T.; Hayashi, N.; Kobayashi, M.; Aoki, N.; Miyao, A.; Mitsuhara, I.; Ichikawa, H.; Komatsu, S.; Hirochika, H.; Kikuchi, S.; et al. A rice calcium-dependent protein kinase OsCPK12 oppositely modulates salt-stress tolerance and blast disease resistance. Plant J. 2012, 69, 26–36. [Google Scholar] [CrossRef]
- Eltayeb, A.E.; Kawano, N.; Badawi, G.H.; Kaminaka, H.; Sanekata, T.; Morishima, I.; Shibahara, T.; Inanaga, S.; Tanaka, K. Enhanced tolerance to ozone and drought stresses in transgenic tobacco overexpressing dehydroascorbate reductase in cytosol. Physiol. Plant. 2006, 127, 57–65. [Google Scholar] [CrossRef]
- Eltayeb, A.E.; Kawano, N.; Badawi, G.H.; Kaminaka, H.; Sanekata, T.; Shibahara, T.; Inanaga, S.; Tanaka, K. Overexpression of monodehydroascorbate reductase in transgenic tobacco confers enhanced tolerance to ozone, salt and polyethylene glycol stresses. Planta 2007, 225, 1255–1264. [Google Scholar] [CrossRef]
- James, R.A.; Blake, C.; Byrt, C.S.; Munns, R. Major genes for Na+ exclusion, Nax1 and Nax2 (wheat HKT1;4 and HKT1;5), decrease Na+ accumulation in bread wheat leaves under saline and waterlogged conditions. J. Exp. Bot. 2011, 62, 2939–2947. [Google Scholar] [CrossRef] [Green Version]
- Vaidyanathan, H.; Sivakumar, P.; Chakrabarty, R.; Thomas, G. Scavenging of reactive oxygen species in NaCl-stressed rice (Oryza sativa L.)—Differential response in salt-tolerant and sensitive varieties. Plant Sci. 2003, 165, 1411–1418. [Google Scholar] [CrossRef]
- Uchida, A.; Jagendorf, A.T.; Hibino, T.; Takabe, T.; Takabe, T. Effects of hydrogen peroxide and nitric oxide on both salt and heat stress tolerance in rice. Plant Sci. 2002, 163, 515–523. [Google Scholar] [CrossRef]
- Ahanger, M.A.; Tomar, N.S.; Tittal, M.; Argal, S.; Agarwal, R.M. Plant growth under water/salt stress: ROS production; antioxidants and significance of added potassium under such conditions. Physiol. Mol. Biol. Plants 2017, 23, 731–744. [Google Scholar] [CrossRef]
- Becana, M.; Dalton, D.A.; Moran, J.F.; Iturbe-Ormaetxe, I.; Matamoros, M.A.; Rubio, M.C. Reactive oxygen species and antioxidants in legume nodules. Physiol. Plant. 2000, 109, 372–381. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Zhang, Q.; Wu, J.; Zheng, X.; Zheng, S.; Sun, X.; Qiu, Q.; Lu, T. Gene Knockout Study Reveals That Cytosolic Ascorbate Peroxidase 2(OsAPX2) Plays a Critical Role in Growth and Reproduction in Rice under Drought, Salt and Cold Stresses. PLoS ONE 2013, 8, e57472. [Google Scholar] [CrossRef] [PubMed]
- Hong, C.-Y.; Hsu, Y.T.; Tsai, Y.-C.; Kao, C.H. Expression of ASCORBATE PEROXIDASE 8 in roots of rice (Oryza sativa L.) seedlings in response to NaCl. J. Exp. Bot. 2007, 58, 3273–3283. [Google Scholar] [CrossRef] [PubMed]
- Kaminaka, H.; Morita, S.; Nakajima, M.; Masumura, T.; Tanaka, K. Gene Cloning and Expression of Cytosolic Glutathione Reductase in Rice (Oryza sativa L.). Plant Cell Physiol. 1998, 39, 1269–1280. [Google Scholar] [CrossRef] [Green Version]
- Wu, T.-M.; Lin, W.-R.; Kao, C.H.; Hong, C.-Y. Gene knockout of glutathione reductase 3 results in increased sensitivity to salt stress in rice. Plant Mol. Biol. 2015, 87, 555–564. [Google Scholar] [CrossRef]
- Chen, T.; Shabala, S.; Niu, Y.; Chen, Z.-H.; Shabala, L.; Meinke, H.; Venkataraman, G.; Pareek, A.; Xu, J.; Zhou, M. Molecular mechanisms of salinity tolerance in rice. Crop J. 2021, 9, 506–520. [Google Scholar] [CrossRef]
- Kumar, V.; Khare, T. Differential growth and yield responses of salt-tolerant and susceptible rice cultivars to individual (Na+ and Cl−) and additive stress effects of NaCl. Acta Physiol. Plant. 2016, 38, 170. [Google Scholar] [CrossRef]
- Chi Lin, C.; Huei Kao, C. Relative importance of Na+, Cl−, and abscisic acid in nacl induced inhibition of root growth of rice seedlings. Plant Soil 2001, 237, 165–171. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, M.; Guo, R.; Shi, D.; Liu, B.; Lin, X.; Yang, C. Effects of salt stress on ion balance and nitrogen metabolism of old and young leaves in rice (Oryza sativa L.). BMC Plant Biol. 2012, 12, 194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blumwald, E. Sodium transport and salt tolerance in plants. Curr. Opin. Cell Biol. 2000, 12, 431–434. [Google Scholar] [CrossRef]
- Roy, S.J.; Negrão, S.; Tester, M. Salt resistant crop plants. Curr. Opin. Biotechnol. 2014, 26, 115–124. [Google Scholar] [CrossRef]
- Pires, I.S.; Negrão, S.; Oliveira, M.M.; Purugganan, M.D. Comprehensive phenotypic analysis of rice (Oryza sativa) response to salinity stress. Physiol. Plant. 2015, 155, 43–54. [Google Scholar] [CrossRef] [Green Version]
- Golldack, D. Molecular Responses of Halophytes to High Salinity. Progress Bot. 2004, 65, 219–234. [Google Scholar]
- Zhu, J.-K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Batelli, G.; Verslues, P.E.; Agius, F.; Qiu, Q.; Fujii, H.; Pan, S.; Schumaker, K.S.; Grillo, S.; Zhu, J.-K. SOS2 Promotes Salt Tolerance in Part by Interacting with the Vacuolar H + -ATPase and Upregulating Its Transport Activity. Mol. Cell. Biol. 2007, 27, 7781–7790. [Google Scholar] [CrossRef] [Green Version]
- Gong, Z. Plant abiotic stress: New insights into the factors that activate and modulate plant responses. J. Integr. Plant Biol. 2021, 63, 429–430. [Google Scholar] [CrossRef]
- Singhal, R.K.; Saha, D.; Skalicky, M.; Mishra, U.N.; Chauhan, J.; Behera, L.P.; Lenka, D.; Chand, S.; Kumar, V.; Dey, P.; et al. Crucial Cell Signaling Compounds Crosstalk and Integrative Multi-Omics Techniques for Salinity Stress Tolerance in Plants. Front. Plant Sci. 2021, 12, 1227. [Google Scholar] [CrossRef]
- Wang, F.; Jing, W.; Zhang, W. The mitogen-activated protein kinase cascade MKK1–MPK4 mediates salt signaling in rice. Plant Sci. 2014, 227, 181–189. [Google Scholar] [CrossRef]
- Kumar, K.; Kumar, M.; Kim, S.R.; Ryu, H.; Cho, Y.G. Insights into genomics of salt stress response in rice. Rice 2013, 6, 27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cramer, G.R.; Urano, K.; Delrot, S.; Pezzotti, M.; Shinozaki, K. Effects of abiotic stress on plants: A systems biology perspective. BMC Plant Biol. 2011, 11, 163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karahalil, B. Overview of Systems Biology and Omics Technologies. Curr. Med. Chem. 2016, 23, 4221–4230. [Google Scholar] [CrossRef] [PubMed]
- Hasin, Y.; Seldin, M.; Lusis, A. Multi-omics approaches to disease. Genome Biol. 2017, 18, 83. [Google Scholar] [CrossRef]
- Yang, Y.; Saand, M.A.; Huang, L.; Abdelaal, W.B.; Zhang, J.; Wu, Y.; Li, J.; Sirohi, M.H.; Wang, F. Applications of Multi-Omics Technologies for Crop Improvement. Front. Plant Sci. 2021, 12, 1846. [Google Scholar] [CrossRef]
- Sasaki, T. The map-based sequence of the rice genome. Nature 2005, 436, 793–800. [Google Scholar] [CrossRef]
- De Leon, T.B.; Linscombe, S.; Subudhi, P.K. Identification and validation of QTLs for seedling salinity tolerance in introgression lines of a salt tolerant rice landrace “Pokkali”. PLoS ONE 2017, 12, e0175361. [Google Scholar] [CrossRef]
- Thomson, M.J. High-Throughput SNP Genotyping to Accelerate Crop Improvement. Plant Breed. Biotechnol. 2014, 2, 195–212. [Google Scholar] [CrossRef]
- Chen, T.; Zhu, Y.; Chen, K.; Shen, C.; Zhao, X.; Shabala, S.; Shabala, L.; Meinke, H.; Venkataraman, G.; Chen, Z.; et al. Identification of new QTL for salt tolerance from rice variety Pokkali. J. Agron. Crop Sci. 2020, 206, 202–213. [Google Scholar] [CrossRef]
- Lin, H.X.; Zhu, M.Z.; Yano, M.; Gao, J.P.; Liang, Z.W.; Su, W.A.; Hu, X.H.; Ren, Z.H.; Chao, D.Y. QTLs for Na+ and K+ uptake of the shoots and roots controlling rice salt tolerance. Theor. Appl. Genet. 2004, 108, 253–260. [Google Scholar] [CrossRef]
- Sanchouli, S.; Neghab, M.; Sabouri, H.; Zare Mehrjerdi, M. Genetic Structure of Salinity Tolerance in Rice at Seedling Stage. J. Genet. Resour. 2019, 5, 22–30. [Google Scholar]
- Rahman, M.A.; Thomson, M.J.; De Ocampo, M.; Egdane, J.A.; Salam, M.A.; Shah-E-Alam, M.; Ismail, A.M. Assessing trait contribution and mapping novel QTL for salinity tolerance using the Bangladeshi rice landrace Capsule. Rice 2019, 12, 63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rahman, M.A.; Bimpong, I.K.; Bizimana, J.B.; Pascual, E.D.; Arceta, M.; Swamy, B.P.M.; Diaw, F.; Rahman, M.S.; Singh, R.K. Mapping QTLs using a novel source of salinity tolerance from Hasawi and their interaction with environments in rice. Rice 2017, 10, 47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ocampo, M.; The, H.V.; Thomson, M.; Mitsuya, S.; Yamauchi, A.; Ismail, A. QTL mapping and candidate gene identification in rice using a Kalarata-Azucena population under salt stress. Res. Sq. 2020, 1–15. [Google Scholar] [CrossRef]
- Puram, V.R.R.; Ontoy, J.; Linscombe, S.; Subudhi, P.K. Genetic Dissection of Seedling Stage Salinity Tolerance in Rice Using Introgression Lines of a Salt Tolerant Landrace Nona Bokra. J. Hered. 2017, 108, 658–670. [Google Scholar] [CrossRef]
