Assessment of the Genetic Structure and Diversity of Soybean (Glycine max L.) Germplasm Using Diversity Array Technology and Single Nucleotide Polymorphism Markers
Abstract
:1. Introduction
2. Results
2.1. Marker Quality and Genome Characterization
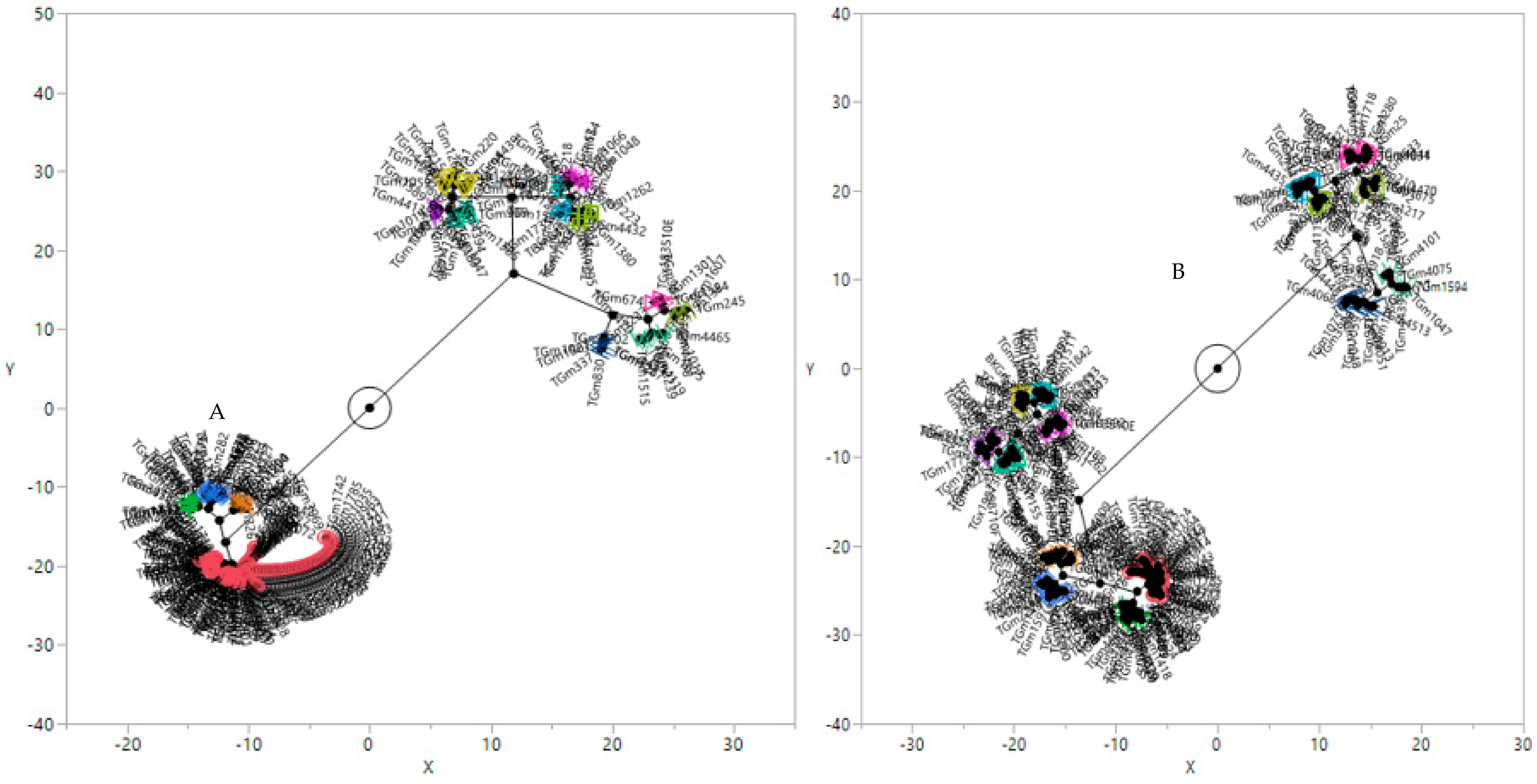
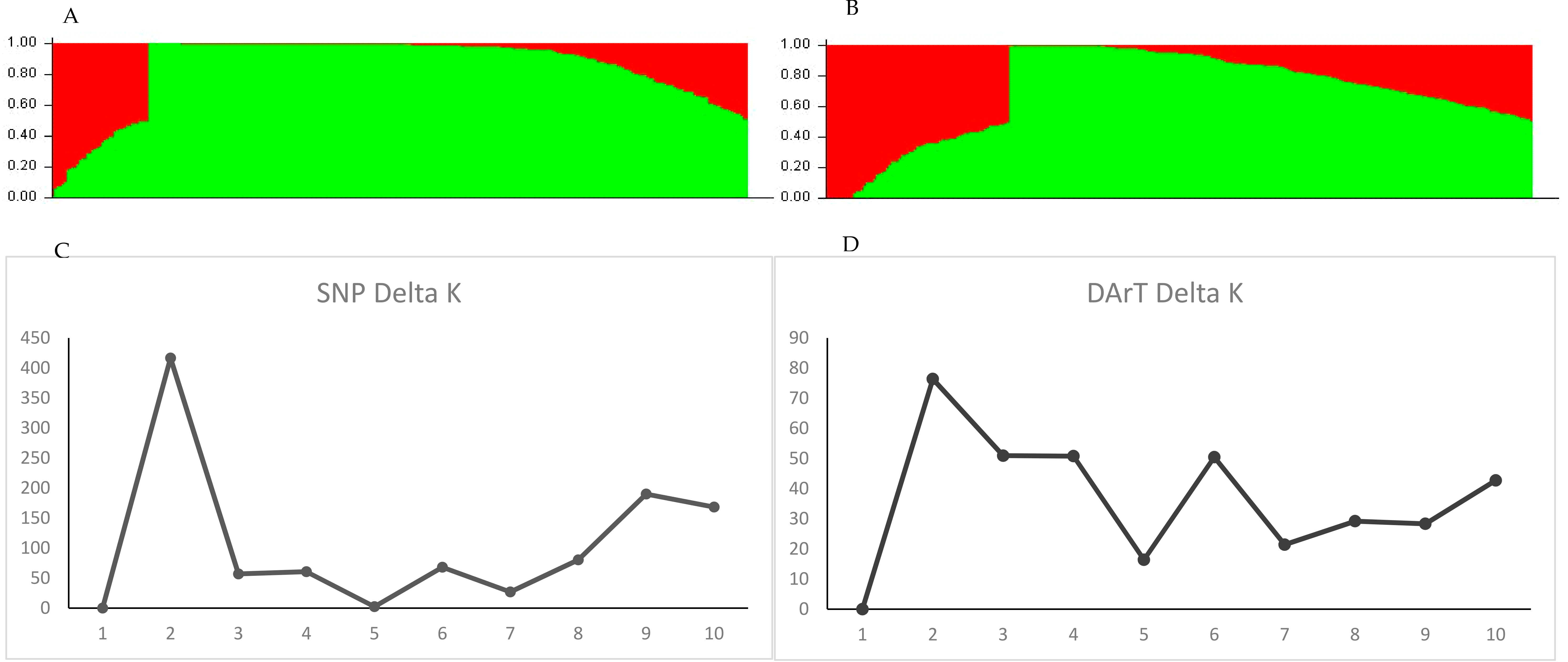
2.2. Population Structure of the Soybean Accessions
2.3. Analysis of Molecular Variance
2.4. Allelic Patterns across the Populations
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. DArT-Based Genotyping by Sequencing
4.3. Statistical Analysis
4.3.1. SNP Filtering and Genome Characterization
4.3.2. Cluster and Population Structure Analyses
4.3.3. Analysis of Molecular Variance
4.3.4. Analysis of Allelic Pattern
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- FAOSTAT Databases: Import and Export Values of Soybean-Based Products; Food Balance Sheets for Soybean 2021. Available online: https://www.fao.org/faostat/en/#data/QCL (accessed on 11 December 2021).