- Wu, F.; Yang, J.; Yu, D.; Xu, P. Identification and Validation a Major QTL from “Sea Rice 86” Seedlings Conferred Salt Tolerance. Agronomy 2020, 10, 410. [Google Scholar] [CrossRef] [Green Version]
- Ammar, M.H.M.; Pandit, A.; Singh, R.K.; Sameena, S.; Chauhan, M.S.; Singh, A.K.; Sharma, P.C.; Gaikwad, K.; Sharma, T.R.; Mohapatra, T.; et al. Mapping of QTLs Controlling Na+, K+ and CI− Ion Concentrations in Salt Tolerant Indica Rice Variety CSR27. J. Plant Biochem. Biotechnol. 2009, 18, 139–150. [Google Scholar] [CrossRef]
- Lang, N.T.; Phuoc, N.T.; Ha, P.T.T.; Buu, B.C. Identifying QTLs Associated and Marker-Assisted Selection for Salinity Tolerance at the Seedling, Vegetative and Reproductive Stages in Rice (Oryza sativa L.). Int. J. Environ. Agric. Biotechnol. 2017, 2, 2927–2935. [Google Scholar] [CrossRef]
- Haque, T.; Elias, S.M.; Razzaque, S.; Biswas, S.; Khan, S.F.; Jewel, G.N.A.; Rahman, M.S.; Juenger, T.E.; Seraj, Z.I. Natural variation in growth and physiology under salt stress in rice: QTL mapping in a Horkuch × IR29 mapping population at seedling and reproductive stages. bioRxiv 2020, 1–30. [Google Scholar] [CrossRef] [Green Version]
- Hossain, H.; Rahman, M.A.; Alam, M.S.; Singh, R.K. Mapping of Quantitative Trait Loci Associated with Reproductive-Stage Salt Tolerance in Rice. J. Agron. Crop Sci. 2015, 201, 17–31. [Google Scholar] [CrossRef]
- Ren, Z.H.; Gao, J.P.; Li, L.G.; Cai, X.L.; Huang, W.; Chao, D.Y.; Zhu, M.Z.; Wang, Z.Y.; Luan, S.; Lin, H.X. A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat. Genet. 2005, 37, 1141–1146. [Google Scholar] [CrossRef] [PubMed]
- Pandit, A.; Rai, V.; Bal, S.; Sinha, S.; Kumar, V.; Chauhan, M.; Gautam, R.K.; Singh, R.; Sharma, P.C.; Singh, A.K.; et al. Combining QTL mapping and transcriptome profiling of bulked RILs for identification of functional polymorphism for salt tolerance genes in rice (Oryza sativa L.). Mol. Genet. Genom. 2010, 284, 121–136. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, J.; Bao, Y.; Wu, Y.; Zhang, H. Quantitative trait loci controlling rice seed germination under salt stress. Euphytica 2011, 178, 297–307. [Google Scholar] [CrossRef]
- Zeng, P.; Zhu, P.; Qian, L.; Qian, X.; Mi, Y.; Lin, Z.; Dong, S.; Aronsson, H.; Zhang, H.; Cheng, J. Identification and fine mapping of qGR6.2, a novel locus controlling rice seed germination under salt stress. BMC Plant Biol. 2021, 21, 36. [Google Scholar] [CrossRef] [PubMed]
- Bonilla, P.; Dvorak, J.; Mackill, D.; Deal, K.; Gregorio, G. RFLP and SSLP mapping of salinity tolerance genes in chromosome 1 of rice (Oryza sativa L.) using recombinant inbred lines. Philipp. Agric. Sci. 2002, 65, 68–76. [Google Scholar]
- Cortés, A.J.; Skeen, P.; Blair, M.W.; Chacón-Sánchez, M.I. Does the Genomic Landscape of Species Divergence in Phaseolus Beans Coerce Parallel Signatures of Adaptation and Domestication? Front. Plant Sci. 2018, 9, 1816. [Google Scholar] [CrossRef] [Green Version]
- Ravinet, M.; Faria, R.; Butlin, R.K.; Galindo, J.; Bierne, N.; Rafajlović, M.; Noor, M.A.F.; Mehlig, B.; Westram, A.M. Interpreting the genomic landscape of speciation: A road map for finding barriers to gene flow. J. Evol. Biol. 2017, 30, 1450–1477. [Google Scholar] [CrossRef] [Green Version]
- Cortés, A.J.; López-Hernández, F.; Osorio-Rodriguez, D. Predicting Thermal Adaptation by Looking Into Populations’ Genomic Past. Front. Genet. 2020, 11, 1093. [Google Scholar] [CrossRef]
- Cortés, A.J.; Blair, M.W. Genotyping by Sequencing and Genome–Environment Associations in Wild Common Bean Predict Widespread Divergent Adaptation to Drought. Front. Plant Sci. 2018, 9, 128. [Google Scholar] [CrossRef] [Green Version]
- Challa, S.; Neelapu, N.R.R. Genome-Wide Association Studies (GWAS) for Abiotic Stress Tolerance in Plants. In Biochemical, Physiological and Molecular Avenues for Combating Abiotic Stress Tolerance in Plants; Elsevier: Amsterdam, The Netherlands, 2018; pp. 135–150. [Google Scholar]
- Yadav, A.K.; Kumar, A.; Grover, N.; Ellur, R.K.; Bollinedi, H.; Krishnan, S.G.; Bhowmick, P.K.; Vinod, K.K.; Nagarajan, M.; Singh, A.K. Genome-Wide Association Study Reveals Marker–Trait Associations for Early Vegetative Stage Salinity Tolerance in Rice. Plants 2021, 10, 559. [Google Scholar] [CrossRef]
- Naveed, S.A.; Zhang, F.; Zhang, J.; Zheng, T.-Q.; Meng, L.-J.; Pang, Y.-L.; Xu, J.-L.; Li, Z.-K. Identification of QTN and candidate genes for Salinity Tolerance at the Germination and Seedling Stages in Rice by Genome-Wide Association Analyses. Sci. Rep. 2018, 8, 6505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, Y.; Zhang, F.; Zhou, Y. The Application of Multi-Locus GWAS for the Detection of Salt-Tolerance Loci in Rice. Front. Plant Sci. 2018, 9, 1464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muthamilarasan, M.; Singh, N.K.; Prasad, M. Multi-omics approaches for strategic improvement of stress tolerance in underutilized crop species: A climate change perspective. Adv. Genet. 2019, 103, 1–38. [Google Scholar] [PubMed]
- Qin, H.; Li, Y.; Huang, R. Advances and Challenges in the Breeding of Salt-Tolerant Rice. Int. J. Mol. Sci. 2020, 21, 8385. [Google Scholar] [CrossRef]
- Garg, P.; Jaiswal, P. Databases and bioinformatics tools for rice research. Curr. Plant Biol. 2016, 7–8, 39–52. [Google Scholar] [CrossRef]
- Zhou, X.; Bai, X.; Xing, Y. A Rice Genetic Improvement Boom by Next Generation Sequencing. Curr. Issues Mol. Biol. 2018, 27, 109–126. [Google Scholar] [CrossRef]
- Prasanna, S.; Jain, S.M. Mutant Resources and Mutagenomics in crop plants. Emirates J. Food Agric. 2017, 29, 651–657. [Google Scholar] [CrossRef] [Green Version]
- Varshney, R.K.; Thundi, M.; May, G.D.; Jackson, S.A. Legume Genomics and Breeding. In Plant Breeding Reviews; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2010; pp. 257–304. [Google Scholar]
- Das, P.; Mishra, M.; Lakra, N.; Singla-Pareek, S.L.; Pareek, A. Mutation breeding: A powerful approach for obtaining abiotic stress tolerant crops and upgrading food security for human nutrition. In Mutagenesis: Exploring Novel Genes and Pathways; Wageningen Academic Publishers: Wageningen, The Netherlands, 2014; pp. 615–621. [Google Scholar]
- Viana, V.E.; Pegoraro, C.; Busanello, C.; Costa de Oliveira, A. Mutagenesis in Rice: The Basis for Breeding a New Super Plant. Front. Plant Sci. 2019, 10, 1326. [Google Scholar] [CrossRef] [Green Version]
- Oladosu, Y.; Rafii, M.Y.; Abdullah, N.; Hussin, G.; Ramli, A.; Rahim, H.A.; Miah, G.; Usman, M. Principle and application of plant mutagenesis in crop improvement: A review. Biotechnol. Biotechnol. Equip. 2016, 30, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Mohapatra, T.; Robin, S.; Sarla, N.; Sheshasayee, M.; Singh, A.K.; Singh, K.; Singh, N.K.; Amitha Mithra, S.V.; Sharma, R.P. EMS Induced Mutants of Upland Rice Variety Nagina22: Generation and Characterization. Proc. Indian Natl. Sci. Acad. 2014, 80, 163. [Google Scholar] [CrossRef]
- Lin, K.C.; Jwo, W.S.; Chandrika, N.N.P.; Wu, T.M.; Lai, M.H.; Wang, C.S.; Hong, C.Y. A rice mutant defective in antioxidant-defense system and sodium homeostasis possesses increased sensitivity to salt stress. Biol. Plant. 2016, 60, 86–94. [Google Scholar] [CrossRef]
- Tu, Y.; Jiang, A.; Gan, L.; Hossain, M.; Zhang, J.; Peng, B.; Xiong, Y.; Song, Z.; Cai, D.; Xu, W.; et al. Genome duplication improves rice root resistance to salt stress. Rice 2014, 7, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakhoda, B.; Leung, H.; Mendioro, M.S.; Mohammadi-nejad, G.; Ismail, A.M. Isolation, characterization, and field evaluation of rice (Oryza sativa L., Var. IR64) mutants with altered responses to salt stress. Field Crops Res. 2012, 127, 191–202. [Google Scholar] [CrossRef]
- Roldán-Arjona, T.; Ariza, R.R. Repair and tolerance of oxidative DNA damage in plants. Mutat. Res. Mutat. Res. 2009, 681, 169–179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, J.; Kim, D.; Lee, M.-C.; Lee, K.; Kim, J.; Kim, S.; Ha, B.-K.; Yun, S.; Kang, S.-Y. Physiological characterization of gamma-ray induced salt tolerant rice mutants. Aust. J. Crop Sci. 2012, 6, 421–429. [Google Scholar]
- Joshi, R.; Prashat, R.; Sharma, P.C.; Singla-Pareek, S.L.; Pareek, A. Physiological characterization of gamma-ray induced mutant population of rice to facilitate biomass and yield improvement under salinity stress. Indian J. Plant Physiol. 2016, 21, 545–555. [Google Scholar] [CrossRef]
- Kumar, P.; Sharma, V.; Yadav, P.; Singh, B. Gamma Ray Irradiation for Crop Protection Against Salt Stress. Def. Life Sci. J. 2017, 2, 292. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Zhou, D.-X. Rice epigenomics and epigenetics: Challenges and opportunities. Curr. Opin. Plant Biol. 2013, 16, 164–169. [Google Scholar] [CrossRef]
- Ferreira, L.J.; Azevedo, V.; Maroco, J.; Oliveira, M.M.; Santos, A.P. Salt Tolerant and Sensitive Rice Varieties Display Differential Methylome Flexibility under Salt Stress. PLoS ONE 2015, 10, e0124060. [Google Scholar] [CrossRef] [Green Version]