- Bisen, A.; Khare, D.; Nair, P.; Tripathi, N. SSR Analysis of 38 Genotypes of Soybean (Glycine max (L.) Merr.) Genetic Diversity in India. Physiol. Mol. Biol. Plants 2015, 21, 109–115. [Google Scholar] [CrossRef] [Green Version]
- Wilson, R.F. Designing Soybeans for 21st Century Markets; Elsevier: Amsterdam, The Netherlands, 2015; ISBN 1630670111. [Google Scholar]
- Salgotra, R.K.; Gupta, B.B.; Bhat, J.A.; Sharma, S. Genetic Diversity and Population Structure of Basmati Rice (Oryza sativa L.) Germplasm Collected from North Western Himalayas Using Trait Linked SSR Markers. PLoS ONE 2015, 10, e0131858. [Google Scholar]
- Hyten, D.L.; Song, Q.; Zhu, Y.; Choi, I.-Y.; Nelson, R.L.; Costa, J.M.; Specht, J.E.; Shoemaker, R.C.; Cregan, P.B. Impacts of Genetic Bottlenecks on Soybean Genome Diversity. Proc. Natl. Acad. Sci. USA 2006, 103, 16666–16671. [Google Scholar] [CrossRef] [Green Version]
- Kofsky, J.; Zhang, H.; Song, B.-H. The Untapped Genetic Reservoir: The Past, Current, and Future Applications of the Wild Soybean (Glycine soja). Front. Plant Sci. 2018, 9, 949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lam, H.-M.; Xu, X.; Liu, X.; Chen, W.; Yang, G.; Wong, F.-L.; Li, M.-W.; He, W.; Qin, N.; Wang, B. Resequencing of 31 Wild and Cultivated Soybean Genomes Identifies Patterns of Genetic Diversity and Selection. Nat. Genet. 2010, 42, 1053–1059. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhao, S.; Ma, J.; Li, D.; Yan, L.; Li, J.; Qi, X.; Guo, X.; Zhang, L.; He, W. Molecular Footprints of Domestication and Improvement in Soybean Revealed by Whole Genome Re-Sequencing. BMC Genom. 2013, 14, 579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Z.; Li, H.; Wen, Z.; Fan, X.; Li, Y.; Guan, R.; Guo, Y.; Wang, S.; Wang, D.; Qiu, L. Comparison of Genetic Diversity between Chinese and American Soybean (Glycine max (L.)) Accessions Revealed by High-Density SNPs.v. Front. Plant Sci. 2017, 8, 2014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeong, S.-C.; Moon, J.-K.; Park, S.-K.; Kim, M.-S.; Lee, K.; Lee, S.R.; Jeong, N.; Choi, M.S.; Kim, N.; Kang, S.-T.; et al. Genetic Diversity Patterns and Domestication Origin of Soybean. Theor. Appl. Genet. 2019, 132, 1179–1193. [Google Scholar] [CrossRef] [Green Version]
- Karikari, B.; Bhat, J.A.; Denwar, N.N.; Zhao, T. Exploring the Genetic Base of the Soybean Germplasm from Africa, America and Asia as Well as Mining of Beneficial Allele for Flowering and Seed Weight. 3 Biotech 2020, 10, 195. [Google Scholar] [CrossRef] [PubMed]
- Foyer, C.H.; Siddique, K.H.M.; Tai, A.P.K.; Anders, S.; Fodor, N.; Wong, F.-L.; Ludidi, N.; Chapman, M.A.; Ferguson, B.J.; Considine, M.J.; et al. Modelling Predicts That Soybean Is Poised to Dominate Crop Production across Africa. Plant. Cell Environ. 2019, 42, 373–385. [Google Scholar] [CrossRef] [PubMed]
- Cobb, J.N.; DeClerck, G.; Greenberg, A.; Clark, R.; McCouch, S. Next-Generation Phenotyping: Requirements and Strategies for Enhancing Our Understanding of Genotype–Phenotype Relationships and Its Relevance to Crop Improvement. Theor. Appl. Genet. 2013, 126, 867–887. [Google Scholar] [CrossRef] [Green Version]