- Chinnusamy, V.; Zhu, J.-K. Epigenetic regulation of stress responses in plants. Curr. Opin. Plant Biol. 2009, 12, 133–139. [Google Scholar] [CrossRef] [Green Version]
- Karan, R.; DeLeon, T.; Biradar, H.; Subudhi, P.K. Salt Stress Induced Variation in DNA Methylation Pattern and Its Influence on Gene Expression in Contrasting Rice Genotypes. PLoS ONE 2012, 7, e40203. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Huang, F.; Qin, Q.; Zhao, X.; Li, Z.; Fu, B. Comparative analysis of DNA methylation changes in two rice genotypes under salt stress and subsequent recovery. Biochem. Biophys. Res. Commun. 2015, 465, 790–796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferreira, L.J.; Donoghue, M.T.A.; Barros, P.; Saibo, N.J.; Santos, A.P.; Oliveira, M.M. Uncovering Differentially Methylated Regions (DMRs) in a Salt-Tolerant Rice Variety under Stress: One Step towards New Regulatory Regions for Enhanced Salt Tolerance. Epigenomes 2019, 3, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Q.; Feng, Q.; Lu, H.; Li, Y.; Wang, A.; Tian, Q.; Zhan, Q.; Lu, Y.; Zhang, L.; Huang, T.; et al. Pan-genome analysis highlights the extent of genomic variation in cultivated and wild rice. Nat. Genet. 2018, 50, 278–284. [Google Scholar] [CrossRef] [Green Version]
- Bayer, P.E.; Golicz, A.A.; Scheben, A.; Batley, J.; Edwards, D. Plant pan-genomes are the new reference. Nat. Plants 2020, 6, 914–920. [Google Scholar] [CrossRef]
- Jaiswal, S.; Gautam, R.K.; Singh, R.K.; Krishnamurthy, S.L.; Ali, S.; Sakthivel, K.; Iquebal, M.A.; Rai, A.; Kumar, D. Harmonizing technological advances in phenomics and genomics for enhanced salt tolerance in rice from a practical perspective. Rice 2019, 12, 89. [Google Scholar] [CrossRef]
- Shi, Y.; Gao, L.; Wu, Z.; Zhang, X.; Wang, M.; Zhang, C.; Zhang, F.; Zhou, Y.; Li, Z. Genome-wide association study of salt tolerance at the seed germination stage in rice. BMC Plant Biol. 2017, 17, 92. [Google Scholar] [CrossRef] [Green Version]
- Guo, T.; Yang, J.; Li, D.; Sun, K.; Luo, L.; Xiao, W.; Wang, J.; Liu, Y.; Wang, S.; Wang, H.; et al. Integrating GWAS, QTL, mapping and RNA-seq to identify candidate genes for seed vigor in rice (Oryza sativa L.). Mol. Breed. 2019, 39, 87. [Google Scholar] [CrossRef]
- Matsuda, F.; Nakabayashi, R.; Yang, Z.; Okazaki, Y.; Yonemaru, J.; Ebana, K.; Yano, M.; Saito, K. Metabolome-genome-wide association study dissects genetic architecture for generating natural variation in rice secondary metabolism. Plant J. 2015, 81, 13–23. [Google Scholar] [CrossRef] [Green Version]
- Raza, A.; Tabassum, J.; Kudapa, H.; Varshney, R.K. Can omics deliver temperature resilient ready-to-grow crops? Crit. Rev. Biotechnol. 2021, 41, 1209–1232. [Google Scholar] [CrossRef]
- McGettigan, P.A. Transcriptomics in the RNA-seq era. Curr. Opin. Chem. Biol. 2013, 17, 4–11. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.C.; Chen, Y. Transcriptomics: Advances and approaches. Sci. China Life Sci. 2013, 56, 960–967. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilhelm, B.T.; Marguerat, S.; Watt, S.; Schubert, F.; Wood, V.; Goodhead, I.; Penkett, C.J.; Rogers, J.; Bähler, J. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature 2008, 453, 1239–1243. [Google Scholar] [CrossRef] [PubMed]
- Schena, M.; Shalon, D.; Davis, R.W.; Brown, P.O. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 1995, 270, 467–470. [Google Scholar] [CrossRef] [Green Version]
- Lowe, R.; Shirley, N.; Bleackley, M.; Dolan, S.; Shafee, T. Transcriptomics technologies. PLoS Comput. Biol. 2017, 13, e1005457. [Google Scholar] [CrossRef] [Green Version]
- Thiemann, A.; Fu, J.; Schrag, T.A.; Melchinger, A.E.; Frisch, M.; Scholten, S. Correlation between parental transcriptome and field data for the characterization of heterosis in Zea mays L. Theor. Appl. Genet. 2010, 120, 401–413. [Google Scholar] [CrossRef] [PubMed]
- Ge, X.; Chen, W.; Song, S.; Wang, W.; Hu, S.; Yu, J. Transcriptomic profiling of mature embryo from an elite super-hybrid rice LYP9 and its parental lines. BMC Plant Biol. 2008, 8, 114. [Google Scholar] [CrossRef] [Green Version]
- Eyidogan, F.; Öz, M.T. Effect of salinity on antioxidant responses of chickpea seedlings. Acta Physiol. Plant. 2007, 29, 485–493. [Google Scholar] [CrossRef]
- Kumari, S.; Panjabinee Sabharwal, V.; Kushwaha, H.R.; Sopory, S.K.; Singla-Pareek, S.L.; Pareek, A. Transcriptome map for seedling stage specific salinity stress response indicates a specific set of genes as candidate for saline tolerance in Oryza sativa L. Funct. Integr. Genom. 2009, 9, 109–123. [Google Scholar] [CrossRef]
- Chao, D.Y.; Luo, Y.H.; Shi, M.; Luo, D.; Lin, H.X. Salt-responsive genes in rice revealed by cDNA microarray analysis. Cell Res. 2005, 15, 796–810. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics in Western Equatoria State. Nat. Rev. Genet. 2009, 10, 57. [Google Scholar] [CrossRef] [PubMed]
- Ozsolak, F.; Milos, P.M. RNA sequencing: Advances, challenges and opportunities. Nat. Rev. Genet. 2011, 12, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Zainal-Abidin, R.-A.; Ruhaizat-Ooi, I.-H.; Harun, S. A Review of Omics Technologies and Bioinformatics to Accelerate Improvement of Papaya Traits. Agronomy 2021, 11, 1356. [Google Scholar] [CrossRef]
- Zhou, Y.; Yang, P.; Cui, F.; Zhang, F.; Luo, X.; Xie, J. Transcriptome analysis of salt stress responsiveness in the seedlings of dongxiang wild rice (Oryza rufipogon Griff.). PLoS ONE 2016, 11, e0146242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chandran, A.K.N.; Kim, J.-W.; Yoo, Y.-H.; Park, H.L.; Kim, Y.-J.; Cho, M.-H.; Jung, K.-H. Transcriptome analysis of rice-seedling roots under soil–salt stress using RNA-Seq method. Plant Biotechnol. Rep. 2019, 13, 567–578. [Google Scholar] [CrossRef]
- Jahan, N.; Lv, Y.; Song, M.; Zhang, Y.; Shang, L.; Lu, Y.; Ye, G.; Qian, Q.; Gao, Z.; Guo, L. Transcriptomic Analysis of Short-Term Salt-Stress Response in Mega Hybrid Rice Seedlings. Agronomy 2021, 11, 1328. [Google Scholar] [CrossRef]
- Gupta, B.; Huang, B. Mechanism of Salinity Tolerance in Plants: Physiological, Biochemical, and Molecular Characterization. Int. J. Genom. 2014, 2014, 701596. [Google Scholar] [CrossRef]
- Huang, X.-Y.; Chao, D.-Y.; Gao, J.-P.; Zhu, M.-Z.; Shi, M.; Lin, H.-X. A previously unknown zinc finger protein, DST, regulates drought and salt tolerance in rice via stomatal aperture control. Genes Dev. 2009, 23, 1805–1817. [Google Scholar] [CrossRef] [Green Version]
- Zhu, N.; Cheng, S.; Liu, X.; Du, H.; Dai, M.; Zhou, D.X.; Yang, W.; Zhao, Y. The R2R3-type MYB gene OsMYB91 has a function in coordinating plant growth and salt stress tolerance in rice. Plant Sci. 2015, 236, 146–156. [Google Scholar] [CrossRef]
- Liu, C.; Mao, B.; Ou, S.; Wang, W.; Liu, L.; Wu, Y.; Chu, C.; Wang, X. OsbZIP71, a bZIP transcription factor, confers salinity and drought tolerance in rice. Plant Mol. Biol. 2014, 84, 19–36. [Google Scholar] [CrossRef]
- Han, M.; Kim, C.Y.; Lee, J.; Lee, S.K.; Jeon, J.S. OsWRKY42 represses OsMTId and induces reactive oxygen species and leaf senescence in rice. Mol. Cells 2014, 37, 532–539. [Google Scholar] [CrossRef] [PubMed]
- Jan, A.; Maruyama, K.; Todaka, D.; Kidokoro, S.; Abo, M.; Yoshimura, E.; Shinozaki, K.; Nakashima, K.; Yamaguchi-Shinozaki, K. OsTZF1, a CCCH-tandem zinc finger protein, confers delayed senescence and stress tolerance in rice by regulating stress-related genes. Plant Physiol. 2013, 161, 1202–1216. [Google Scholar] [CrossRef] [Green Version]
- Song, S.Y.; Chen, Y.; Chen, J.; Dai, X.Y.; Zhang, W.H. Physiological mechanisms underlying OsNAC5-dependent tolerance of rice plants to abiotic stress. Planta 2011, 234, 331–345. [Google Scholar] [CrossRef] [PubMed]
- Lan, X.; Hoang, T.; Ngoc, D.; Nhi, H.; Binh, N.; Thu, A.; Thao, N.P.; Tran, L.P. Transcription Factors and Their Roles in Signal Transduction in Plants under Abiotic Stresses. Curr. Genom. 2017, 483–497. [Google Scholar]
- Kumar, D.; Das, P.K.; Singha, C.; Sarmah, B.K. Mining and Characterizing the SSR Markers for Black Rice Using the Illumina Sequencing Platform. Preprints 2020, 2020030119. [Google Scholar] [CrossRef]
- Alisoltani, A.; Shiran, B.; Sarvestani, N.R.; Fallahi, H.; Feto, N.; Ebrahimie, E. Changes in Microsatellite Motifs in Response to Abiotic Stresses: A Case Study Using Wheat and Rice RNA-sequencing Data. Asian J. Sci. Res. 2018, 11, 12–21. [Google Scholar] [CrossRef]
- Wang, J.; Zhu, J.; Zhang, Y.; Fan, F.; Li, W.; Wang, F.; Zhong, W.; Wang, C.; Yang, J. Comparative transcriptome analysis reveals molecular response to salinity stress of salt-tolerant and sensitive genotypes of indica rice at seedling stage. Sci. Rep. 2018, 8, 2085. [Google Scholar] [CrossRef] [Green Version]
- Cartagena, J.A.; Yao, Y.; Mitsuya, S.; Tsuge, T. Comparative transcriptome analysis of root types in salt tolerant and sensitive rice varieties in response to salinity stress. Physiol. Plant. 2021, 173, 1629–1642. [Google Scholar] [CrossRef]