- Holbrook, C.C.; Stalker, H.T. Peanut Breeding and Genetic Resources. Plant Breed. Rev. 2003, 22, 297–356. [Google Scholar]
- Fischer, M.C.; Rellstab, C.; Leuzinger, M.; Roumet, M.; Gugerli, F.; Shimizu, K.K.; Holderegger, R.; Widmer, A. Estimating Genomic Diversity and Population Differentiation—An Empirical Comparison of Microsatellite and SNP Variation in Arabidopsis Halleri. BMC Genom. 2017, 18, 69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sansaloni, C.; Petroli, C.; Jaccoud, D.; Carling, J.; Detering, F.; Grattapaglia, D.; Kilian, A. Diversity Arrays Technology (DArT) and next-Generation Sequencing Combined: Genome-Wide, High Throughput, Highly Informative Genotyping for Molecular Breeding of Eucalyptus. In Proceedings of the BMC Proceedings; BioMed Central: London, UK, 2011; Volume 5, pp. 1–2. [Google Scholar]
- Poland, J.A.; Rife, T.W. Genotyping-by-sequencing for Plant Breeding and Genetics. Plant Genome 2012, 5, 92–102. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Zavala, C.; Ortega, N.; Petroli, C.; Franco, J.; Burgueño, J.; Costich, D.E.; Hearne, S.J. The Development of Quality Control Genotyping Approaches: A Case Study Using Elite Maize Lines. PLoS ONE 2016, 11, e0157236. [Google Scholar]
- Yang, X.; Ren, R.; Ray, R.; Xu, J.; Li, P.; Zhang, M.; Liu, G.; Yao, X.; Kilian, A. Genetic Diversity and Population Structure of Core Watermelon (Citrullus lanatus) Genotypes Using DArTseq-Based SNPs. Plant Genet. Resour. 2016, 14, 226–233. [Google Scholar] [CrossRef]
- Denwar, N.N.; Awuku, F.J.; Diers, B.; Addae-Frimpomaah, F.; Chigeza, G.; Oteng-Frimpong, R.; Puozaa, D.K.; Barnor, M.T. Genetic Diversity, Population Structure and Key Phenotypic Traits Driving Variation within Soyabean (Glycine max) Collection in Ghana. Plant Breed. 2019, 138, 577–587. [Google Scholar] [CrossRef]
- Chigeza, G.; Boahen, S.; Gedil, M.; Agoyi, E.; Mushoriwa, H.; Denwar, N.; Gondwe, T.; Tesfaye, A.; Kamara, A.; Alamu, O.E.; et al. Public Sector Soybean (Glycine max) Breeding: Advances in Cultivar Development in the African Tropics. Plant Breed. 2019, 138, 455–464. [Google Scholar] [CrossRef] [Green Version]
- Beissinger, T.M.; Hirsch, C.N.; Sekhon, R.S.; Foerster, J.M.; Johnson, J.M.; Muttoni, G.; Vaillancourt, B.; Buell, C.R.; Kaeppler, S.M.; de Leon, N. Marker Density and Read Depth for Genotyping Populations Using Genotyping-by-Sequencing. Genetics 2013, 193, 1073–1081. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deschamps, S.; Llaca, V.; May, G.D. Genotyping-by-Sequencing in Plants. Biology 2012, 1, 460–483. [Google Scholar] [CrossRef] [Green Version]
- Adeboye, K.A.; Oyedeji, O.E.; Alqudah, A.M.; Börner, A.; Oduwaye, O.; Adebambo, O.; Daniel, I.O. Genetic Structure and Diversity of Upland Rice Germplasm Using Diversity Array Technology (DArT)-Based Single Nucleotide Polymorphism (SNP) Markers. Plant Genet. Resour. 2020, 18, 343–350. [Google Scholar] [CrossRef]
- Pootakham, W.; Shearman, J.R.; Ruang-Areerate, P.; Sonthirod, C.; Sangsrakru, D.; Jomchai, N.; Yoocha, T.; Triwitayakorn, K.; Tragoonrung, S.; Tangphatsornruang, S. Large-Scale SNP Discovery through RNA Sequencing and SNP Genotyping by Targeted Enrichment Sequencing in Cassava (Manihot esculenta Crantz). PLoS ONE 2014, 9, e116028. [Google Scholar] [CrossRef] [Green Version]