- Li, Y.; Zheng, Y.; Vemireddy, L.R.; Panda, S.K.; Jose, S.; Ranjan, A.; Panda, P.; Govindan, G.; Cui, J.; Wei, K.; et al. Comparative transcriptome and translatome analysis in contrasting rice genotypes reveals differential mRNA translation in salt-tolerant Pokkali under salt stress. BMC Genom. 2018, 19, 935. [Google Scholar] [CrossRef] [Green Version]
- Cho, K.; Shibato, J.; Agrawal, G.K.; Jung, Y.-H.; Kubo, A.; Jwa, N.-S.; Tamogami, S.; Satoh, K.; Kikuchi, S.; Higashi, T.; et al. Integrated Transcriptomics, Proteomics, and Metabolomics Analyses To Survey Ozone Responses in the Leaves of Rice Seedling. J. Proteome Res. 2008, 7, 2980–2998. [Google Scholar] [CrossRef]
- Aslam, B.; Basit, M.; Nisar, M.A.; Khurshid, M.; Rasool, M.H. Proteomics: Technologies and Their Applications. J. Chromatogr. Sci. 2017, 55, 182–196. [Google Scholar] [CrossRef] [Green Version]
- Yates, J.R.; Ruse, C.I.; Nakorchevsky, A. Proteomics by Mass Spectrometry: Approaches, Advances, and Applications. Annu. Rev. Biomed. Eng. 2009, 11, 49–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, J.; Rampitsch, C.; Bykova, N.V. Advances in plant proteomics toward improvement of crop productivity and stress resistancex. Front. Plant Sci. 2015, 6, 209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, D.-G.; Woong Park, K.; Young An, J.; Geol Sohn, Y.; Ki Ha, J.; Yoon Kim, H.; Won Bae, D.; Hee Lee, K.; Jun Kang, N.; Lee, B.-H.; et al. Proteomics analysis of salt-induced leaf proteins in two rice germplasms with different salt sensitivity. Can. J. Plant Sci. 2011, 91, 337–349. [Google Scholar] [CrossRef]
- Sarhadi, E.; Bazargani, M.M.; Sajise, A.G.; Abdolahi, S.; Vispo, N.A.; Arceta, M.; Nejad, G.M.; Singh, R.K.; Salekdeh, G.H. Proteomic analysis of rice anthers under salt stress. Plant Physiol. Biochem. 2012, 58, 280–287. [Google Scholar] [CrossRef]
- Song, Y.; Zhang, C.; Ge, W.; Zhang, Y.; Burlingame, A.L.; Guo, Y. Identification of NaCl stress-responsive apoplastic proteins in rice shoot stems by 2D-DIGE. J. Proteom. 2011, 74, 1045–1067. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.W.; Rakwal, R.; Agrawal, G.K.; Jung, Y.H.; Shibato, J.; Jwa, N.S.; Iwahashi, Y.; Iwahashi, H.; Kim, D.H.; Shim, I.S.; et al. A hydroponic rice seedling culture model system for investigating proteome of salt stress in rice leaf. Electrophoresis 2005, 26, 4521–4539. [Google Scholar] [CrossRef]
- Sengupta, S.; Majumder, A.L. Insight into the salt tolerance factors of a wild halophytic rice, Porteresia coarctata: A physiological and proteomic approach. Planta 2009, 229, 911–929. [Google Scholar] [CrossRef]
- Naqvi, S.M.S.; Raza, S.Q.; Hyder, M.Z.; Ozalp, C.V.; Oktem, H.A.; Yucel, M. Sub-cellular distribution of two salt-induced peptides in roots of Oryza sativa L. var Nonabokra. Afr. J. Biotechnol. 2009, 8, 4613–4617. [Google Scholar]
- Zhang, L.; Tian, L.H.; Zhao, J.F.; Song, Y.; Zhang, C.J.; Guo, Y. Identification of an apoplastic protein involved in the initial phase of salt stress response in rice root by two-dimensional electrophoresis. Plant Physiol. 2009, 149, 916–928. [Google Scholar] [CrossRef] [Green Version]
- Nam, M.H.; Huh, S.M.; Kim, K.M.; Park, W.J.; Seo, J.B.; Cho, K.; Kim, D.Y.; Kim, B.G.; Yoon, I.S. Comparative proteomic analysis of early salt stress-responsive proteins in roots of SnRK2 transgenic rice. Proteome Sci. 2012, 10, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dooki, A.D.; Mayer-Posner, F.J.; Askari, H.; Zaiee, A.A.; Salekdeh, G.H. Proteomic responses of rice young panicles to salinity. Proteomics 2006, 6, 6498–6507. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Song, Y. Differential proteomic analysis of apoplastic proteins during initial phase of salt stress in rice. Plant Signal. Behav. 2009, 4, 121–122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.-J.; Yang, M.-F.; Zhu, Y.; Liang, Y.; Shen, S.-H. Proteomic Analysis of Salt Stress Responses in Rice Shoot. J. Plant Biol. 2011, 54, 384–395. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, B.; Li, J.; Song, Z.; Lu, B.; Chi, M.; Yang, B.; Liu, J.; Lam, Y.W.; Li, J.; et al. Salt-response analysis in two rice cultivars at seedling stage. Acta Physiol. Plant. 2017, 39, 215. [Google Scholar] [CrossRef]
- Liu, C.-W.; Hsu, Y.-K.; Cheng, Y.-H.; Yen, H.-C.; Wu, Y.-P.; Wang, C.-S.; Lai, C.-C. Proteomic analysis of salt-responsive ubiquitin-related proteins in rice roots. Rapid Commun. Mass Spectrom. 2012, 26, 1649–1660. [Google Scholar] [CrossRef]
- Wen, F.; Zhang, Z.; Bai, T.; Xu, Q.; Pan, Y. Proteomics reveals the effects of gibberellic acid (GA3) on salt-stressed rice (Oryza sativa L.) shoots. Plant Sci. 2010, 178, 170–175. [Google Scholar] [CrossRef]
- Kosová, K.; Vítámvás, P.; Prášil, I.T.; Renaut, J. Plant proteome changes under abiotic stress—Contribution of proteomics studies to understanding plant stress response. J. Proteom. 2011, 74, 1301–1322. [Google Scholar] [CrossRef]
- Li, X.-J.; Yang, M.-F.; Chen, H.; Qu, L.-Q.; Chen, F.; Shen, S.-H. Abscisic acid pretreatment enhances salt tolerance of rice seedlings: Proteomic evidence. Biochim. Biophys. Acta—Proteins Proteom. 2010, 1804, 929–940. [Google Scholar] [CrossRef]
- Xu, E.; Chen, M.; He, H.; Zhan, C.; Cheng, Y.; Zhang, H.; Wang, Z. Proteomic Analysis Reveals Proteins Involved in Seed Imbibition under Salt Stress in Rice. Front. Plant Sci. 2017, 7, 2006. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Lan, H.; Fang, H.; Huang, X.; Zhang, H.; Huang, J. Quantitative Proteomic Analysis of the Rice (Oryza sativa L.) Salt Response. PLoS ONE 2015, 10, e0120978. [Google Scholar]
- Ruan, C.J.; Teixeira Da Silva, J.A. Metabolomics: Creating new potentials for unraveling the mechanisms in response to salt and drought stress and for the biotechnological improvement of xero-halophytes. Crit. Rev. Biotechnol. 2011, 31, 153–169. [Google Scholar] [CrossRef] [PubMed]
- Udomchalothorn, T.; Maneeprasobsuk, S.; Bangyeekhun, E.; Boon-Long, P.; Chadchawan, S. The role of the bifunctional enzyme, fructose-6-phosphate-2-kinase/fructose-2,6-bisphosphatase, in carbon partitioning during salt stress and salt tolerance in Rice (Oryza sativa L.). Plant Sci. 2009, 176, 334–341. [Google Scholar] [CrossRef]
- Hussain, S.; Zhu, C.; Bai, Z.; Huang, J.; Zhu, L.; Cao, X.; Nanda, S.; Hussain, S.; Riaz, A.; Liang, Q.; et al. iTRAQ-Based Protein Profiling and Biochemical Analysis of Two Contrasting Rice Genotypes Revealed Their Differential Responses to Salt Stress. Int. J. Mol. Sci. 2019, 20, 547. [Google Scholar] [CrossRef] [Green Version]
- López-Cristoffanini, C.; Bundó, M.; Serrat, X.; San Segundo, B.; López-Carbonell, M.; Nogués, S. A comprehensive study of the proteins involved in salinity stress response in roots and shoots of the FL478 genotype of rice (Oryza sativa L. ssp. indica). Crop J. 2021, 9, 1154–1168. [Google Scholar] [CrossRef]
- Xiong, E.; Zhang, C.; Ye, C.; Jiang, Y.; Zhang, Y.; Chen, F.; Dong, G.; Zeng, D.; Yu, Y.; Wu, L. iTRAQ-based proteomic analysis provides insights into the molecular mechanisms of rice formyl tetrahydrofolate deformylase in salt response. Planta 2021, 254, 76. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Ford, K.L.; Roessner, U.; Natera, S.; Cassin, A.M.; Patterson, J.H.; Bacic, A. Rice suspension cultured cells are evaluated as a model system to study salt responsive networks in plants using a combined proteomic and metabolomic profiling approach. Proteomics 2013, 13, 2046–2062. [Google Scholar] [CrossRef]
- Baharum, S.N.; Azizan, K.A. Metabolomics in Systems Biology. Adv. Exp. Med. Biol. 2018, 1102, 51–68. [Google Scholar]
- Fahimirad, S.; Ghorbanpour, M. Omics Approaches in Developing Abiotic Stress Tolerance in Rice (Oryza sativa L.); Elsevier Inc.: Amsterdam, The Netherlands, 2019; ISBN 9780128143322. [Google Scholar]
- Pandey, M.K.; Roorkiwal, M.; Singh, V.K.; Ramalingam, A.; Kudapa, H.; Thudi, M.; Chitikineni, A.; Rathore, A.; Varshney, R.K. Emerging Genomic Tools for Legume Breeding: Current Status and Future Prospects. Front. Plant Sci. 2016, 7, 455. [Google Scholar] [CrossRef] [Green Version]
- Royuela, M.; Gonzalez, A.; Gonzalez, E.M.; Arrese-Igor, C.; Aparicio-Tejo, P.M.; Gonzalez-Murua, C. Physiological consequences of continuous, sublethal imazethapyr supply to pea plants. J. Plant Physiol. 2000, 157, 345–354. [Google Scholar] [CrossRef]
- Kusano, M.; Yang, Z.; Okazaki, Y.; Nakabayashi, R.; Fukushima, A.; Saito, K. Using metabolomic approaches to explore chemical diversity in rice. Mol. Plant 2015, 8, 58–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lindon, J.C.; Nicholson, J.K. Analytical technologies for metabonomics and metabolomics, and multi-omic information recovery. TrAC Trends Anal. Chem. 2008, 27, 194–204. [Google Scholar] [CrossRef]
- Arbona, V.; Manzi, M.; de Ollas, C.; Gómez-Cadenas, A. Metabolomics as a tool to investigate abiotic stress tolerance in plants. Int. J. Mol. Sci. 2013, 14, 4885–4911. [Google Scholar] [CrossRef]
- Shulaev, V.; Cortes, D.; Miller, G.; Mittler, R. Metabolomics for plant stress response. Physiol. Plant. 2008, 132, 199–208. [Google Scholar] [CrossRef]
- Ruan, C.-J.; da Silva, J.A.T.; Mopper, S.; Qin, P.; Lutts, S. Halophyte Improvement for a Salinized World. CRC. Crit. Rev. Plant Sci. 2010, 29, 329–359. [Google Scholar] [CrossRef]
- Krishnan, P.; Kruger, N.J.; Ratcliffe, R.G. Metabolite fingerprinting and profiling in plants using NMR. J. Exp. Bot. 2005, 56, 255–265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.K.; Choi, Y.H.; Verpoorte, R. NMR-based metabolomic analysis of plants. Nat. Protoc. 2010, 5, 536–549. [Google Scholar] [CrossRef]
- Goodacre, R.; York, E.V.; Heald, J.K.; Scott, I.M. Chemometric discrimination of unfractionated plant extracts analyzed by electrospray mass spectrometry. Phytochemistry 2003, 62, 859–863. [Google Scholar] [CrossRef]
- Kaplan, F.; Kopka, J.; Haskell, D.W.; Zhao, W.; Schiller, K.C.; Gatzke, N.; Sung, D.Y.; Guy, C.L.; Molecular, P.; Program, C.B.; et al. Exploring the Temperature-Stress Metabolome. Plant Physiol. 2004, 136, 4159–4168. [Google Scholar] [CrossRef] [Green Version]
- Shulaev, V. Metabolomics technology and bioinformatics. Brief. Bioinform. 2006, 7, 128–139. [Google Scholar] [CrossRef]
- Hirai, M.Y.; Yano, M.; Goodenowe, D.B.; Kanaya, S.; Kimura, T.; Awazuhara, M.; Arita, M.; Fujiwara, T.; Saito, K. Integration of transcriptomics and metabolomics for understanding of global responses to nutritional stresses in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2004, 101, 10205–10210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, H.E.; Broadhurst, D.; Goodacre, R.; Smith, A.R. Metabolic fingerprinting of salt-stressed tomatoes. Phytochemistry 2003, 62, 919–928. [Google Scholar] [CrossRef]
- Alseekh, S.; Aharoni, A.; Brotman, Y.; Contrepois, K.; D’Auria, J.; Ewald, J.; Ewald, J.C.; Fraser, P.D.; Giavalisco, P.; Hall, R.D.; et al. Mass spectrometry-based metabolomics: A guide for annotation, quantification and best reporting practices. Nat. Methods 2021, 18, 747–756. [Google Scholar] [CrossRef] [PubMed]
- Roessner, U.; Wagner, C.; Kopka, J.; Trethewey, R.N.; Willmitzer, L. Simultaneous analysis of metabolites in potato tuber by gas chromatography-mass spectrometry. Plant J. 2000, 23, 131–142. [Google Scholar] [CrossRef]
- Moons, A.; Bauw, G.; Prinsen, E.; Van Montagu, M.; Van Der Straeten, D. Molecular and physiological responses to abscisic acid and salts in roots of salt-sensitive and salt-tolerant indica rice varieties. Plant Physiol. 1995, 107, 177–186. [Google Scholar] [CrossRef] [Green Version]
- Shu, X.L.; Frank, T.; Shu, Q.Y.; Engel, K.H. Metabolite profiling of germinating rice seeds. J. Agric. Food Chem. 2008, 56, 11612–11620. [Google Scholar] [CrossRef]
- Wakasa, K.; Hasegawa, H.; Nemoto, H.; Matsuda, F.; Miyazawa, H.; Tozawa, Y.; Morino, K.; Komatsu, A.; Yamada, T.; Terakawa, T.; et al. High-level tryptophan accumulation in seeds of transgenic rice and its limited effects on agronomic traits and seed metabolite profile. J. Exp. Bot. 2006, 57, 3069–3078. [Google Scholar] [CrossRef] [Green Version]
- Tarpley, L.; Duran, A.L.; Kebrom, T.H.; Sumner, L.W. Biomarker metabolites capturing the metabolite variance present in a rice plant developmental period. BMC Plant Biol. 2005, 5, 8. [Google Scholar] [CrossRef] [Green Version]
- Kusano, M.; Fukushima, A.; Kobayashi, M.; Hayashi, N.; Jonsson, P.; Moritz, T.; Ebana, K.; Saito, K. Application of a metabolomic method combining one-dimensional and two-dimensional gas chromatography-time-of-flight/mass spectrometry to metabolic phenotyping of natural variants in rice. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2007, 855, 71–79. [Google Scholar] [CrossRef]
- Zuther, E.; Koehl, K.; Kopka, J. Comparative metabolome analysis of the salt response in breeding cultivars of rice. In Advances in Molecular Breeding Toward Drought and Salt Tolerant Crops; Springer: Dordrecht, The Netherlands, 2007; ISBN 9781402055775. [Google Scholar]
- Nam, M.H.; Bang, E.; Kwon, T.Y.; Kim, Y.; Kim, E.H.; Cho, K.; Park, W.J.; Kim, B.G.; Yoon, I.S. Metabolite profiling of diverse rice germplasm and identification of conserved metabolic markers of rice roots in response to long-term mild salinity stress. Int. J. Mol. Sci. 2015, 16, 21959–21974. [Google Scholar] [CrossRef] [Green Version]
- Fumagalli, E.; Baldoni, E.; Abbruscato, P.; Piffanelli, P.; Genga, A.; Lamanna, R.; Consonni, R. NMR techniques coupled with multivariate statistical analysis: Tools to analyse Oryza sativa metabolic content under stress conditions. J. Agron. Crop Sci. 2009, 195, 77–88. [Google Scholar] [CrossRef]
- Zhao, X.; Wang, W.; Zhang, F.; Deng, J.; Li, Z.; Fu, B. Comparative metabolite profiling of two rice genotypes with contrasting salt stress tolerance at the seedling stage. PLoS ONE 2014, 9, e108020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hakim, M.A.; Juraimi, A.S.; Hanafi, M.M.; Ismail, M.R.; Selamat, A.; Rafii, M.Y.; Latif, M.A. Biochemical and anatomical changes and yield reduction in rice (Oryza sativa L.) under varied salinity regimes. Biomed Res. Int. 2014, 2014, 208584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, P.; De, B. Metabolomics analysis of rice responses to salinity stress revealed elevation of serotonin, and gentisic acid levels in leaves of tolerant varieties. Plant Signal. Behav. 2017, 12, e1335845. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Wang, C.; Zhu, S.; Wang, W.; Xu, J.; Zhao, X. Characterizing the metabolites related to rice salt tolerance with introgression lines exhibiting contrasting performances in response to saline conditions. Plant Growth Regul. 2020, 92, 157–167. [Google Scholar] [CrossRef]
- Wang, W.; Zhao, X.; Li, M.; Huang, L.; Xu, J.; Zhang, F.; Cui, Y.; Fu, B.; Li, Z. Complex molecular mechanisms underlying seedling salt tolerance in rice revealed by comparative transcriptome and metabolomic profiling. J. Exp. Bot. 2016, 67, 405–419. [Google Scholar] [CrossRef] [Green Version]
- Xie, Z.; Wang, J.; Wang, W.; Wang, Y.; Xu, J.; Li, Z.; Zhao, X.; Fu, B. Integrated Analysis of the Transcriptome and Metabolome Revealed the Molecular Mechanisms Underlying the Enhanced Salt Tolerance of Rice Due to the Application of Exogenous Melatonin. Front. Plant Sci. 2021, 11, 2277. [Google Scholar] [CrossRef]
- Wanichthanarak, K.; Boonchai, C.; Kojonna, T.; Chadchawan, S.; Sangwongchai, W.; Thitisaksakul, M. Deciphering rice metabolic flux reprograming under salinity stress via in silico metabolic modeling. Comput. Struct. Biotechnol. J. 2020, 18, 3555–3566. [Google Scholar] [CrossRef]
- Wang, Y.; Huang, L.; Du, F.; Wang, J.; Zhao, X.; Li, Z.; Wang, W.; Xu, J.; Fu, B. Comparative transcriptome and metabolome profiling reveal molecular mechanisms underlying OsDRAP1-mediated salt tolerance in rice. Sci. Rep. 2021, 11, 5166. [Google Scholar] [CrossRef]
- Kim, S.; Jeong, H.; Jung, K. Integrating omics analysis of salt stress-responsive genes in rice. Genes Genom. 2015, 37, 645–655. [Google Scholar] [CrossRef]
- Scossa, F.; Alseekh, S.; Fernie, A.R. Integrating multi-omics data for crop improvement. J. Plant Physiol. 2021, 257, 153352. [Google Scholar] [CrossRef] [PubMed]
- Houle, D.; Govindaraju, D.R.; Omholt, S. Phenomics: The next challenge. Nat. Rev. Genet. 2010, 11, 855–866. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Zhang, Y.; Du, J.; Guo, X.; Wen, W.; Gu, S.; Wang, J.; Fan, J. Crop phenomics: Current status and perspectives. Front. Plant Sci. 2019, 10, 714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gautam, R.K.; Singh, P.K.; Singh, A.K.; Ahmed, S.K.Z.; Kumar, N.; Rao, S.S.; Velmurugan, A.; Roy, S.D. Identification and dissemination of salt tolerant rice varieties through farmer’ s participation in Andaman and Nicobar Islands. J. Andaman Sci. Assoc. 2014, 19, 136–141. [Google Scholar]
- Campbell, M.T.; Knecht, A.C.; Berger, B.; Brien, C.J.; Wang, D.; Walia, H. Integrating image-based phenomics and association analysis to dissect the genetic architecture of temporal salinity responses in rice. Plant Physiol. 2015, 168, 1476–1489. [Google Scholar] [CrossRef] [Green Version]
- Al-Tamimi, N.; Brien, C.; Oakey, H.; Berger, B.; Saade, S.; Ho, Y.S.; Schmöckel, S.M.; Tester, M.; Negraõ, S. Salinity tolerance loci revealed in rice using high-throughput non-invasive phenotyping. Nat. Commun. 2016, 7, 13342. [Google Scholar] [CrossRef] [Green Version]
- Sytar, O.; Brestic, M.; Zivcak, M.; Olsovska, K.; Kovar, M.; Shao, H.; He, X. Applying hyperspectral imaging to explore natural plant diversity towards improving salt stress tolerance. Sci. Total Environ. 2017, 578, 90–99. [Google Scholar] [CrossRef]
- Awada, L.; Phillips, P.W.B.; Smyth, S.J. The adoption of automated phenotyping by plant breeders. Euphytica 2018, 214, 148. [Google Scholar] [CrossRef] [Green Version]
- Rebetzke, G.J.; Jimenez-Berni, J.; Fischer, R.A.; Deery, D.M.; Smith, D.J. Review: High-throughput phenotyping to enhance the use of crop genetic resources. Plant Sci. 2019, 282, 40–48. [Google Scholar] [CrossRef]