- Yang, G.; Chen, S.; Chen, L.; Sun, K.; Huang, C.; Zhou, D.; Huang, Y.; Wang, J.; Liu, Y.; Wang, H. Development of a Core SNP Arrays Based on the KASP Method for Molecular Breeding of Rice. Rice 2019, 12, 21. [Google Scholar] [CrossRef] [PubMed]
- Anderson, C.A.; Pettersson, F.H.; Clarke, G.M.; Cardon, L.R.; Morris, A.P.; Zondervan, K.T. Data Quality Control in Genetic Case-Control Association Studies. Nat. Protoc. 2010, 5, 1564–1573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eltaher, S.; Sallam, A.; Belamkar, V.; Emara, H.A.; Nower, A.A.; Salem, K.F.M.; Poland, J.; Baenziger, P.S. Genetic Diversity and Population Structure of F3: 6 Nebraska Winter Wheat Genotypes Using Genotyping-by-Sequencing. Front. Genet. 2018, 9, 76. [Google Scholar] [CrossRef] [PubMed]
- Wright, S. Evolution and the Genetics of Populations, Volume 4: Variability within and among Natural Populations; University of Chicago Press: Chicago, IL, USA, 1984; Volume 4, ISBN 0226910415. [Google Scholar]
- Zhang, S.; Li, B.; Chen, Y.; Shaibu, A.S.; Zheng, H.; Sun, J. Molecular-Assisted Distinctness and Uniformity Testing Using SLAF-Sequencing Approach in Soybean. Genes 2020, 11, 175. [Google Scholar] [CrossRef] [Green Version]
- Luo, Z.; Brock, J.; Dyer, J.M.; Kutchan, T.; Schachtman, D.; Augustin, M.; Ge, Y.; Fahlgren, N.; Abdel-Haleem, H. Genetic Diversity and Population Structure of a Camelina sativa Spring Panel. Front. Plant Sci. 2019, 10, 184. [Google Scholar] [CrossRef] [Green Version]
- Islam, M.Z.; Khalequzzaman, M.; Prince, M.; Siddique, M.A.; Rashid, E.; Ahmed, M.S.U.; Pittendrigh, B.R.; Ali, M.P. Diversity and Population Structure of Red Rice Germplasm in Bangladesh. PLoS ONE 2018, 13, e0196096. [Google Scholar] [CrossRef] [Green Version]
- Buckler IV, E.S.; Thornsberry, J.M. Plant Molecular Diversity and Applications to Genomics. Curr. Opin. Plant Biol. 2002, 5, 107–111. [Google Scholar] [CrossRef]
- Breseghello, F.; Sorrells, M.E. Association Mapping of Kernel Size and Milling Quality in Wheat (Triticum aestivum L.) Cultivars. Genetics 2006, 172, 1165–1177. [Google Scholar] [CrossRef] [Green Version]
- Arora, A.; Kundu, S.; Dilbaghi, N.; Sharma, I.; Tiwari, R. Population Structure and Genetic Diversity among Indian Wheat Varieties Using Microsatellite (SSR) Markers. Aust. J. Crop Sci. 2014, 8, 1281–1289. [Google Scholar]
- Kilian, A.; Wenzl, P.; Huttner, E.; Carling, J.; Xia, L.; Blois, H.; Caig, V.; Heller-Uszynska, K.; Jaccoud, D.; Hopper, C.; et al. Diversity Arrays Technology: A Generic Genome Profiling Technology on Open Platforms. Methods Mol. Biol. 2012, 888, 67–89. [Google Scholar] [CrossRef] [PubMed]