- Sozzani, R.; Busch, W.; Spalding, E.P.; Benfey, P.N. Advanced imaging techniques for the study of plant growth and development. Trends Plant Sci. 2014, 19, 304–310. [Google Scholar] [CrossRef] [Green Version]
- Paproki, A.; Sirault, X.; Berry, S.; Furbank, R.; Fripp, J. A novel mesh processing based technique for 3D plant analysis. BMC Plant Biol. 2012, 12, 63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berger, B.; de Regt, B.; Tester, M. Trait Dissection of Salinity Tolerance with Plant Phenomics. In Plant Salt Tolerance; Humana Press: Totowa, NJ, USA, 2012; pp. 399–413. [Google Scholar]
- Großkinsky, D.K.; Syaifullah, S.J.; Roitsch, T. Integration of multi-omics techniques and physiological phenotyping within a holistic phenomics approach to study senescence in model and crop plants. J. Exp. Bot. 2018, 69, 825–844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Humplík, J.F.; Lazár, D.; Husičková, A.; Spíchal, L. Automated phenotyping of plant shoots using imaging methods for analysis of plant stress responses—A review. Plant Methods 2015, 11, 29. [Google Scholar] [CrossRef] [Green Version]
- Das, P.; Nutan, K.K.; Singla-Pareek, S.L.; Pareek, A. Understanding salinity responses and adopting ‘omics-based’ approaches to generate salinity tolerant cultivars of rice. Front. Plant Sci. 2015, 6, 712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hairmansis, A.; Berger, B.; Tester, M.; Roy, S.J. Image-based phenotyping for non-destructive screening of different salinity tolerance traits in rice. Rice 2014, 7, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siddiqui, Z.S.; Cho, J.I.; Park, S.H.; Kwon, T.R.; Ahn, B.O.; Lee, G.S.; Jeong, M.J.; Kim, K.W.; Lee, S.K.; Park, S.C. Phenotyping of rice in salt stress environment using high-throughput infrared imaging. Acta Bot. Croat. 2014, 73, 149–158. [Google Scholar] [CrossRef] [Green Version]
- Iyer-Pascuzzi, A.S.; Symonova, O.; Mileyko, Y.; Hao, Y.; Belcher, H.; Harer, J.; Weitz, J.S.; Benfey, P.N. Imaging and Analysis Platform for Automatic Phenotyping and Trait Ranking of Plant Root Systems. Plant Physiol. 2010, 152, 1148–1157. [Google Scholar] [CrossRef] [Green Version]
- Rogers, E.D.; Monaenkova, D.; Mijar, M.; Nori, A.; Goldman, D.I.; Benfey, P.N. X-ray Computed Tomography Reveals the Response of Root System Architecture to Soil Texture. Plant Physiol. 2016, 171, 2028–2040. [Google Scholar] [CrossRef] [Green Version]
- Topp, C.N.; Iyer-Pascuzzi, A.S.; Anderson, J.T.; Lee, C.-R.; Zurek, P.R.; Symonova, O.; Zheng, Y.; Bucksch, A.; Mileyko, Y.; Galkovskyi, T.; et al. 3D phenotyping and quantitative trait locus mapping identify core regions of the rice genome controlling root architecture. Proc. Natl. Acad. Sci. USA 2013, 110, E1695–E1704. [Google Scholar] [CrossRef] [Green Version]
- Yichie, Y.; Brien, C.; Berger, B.; Roberts, T.H.; Atwell, B.J. Salinity tolerance in Australian wild Oryza species varies widely and matches that observed in O. sativa. Rice 2018, 11, 66. [Google Scholar] [CrossRef]
- Singh, R.K.; Ahmadizadeh, M.; Vispo, N.A. QTL identification for reproductive-stage salinity tolerance in rice using novel phenotyping technique. In Proceedings of the 4th International Conference on Plant Genomics, Brisbane, Australia, 14–15 July 2016. [Google Scholar]
- Campbell, M.T.; Du, Q.; Liu, K.; Brien, C.J.; Berger, B.; Zhang, C.; Walia, H. A Comprehensive Image-based Phenomic Analysis Reveals the Complex Genetic Architecture of Shoot Growth Dynamics in Rice (Oryza sativa). Plant Genome 2017, 10. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Guo, Z.; Huang, C.; Duan, L.; Chen, G.; Jiang, N.; Fang, W.; Feng, H.; Xie, W.; Lian, X.; et al. Combining high-throughput phenotyping and genome-wide association studies to reveal natural genetic variation in rice. Nat. Commun. 2014, 5, 5087. [Google Scholar] [CrossRef] [PubMed]
- Roitsch, T.; Cabrera-Bosquet, L.; Fournier, A.; Ghamkhar, K.; Jiménez-Berni, J.; Pinto, F.; Ober, E.S. Review: New sensors and data-driven approaches—A path to next generation phenomics. Plant Sci. 2019, 282, 2–10. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, H.; Kawahara, Y.; Sakai, H.; Kanamori, H.; Wakimoto, H.; Yamagata, H.; Oono, Y.; Wu, J.; Ikawa, H.; Itoh, T.; et al. Massive parallel sequencing of mRNA in identification of unannotated salinity stress-inducible transcripts in rice (Oryza sativa L.). BMC Genom. 2010, 11, 683. [Google Scholar] [CrossRef] [Green Version]
- Mirdar Mansuri, R.; Shobbar, Z.-S.; Babaeian Jelodar, N.; Ghaffari, M.; Mohammadi, S.M.; Daryani, P. Salt tolerance involved candidate genes in rice: An integrative meta-analysis approach. BMC Plant Biol. 2020, 20, 452. [Google Scholar] [CrossRef]
- Lou, D.; Wang, H.; Yu, D. The sucrose non-fermenting-1-related protein kinases SAPK1 and SAPK2 function collaboratively as positive regulators of salt stress tolerance in rice. BMC Plant Biol. 2018, 18, 203. [Google Scholar] [CrossRef] [Green Version]
- Zhang, A.; Liu, Y.; Wang, F.; Li, T.; Chen, Z.; Kong, D.; Bi, J.; Zhang, F.; Luo, X.; Wang, J.; et al. Enhanced rice salinity tolerance via CRISPR/Cas9-targeted mutagenesis of the OsRR22 gene. Mol. Breed. 2019, 39, 47. [Google Scholar] [CrossRef] [Green Version]
- Das, G.; Patra, J.K.; Baek, K.H. Insight into MAS: A molecular tool for development of stress resistant and quality of rice through gene stacking. Front. Plant Sci. 2017, 8, 985. [Google Scholar] [CrossRef] [Green Version]
- Hoang, T.M.L.; Tran, T.N.; Nguyen, T.K.T.; Williams, B.; Wurm, P.; Bellairs, S.; Mundree, S. Improvement of salinity stress tolerance in rice: Challenges and opportunities. Agronomy 2016, 6, 54. [Google Scholar] [CrossRef]
- Singh, A.K.; Gopalakrishnan, S.; Singh, V.P.; Prabhu, K.V.; Mohapatra, T.; Singh, N.K.; Sharma, T.R.; Nagarajan, M.; Vinod, K.K.; Singh, D.; et al. Marker assisted selection: A paradigm shift in Basmati breeding. Indian J. Genet. Plant Breed. 2011, 71, 120–128. [Google Scholar]
- Huyen, L.T.N.; Cuc, L.M.; Ismail, A.M.; Ham, L.H. Introgression the Salinity Tolerance QTLs Saltol into AS996, the Elite Rice Variety of Vietnam. Am. J. Plant Sci. 2012, 3, 981–987. [Google Scholar] [CrossRef] [Green Version]
- Linh, L.H.; Linh, T.H.; Xuan, T.D.; Ham, L.H.; Ismail, A.M.; Khanh, T.D. Molecular Breeding to Improve Salt Tolerance of Rice (Oryza sativa L.) in the Red River Delta of Vietnam. Int. J. Plant Genom. 2012, 2012, 949038. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vu, H.T.T.; Le, D.D.; Ismail, A.M.; Le, H.H. Marker-assisted backcrossing (MABC) for improved salinity tolerance in rice (“Oryza sativa” L.) to cope with climate change in Vietnam. Aust. J. Crop Sci. 2012, 6, 1649–1654. [Google Scholar]
- Hoque, A.B.M.Z.; Haque, A.; Sarker, R.A.; Rahman, A. Marker-Assisted introgression of saltol locus into genetic background of brri dhan49. Int. J. Biosci. 2015, 6, 71–80. [Google Scholar]
- Muthu, V.; Abbai, R.; Nallathambi, J.; Rahman, H.; Ramasamy, S.; Kambale, R.; Thulasinathan, T.; Ayyenar, B.; Muthurajan, R. Pyramiding QTLs controlling tolerance against drought, salinity, and submergence in rice through marker assisted breeding. PLoS ONE 2020, 15, e0227421. [Google Scholar] [CrossRef] [Green Version]
- Reddy, S.S.S.; Singh, B.; Peter, A.J.; Venkateswar Rao, T. Production of transgenic local rice cultivars (Oryza sativa L.) for improved drought tolerance using Agrobacterium mediated transformation. Saudi J. Biol. Sci. 2018, 25, 1535–1545. [Google Scholar] [CrossRef] [Green Version]
- Low, L.-Y.; Yang, S.-K.; Kok, D.-X.A.; Ong-Abdullah, J.; Tan, N.-P.; Lai, K.-S. Transgenic Plants: Gene Constructs, Vector and Transformation Method. In New Visions in Plant Science; InTech: London, UK, 2018; pp. 41–61. [Google Scholar]
- Mohammed, S.; Samad, A.A.; Rahmat, Z. Agrobacterium-Mediated Transformation of Rice: Constraints and Possible Solutions. Rice Sci. 2019, 26, 133–146. [Google Scholar] [CrossRef]
- Sood, P.; Bhattacharya, A.; Sood, A. Problems and possibilities of monocot transformation. Biol. Plant. 2011, 55, 1–15. [Google Scholar] [CrossRef]
- Hwang, H.-H.; Yu, M.; Lai, E.-M. Agrobacterium-Mediated Plant Transformation: Biology and Applications. Arab. Book 2017, 15, e0186. [Google Scholar] [CrossRef] [Green Version]
- Manimaran, P.; Kumar, G.R.; Reddy, M.R.; Jain, S.; Rao, T.B.; Mangrauthia, S.K.; Sundaram, R.M.; Ravichandran, S.; Balachandran, S.M. Infection of Early and Young Callus Tissues of Indica Rice BPT 5204 Enhances Regeneration and Transformation Efficiency. Rice Sci. 2013, 20, 415–426. [Google Scholar] [CrossRef]
- Tan, L.W.; Rahman, Z.A.; Goh, H.H.; Hwang, D.J.; Ismail, I.; Zainal, Z. Production of transgenic rice (indica cv. MR219) overexpressing ABP57 gene through Agrobacterium-mediated transformation. Sains Malays. 2017, 46, 703–711. [Google Scholar] [CrossRef]
- Sahoo, K.K.; Tripathi, A.K.; Pareek, A.; Sopory, S.K.; Singla-Pareek, S.L. An improved protocol for efficient transformation and regeneration of diverse indica rice cultivars. Plant Methods 2011, 7, 49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fukuda, A.; Nakamura, A.; Tagiri, A.; Tanaka, H.; Miyao, A.; Tanaka, Y. Function, Intracellular Localization and the Importance in Salt Tolerance of a Vacuolar Na+/H+ Antiporter from Rice. Plant Cell Physiol. 2004, 45, 146–159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roy, M.; Wu, R. Arginine decarboxylase transgene expression and analysis of environmental stress tolerance in transgenic rice. Plant Sci. 2001, 160, 869–875. [Google Scholar] [CrossRef]
- Oh, S.-J.; Song, S.I.; Kim, Y.S.; Jang, H.-J.; Kim, S.Y.; Kim, M.; Kim, Y.-K.; Nahm, B.H.; Kim, J.-K. Arabidopsis CBF3/DREB1A and ABF3 in Transgenic Rice Increased Tolerance to Abiotic Stress without Stunting Growth. Plant Physiol. 2005, 138, 341–351. [Google Scholar] [CrossRef] [Green Version]
- Mohanty, A.; Kathuria, H.; Ferjani, A.; Sakamoto, A.; Mohanty, P.; Murata, N.; Tyagi, A. Transgenics of an elite indica rice variety Pusa Basmati 1 harbouring the codA gene are highly tolerant to salt stress. Theor. Appl. Genet. 2002, 106, 51–57. [Google Scholar] [CrossRef]
- Xu, D.; Duan, X.; Wang, B.; Hong, B.; Ho, T.; Wu, R. Expression of a Late Embryogenesis Abundant Protein Gene, HVA1, from Barley Confers Tolerance to Water Deficit and Salt Stress in Transgenic Rice. Plant Physiol. 1996, 110, 249–257. [Google Scholar] [CrossRef] [Green Version]
- Xiong, L.; Yang, Y. Disease Resistance and Abiotic Stress Tolerance in Rice Are Inversely Modulated by an Abscisic Acid–Inducible Mitogen-Activated Protein Kinase. Plant Cell 2003, 15, 745–759. [Google Scholar] [CrossRef] [Green Version]
- Zhao, F.; Guo, S.; Zhang, H.; Zhao, Y. Expression of yeast SOD2 in transgenic rice results in increased salt tolerance. Plant Sci. 2006, 170, 216–224. [Google Scholar] [CrossRef]
- Jang, I.-C.; Oh, S.-J.; Seo, J.-S.; Choi, W.-B.; Song, S.I.; Kim, C.H.; Kim, Y.S.; Seo, H.-S.; Do Choi, Y.; Nahm, B.H.; et al. Expression of a Bifunctional Fusion of the Escherichia coli Genes for Trehalose-6-Phosphate Synthase and Trehalose-6-Phosphate Phosphatase in Transgenic Rice Plants Increases Trehalose Accumulation and Abiotic Stress Tolerance without Stunting Growth. Plant Physiol. 2003, 131, 516–524. [Google Scholar] [CrossRef] [Green Version]
- Liao, Y.-D.; Lin, K.-H.; Chen, C.-C.; Chiang, C.-M. Oryza sativa protein phosphatase 1a (OsPP1a) involved in salt stress tolerance in transgenic rice. Mol. Breed. 2016, 36, 22. [Google Scholar] [CrossRef]
- Park, S.-I.; Kim, J.-J.; Shin, S.-Y.; Kim, Y.-S.; Yoon, H.-S. ASR Enhances Environmental Stress Tolerance and Improves Grain Yield by Modulating Stomatal Closure in Rice. Front. Plant Sci. 2020, 10, 1752. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.W.; Tan, C.S.; Rahman, Z.A.; Goh, H.-H.; Ismail, I.; Zainal, Z. Microarray dataset of transgenic rice overexpressing Abp57. Data Brief 2017, 14, 267–271. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Guo, L.; Guo, C.; Wang, L.; Chen, L. Over-expression of a DUF1644 protein gene, SIDP361, enhances tolerance to salt stress in transgenic rice. J. Plant Biol. 2016, 59, 62–73. [Google Scholar] [CrossRef]
- Sahoo, R.K.; Ansari, M.W.; Tuteja, R.; Tuteja, N. OsSUV3 transgenic rice maintains higher endogenous levels of plant hormones that mitigates adverse effects of salinity and sustains crop productivity. Rice 2014, 7, 17. [Google Scholar] [CrossRef] [Green Version]
- Nath, M.; Yadav, S.; Kumar Sahoo, R.; Passricha, N.; Tuteja, R.; Tuteja, N. PDH45 transgenic rice maintain cell viability through lower accumulation of Na+, ROS and calcium homeostasis in roots under salinity stress. J. Plant Physiol. 2016, 191, 1–11. [Google Scholar] [CrossRef]
- Farhat, S.; Jain, N.; Singh, N.; Sreevathsa, R.; Dash, P.K.; Rai, R.; Yadav, S.; Kumar, P.; Sarkar, A.K.; Jain, A.; et al. CRISPR-Cas9 directed genome engineering for enhancing salt stress tolerance in rice. Semin. Cell Dev. Biol. 2019, 96, 91–99. [Google Scholar] [CrossRef]
- Shan, Q.; Wang, Y.; Li, J.; Zhang, Y.; Chen, K.; Liang, Z.; Zhang, K.; Liu, J.; Xi, J.J.; Qiu, J.-L.; et al. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 2013, 31, 686–688. [Google Scholar] [CrossRef]
- Miao, J.; Guo, D.; Zhang, J.; Huang, Q.; Qin, G.; Zhang, X.; Wan, J.; Gu, H.; Qu, L.-J. Targeted mutagenesis in rice using CRISPR-Cas system. Cell Res. 2013, 23, 1233–1236. [Google Scholar] [CrossRef] [Green Version]
- Yin, X.; Biswal, A.K.; Dionora, J.; Perdigon, K.M.; Balahadia, C.P.; Mazumdar, S.; Chater, C.; Lin, H.-C.; Coe, R.A.; Kretzschmar, T.; et al. CRISPR-Cas9 and CRISPR-Cpf1 mediated targeting of a stomatal developmental gene EPFL9 in rice. Plant Cell Rep. 2017, 36, 745–757. [Google Scholar] [CrossRef]
- Xu, R.; Li, H.; Qin, R.; Li, J.; Qiu, C.; Yang, Y.; Ma, H. Generation of inheritable and “transgene clean” targeted genome-modified rice in later generations using the CRISPR/Cas9 system. Sci. Rep. 2015, 9, 11491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duan, Y.B.; Li, J.; Ying, R.; Rong, Q.; Xu, F.; Li, H. Identification of a regulatory element responsible for salt induction of rice OsRAV2 through ex situ and in situ promoter analysis. Plant Mol. Biol. 2015, 90, 49–62. [Google Scholar] [CrossRef] [PubMed]
- Tak, Y.G.; Farnham, P.J. Making sense of GWAS: Using epigenomics and genome engineering to understand the functional relevance of SNPs in non-coding regions of the human genome. Epigenet. Chromatin 2015, 8, 57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vidalis, A.; Živković, D.; Wardenaar, R.; Roquis, D.; Tellier, A.; Johannes, F. Methylome evolution in plants. Genome Biol. 2016, 17, 264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Cimen, E.; Singh, N.; Buckler, E. Deep learning for plant genomics and crop improvement. Curr. Opin. Plant Biol. 2020, 54, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Reel, P.S.; Reel, S.; Pearson, E.; Trucco, E.; Jefferson, E. Using machine learning approaches for multi-omics data analysis: A review. Biotechnol. Adv. 2021, 49, 107739. [Google Scholar] [CrossRef]
- Yoosefzadeh-Najafabadi, M.; Earl, H.J.; Tulpan, D.; Sulik, J.; Eskandari, M. Application of Machine Learning Algorithms in Plant Breeding: Predicting Yield From Hyperspectral Reflectance in Soybean. Front. Plant Sci. 2021, 11, 2169. [Google Scholar] [CrossRef]
- Lan, H.; Carson, R.; Provart, N.J.; Bonner, A.J. Combining classifiers to predict gene function in Arabidopsis thaliana using large-scale gene expression measurements. BMC Bioinform. 2007, 8, 358. [Google Scholar] [CrossRef] [Green Version]
- Simopoulos, C.M.A.; Weretilnyk, E.A.; Golding, G.B. Prediction of plant lncRNA by ensemble machine learning classifiers. BMC Genom. 2018, 19, 316. [Google Scholar] [CrossRef] [Green Version]
- Ancillo, G.; Gadea, J.; Forment, J.; Guerri, J.; Navarro, L. Class prediction of closely related plant varieties using gene expression profiling. J. Exp. Bot. 2007, 58, 1927–1933. [Google Scholar] [CrossRef] [Green Version]
- Badillo, S.; Banfai, B.; Birzele, F.; Davydov, I.I.; Hutchinson, L.; Kam-Thong, T.; Siebourg-Polster, J.; Steiert, B.; Zhang, J.D. An Introduction to Machine Learning. Clin. Pharmacol. Ther. 2020, 107, 871–885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, K.; Abdullah, A.A.; Huang, M.; Nishioka, T.; Altaf-Ul-Amin, M.; Kanaya, S. Novel Approach to Classify Plants Based on Metabolite-Content Similarity. Biomed Res. Int. 2017, 2017, 5296729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Yapa, M.M.; Hua, Z. A Machine Learning Approach to Prioritizing Functionally Active F-box Members in Arabidopsis thaliana. Front. Plant Sci. 2021, 12, 639253. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Xin, M.; Feldmann, K.A.; Wang, X. Machine Learning–Based Differential Network Analysis: A Study of Stress-Responsive Transcriptomes in Arabidopsis. Plant Cell 2014, 26, 520–537. [Google Scholar] [CrossRef] [Green Version]
- Xulu, S.; Peerbhay, K.; Gebreslasie, M.; Ismail, R. Unsupervised Clustering of Forest Response to Drought Stress in Zululand Region, South Africa. Forests 2019, 10, 531. [Google Scholar] [CrossRef] [Green Version]
- Rico-Chávez, A.K.; Franco, J.A.; Fernandez-Jaramillo, A.A.; Contreras-Medina, L.M.; Guevara-González, R.G.; Hernandez-Escobedo, Q. Machine Learning for Plant Stress Modeling: A Perspective towards Hormesis Management. Plants 2022, 11, 970. [Google Scholar] [CrossRef]
- Das, B.; Manohara, K.K.; Mahajan, G.R.; Sahoo, R.N. Spectroscopy based novel spectral indices, PCA- and PLSR-coupled machine learning models for salinity stress phenotyping of rice. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2020, 229, 117983. [Google Scholar] [CrossRef]
- Sanchez, R.A.; Hall, A.J.; Trapani, N.; de Hunau, R.C. Effects of water stress on the chlorophyll content, nitrogen level and photosynthesis of leaves of two maize genotypes. Photosynth. Res. 1983, 4, 35–47. [Google Scholar] [CrossRef]