- Shaibu, A.; Sneller, C.; Motagi, B.; Chepkoech, J.; Chepngetich, M.; Miko, Z.; Isa, A.M.; Ajeigbe, H.A.; Mohammed, S.G. Genome-Wide Detection of SNP Markers Associated with Four Physiological Traits in Groundnut (Arachis hypogaea L.) Mini Core Collection. Agronomy 2020, 10, 192. [Google Scholar] [CrossRef] [Green Version]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for Association Mapping of Complex Traits in Diverse Samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of Population Structure Using Multilocus Genotype Data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Earl, D.A. STRUCTURE HARVESTER: A Website and Program for Visualizing STRUCTURE Output and Implementing the Evanno Method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the Number of Clusters of Individuals Using the Software STRUCTURE: A Simulation Study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [Green Version]
- Peakall, R.O.D.; Smouse, P.E. GENALEX 6: Genetic Analysis in Excel. Population Genetic Software for Teaching and Research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]

| Cluster Value | Mean Value of Fst | Expected Heterozygosity | Net Nucleotide Distance | |
|---|---|---|---|---|
| SNP | ||||
| Q1 | 0.17 | 0.23 | 0.45 | 0.10 |
| Q2 | 0.83 | 0.40 | 0.33 | 0.10 |
| DArT | ||||
| Q1 | 0.32 | 0.32 | 0.27 | 0.09 |
| Q2 | 0.63 | 0.19 | 0.30 | 0.09 |
| Source | df | SS | MS | Estimated Variance | % |
|---|---|---|---|---|---|
| Among Pops | 13 | 27,640.82 | 2126.227 | 40.45 | 3% |
| Within Pops | 267 | 391,863.18 | 1467.65 | 1467.65 | 97% |
| Total | 280 | 419,504.00 | 1508.10 | 100% | |
| Stat | Value | P(rand ≥ data) | |||
| PhiPT | 0.027 | 0.001 | |||
| Nm (Haploid) | 18.02 |
| Source | df | SS | MS | Estimated Variance | % |
|---|---|---|---|---|---|
| Among Pops | 13 | 15,980.95 | 1229.30 | 22.21 | 2% |
| Within Pops | 267 | 231,682.11 | 867.72 | 867.72 | 98% |
| Total | 280 | 247,663.06 | 889.93 | 100% | |
| Stat | Value | P(rand ≥ data) | |||
| PhiPT | 0.025 | 0.001 | |||
| Nm (Haploid) | 19.54 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shaibu, A.S.; Ibrahim, H.; Miko, Z.L.; Mohammed, I.B.; Mohammed, S.G.; Yusuf, H.L.; Kamara, A.Y.; Omoigui, L.O.; Karikari, B. Assessment of the Genetic Structure and Diversity of Soybean (Glycine max L.) Germplasm Using Diversity Array Technology and Single Nucleotide Polymorphism Markers. Plants 2022, 11, 68. https://doi.org/10.3390/plants11010068
Shaibu AS, Ibrahim H, Miko ZL, Mohammed IB, Mohammed SG, Yusuf HL, Kamara AY, Omoigui LO, Karikari B. Assessment of the Genetic Structure and Diversity of Soybean (Glycine max L.) Germplasm Using Diversity Array Technology and Single Nucleotide Polymorphism Markers. Plants. 2022; 11(1):68. https://doi.org/10.3390/plants11010068
Chicago/Turabian StyleShaibu, Abdulwahab S., Hassan Ibrahim, Zainab L. Miko, Ibrahim B. Mohammed, Sanusi G. Mohammed, Hauwa L. Yusuf, Alpha Y. Kamara, Lucky O. Omoigui, and Benjamin Karikari. 2022. "Assessment of the Genetic Structure and Diversity of Soybean (Glycine max L.) Germplasm Using Diversity Array Technology and Single Nucleotide Polymorphism Markers" Plants 11, no. 1: 68. https://doi.org/10.3390/plants11010068
APA StyleShaibu, A. S., Ibrahim, H., Miko, Z. L., Mohammed, I. B., Mohammed, S. G., Yusuf, H. L., Kamara, A. Y., Omoigui, L. O., & Karikari, B. (2022). Assessment of the Genetic Structure and Diversity of Soybean (Glycine max L.) Germplasm Using Diversity Array Technology and Single Nucleotide Polymorphism Markers. Plants, 11(1), 68. https://doi.org/10.3390/plants11010068