- Zahid, A.; Dashtipour, K.; Abbas, H.T.; Ben Mabrouk, I.; Al-Hasan, M.; Ren, A.; Imran, M.A.; Alomainy, A.; Abbasi, Q.H. Machine learning enabled identification and real-time prediction of living plants’ stress using terahertz waves. Def. Technol. 2022, in press. [Google Scholar] [CrossRef]
- Jafari, M.; Shahsavar, A. The application of artificial neural networks in modeling and predicting the effects of melatonin on morphological responses of citrus to drought stress. PLoS ONE 2020, 15, e0240427. [Google Scholar] [CrossRef]
- Beltramo, T.; Klocke, M.; Hitzmann, B. Prediction of the biogas production using GA and ACO input features selection method for ANN model. Inf. Process. Agric. 2019, 6, 349–356. [Google Scholar] [CrossRef]
- Arab, M.M.; Yadollahi, A.; Ahmadi, H.; Eftekhari, M.; Maleki, M. Mathematical Modeling and Optimizing of in Vitro Hormonal Combination for G × N15 Vegetative Rootstock Proliferation Using Artificial Neural Network-Genetic Algorithm (ANN-GA). Front. Plant Sci. 2017, 8, 1853. [Google Scholar] [CrossRef] [Green Version]
- Ly, D.; Huet, S.; Gauffreteau, A.; Rincent, R.; Touzy, G.; Mini, A.; Jannink, J.-L.; Cormier, F.; Paux, E.; Lafarge, S.; et al. Whole-genome prediction of reaction norms to environmental stress in bread wheat (Triticum aestivum L.) by genomic random regression. Field Crops Res. 2018, 216, 32–41. [Google Scholar] [CrossRef]
- Azimi, S.; Kaur, T.; Gandhi, T.K. A deep learning approach to measure stress level in plants due to Nitrogen deficiency. Measurement 2021, 173, 108650. [Google Scholar] [CrossRef]
- Montesinos-López, O.A.; Montesinos-López, J.C.; Singh, P.; Lozano-Ramirez, N.; Barrón-López, A.; Montesinos-López, A.; Crossa, J. A Multivariate Poisson Deep Learning Model for Genomic Prediction of Count Data. G3—Genes Genomes Genet. 2020, 10, 4177–4190. [Google Scholar] [CrossRef] [PubMed]
- Montesinos-López, O.A.; Montesinos-López, A.; Crossa, J.; Toledo, F.H.; Montesinos-López, J.C.; Singh, P.; Juliana, P.; Salinas-Ruiz, J. A Bayesian Poisson-lognormal Model for Count Data for Multiple-Trait Multiple-Environment Genomic-Enabled Prediction. G3—Genes Genomes Genet. 2017, 7, 1595–1606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anandkumar, M. Texton Features and Deep Belief Network for Leaf Disease Classification. Multimed. Res. 2020, 3, 43–50. [Google Scholar] [CrossRef]
- Rachmatia, H.; Kusuma, W.A.; Hasibuan, L.S. Prediction of maize phenotype based on whole-genome single nucleotide polymorphisms using deep belief networks. J. Phys. Conf. Ser. 2017, 835, 012003. [Google Scholar] [CrossRef] [Green Version]
- Yalcin, H. Plant Recognition based on Deep Belief Network Classifier and Combination of Local Features. In Proceedings of the 29th Signal Processing and Communications Applications Conference (SIU), Istanbul, Turkey, 9–11 June 2021; pp. 1–4. [Google Scholar]
- Rodziewicz, P.; Swarcewicz, B.; Chmielewska, K.; Wojakowska, A.; Stobiecki, M. Influence of abiotic stresses on plant proteome and metabolome changes. Acta Physiol. Plant. 2014, 36, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Mirdar Mansuri, R.; Shobbar, Z.-S.; Babaeian Jelodar, N.; Ghaffari, M.R.; Nematzadeh, G.-A.; Asari, S. Dissecting molecular mechanisms underlying salt tolerance in rice: A comparative transcriptional profiling of the contrasting genotypes. Rice 2019, 12, 13. [Google Scholar] [CrossRef]
- Wang, X. Protein and Proteome Atlas for Plants under Stresses: New Highlights and Ways for Integrated Omics in Post-Genomics Era. Int. J. Mol. Sci. 2019, 20, 5222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, J.; Li, Y.; Han, G.; Song, J.; Wang, B. NaCl markedly improved the reproductive capacity of the euhalophyte Suaeda salsa. Funct. Plant Biol. 2018, 45, 350. [Google Scholar] [CrossRef] [PubMed]
- Ho, W.W.H.; Hill, C.B.; Doblin, M.S.; Shelden, M.C.; van de Meene, A.; Rupasinghe, T.; Bacic, A.; Roessner, U. Integrative Multi-omics Analyses of Barley Rootzones under Salinity Stress Reveal Two Distinctive Salt Tolerance Mechanisms. Plant Commun. 2020, 1, 100031. [Google Scholar] [CrossRef] [PubMed]
- Jia, L.; Xie, L.; Lao, S.; Zhu, Q.-H.; Fan, L. Rice bioinformatics in the genomic era: Status and perspectives. Crop J. 2021, 9, 609–621. [Google Scholar] [CrossRef]
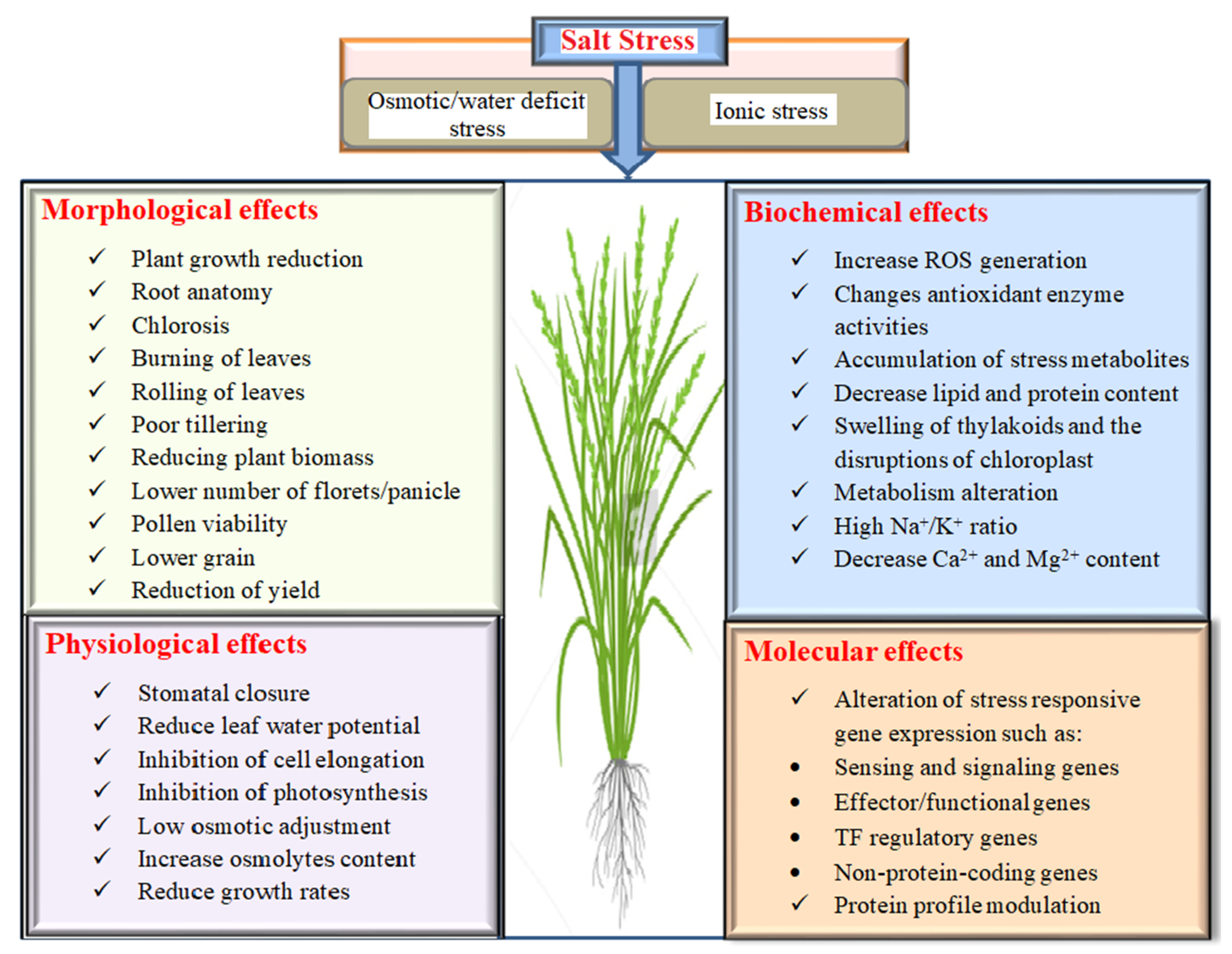
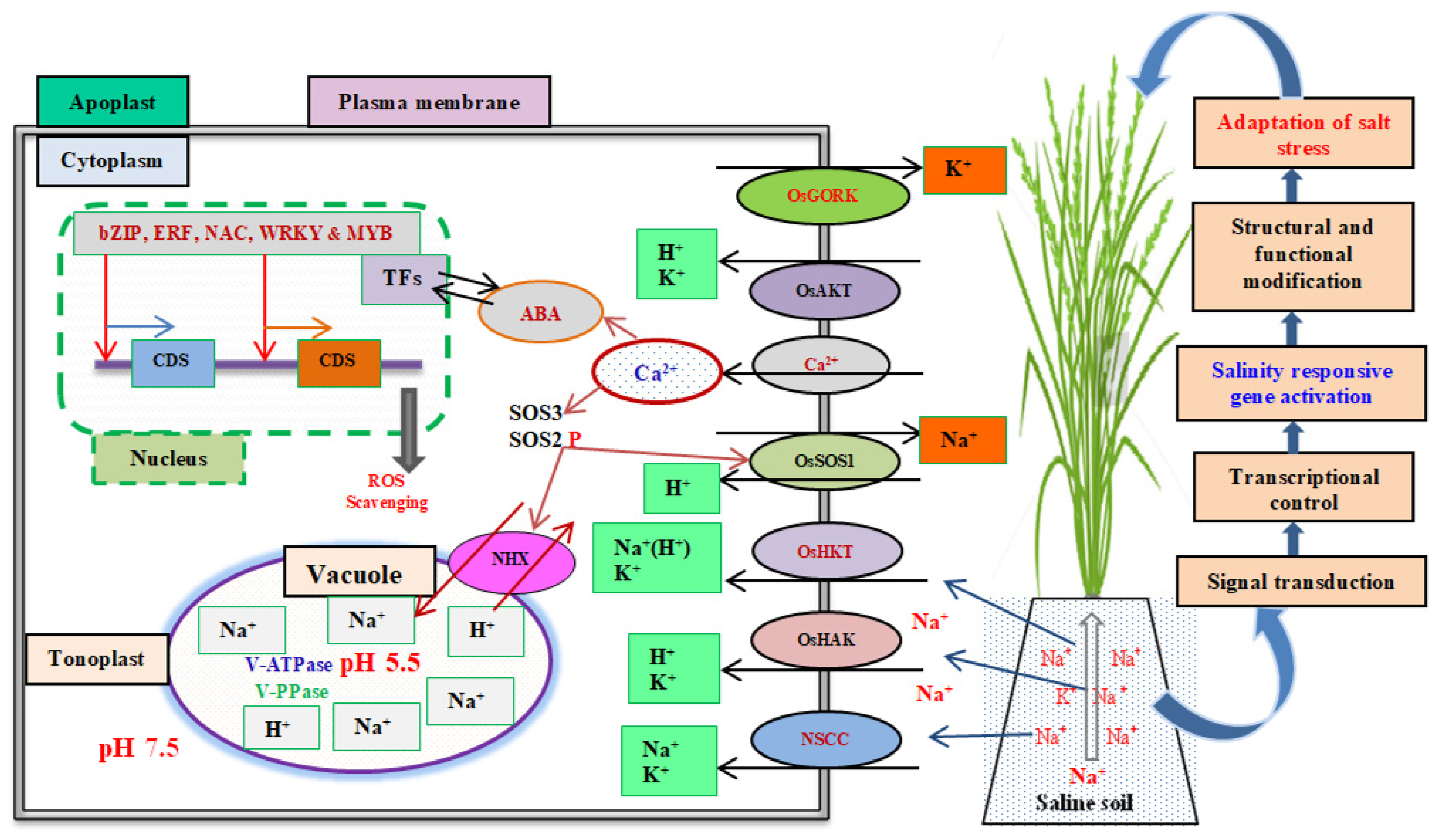

| Database | Description | Web Tool/URL |
|---|---|---|
| RAP-DB | Rice genomics database | https://rapdb.dna.affrc.go.jp (accessed on 10 October 2021) |
| RiceXPro | Expression profile database of rice | https://ricexpro.dna.affrc.go.jp (accessed on 10 October 2021) |
| NCBI GEO | National Center for Biotechnology Information Gene Expression Omnibus | https://ncbi.nlm.nih.gov/geo (accessed on 10 October 2021) |
| QlicRice | Stress related QTLs data mining tool | https://nabg.iasri.res.in:8080/qlic-rice (accessed on 10 October 2021) |
| STIFDB2 | Plant stress-related data mining tool | https://caps.ncbs.res.in/stifdb2 (accessed on 10 October 2021) |
| TENOR | Comprehensive mRNA-seq database of rice under environmental stress conditions | https://tenor.dna.affrc.go.jp (accessed on 10 October 2021) |
| Genevestgator | Transcriptomics database for investigating gene expression in a wide range of biological situations | https://genevestigator.com (accessed on 2 September 2021) |
| CSRDB | Small RNA database for cereals | https://sundarlab.ucdavis.edu/smrnas (accessed on 10 October 2021) |
| RiceSRTFDB | Rice stress-related TF database | https://nipgr.res.in/RiceSRTFDB (accessed on 10 October 2021) |
| Stress2TF | A manually curated database of transcription factor regulation in plants response to stress | https://csgenomics.ahau.edu.cn/Stress2TF (accessed on 10 October 2021) |
| PSPDB | Stress-related protein database for plants | https://bioclues.org/pspdb (accessed on 10 October 2021) |
| OryzaGenome | Integrated biological and genomics database | https://viewer.shigen.info/oryzagenome2detail (accessed on 15 October 2021) |
| Ricebase | Combining molecular marker, pedigree and whole-genome-based data tool | https://ricebase.org (accessed on 10 October 2021) |
| Gramene | A comprehensive data library for comparative genomics studies | https://gramene.org (accessed on 15 October 2021) |
| Phytozome | Plant Comparative Genomics Portal | https://phytozome.net (accessed on 12 February 2020) |
| Ensembl Plants | Integrated tool for plant genomics data mining, interpreting and visualising | https://plants.ensembl.org (accessed on 12 February 2020) |
| PlantPReS | Plant proteome database | https://proteome.ir (accessed on 17 October 2021) |
| Plant Reactome | Genome, transcriptome, proteome and integrated metabolic pathways | https://plants.reactome.org (accessed on 12 February 2020) |
| PlantGDB | Resources for plant genomics | https://plantgdb.org (accessed on 17 October 2021) |
| GabiPD | Integrative omics database | https://gabipd.org (accessed on 17 October 2021) |
| PMND | A vast network of databases on plant metabolic pathways | https://plantcyc.org (accessed on 17 October 2021) |
| RicyerDB | Integrated genomics and proteomics database | https://server.malab.cn/Ricyer (accessed on 13 October 2021) |
| CARMO | Integrative omics database | https://bioinfo.sibs.ac.cn/carmo (accessed on 13 October 2021) |
| PTools | Integrative omics database | https://omictools.com/ptools/tool (accessed on 13 October 2021) |
| Gromacs | Database of genomics, proteomics and metabolomics | https://omictools.com/gromacs/tool (accessed on 13 October 2021) |
| STRING | PPI network analysis containing functional association | https://string-db.org (accessed on 9 January 2020) |
| PANTHER | Analysis of proteins based on evolutionary relationships | https://pantherdb.org (accessed on 13 October 2021) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ullah, M.A.; Abdullah-Zawawi, M.-R.; Zainal-Abidin, R.-A.; Sukiran, N.L.; Uddin, M.I.; Zainal, Z. A Review of Integrative Omic Approaches for Understanding Rice Salt Response Mechanisms. Plants 2022, 11, 1430. https://doi.org/10.3390/plants11111430
Ullah MA, Abdullah-Zawawi M-R, Zainal-Abidin R-A, Sukiran NL, Uddin MI, Zainal Z. A Review of Integrative Omic Approaches for Understanding Rice Salt Response Mechanisms. Plants. 2022; 11(11):1430. https://doi.org/10.3390/plants11111430
Chicago/Turabian StyleUllah, Mohammad Asad, Muhammad-Redha Abdullah-Zawawi, Rabiatul-Adawiah Zainal-Abidin, Noor Liyana Sukiran, Md Imtiaz Uddin, and Zamri Zainal. 2022. "A Review of Integrative Omic Approaches for Understanding Rice Salt Response Mechanisms" Plants 11, no. 11: 1430. https://doi.org/10.3390/plants11111430
APA StyleUllah, M. A., Abdullah-Zawawi, M.-R., Zainal-Abidin, R.-A., Sukiran, N. L., Uddin, M. I., & Zainal, Z. (2022). A Review of Integrative Omic Approaches for Understanding Rice Salt Response Mechanisms. Plants, 11(11), 1430. https://doi.org/10.3390/plants11111430