T6SS Accessory Proteins, Including DUF2169 Domain-Containing Protein and Pentapeptide Repeats Protein, Contribute to Bacterial Virulence in T6SS Group_5 of Burkholderia glumae BGR1
Abstract
1. Introduction
2. Results
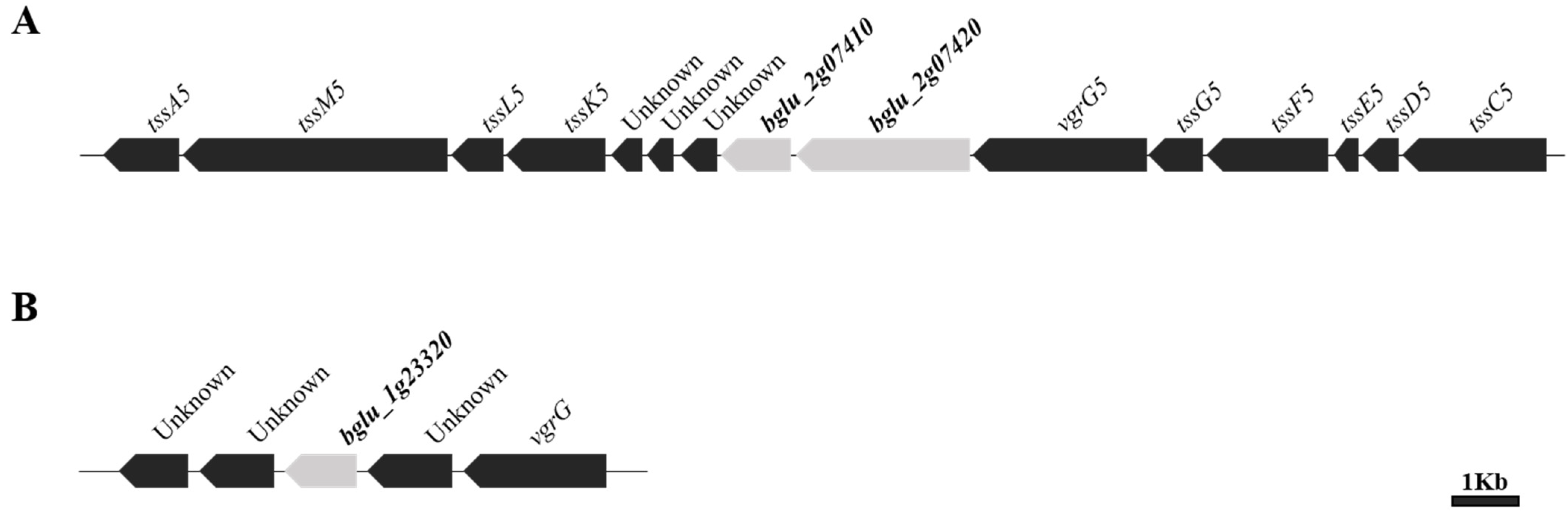
2.1. Investigation and Analysis of the Genes Containing the DUF2169 Domain and Its Adjacent Gene in the Genome of Burkholderia glumae BGR1
2.2. Construction of Markerless Deletion Mutants to Evaluate Whether the Putative T6SS Accessory Protein Affects the Function of T6SS Group_5 in B. glumae BGR1
2.3. The DUF2169 Domain Encoded by the N-terminus of bglu_2g07420 inside the Gene Cluster of T6SS Group_5 Is Involved in Bacterial Virulence in Rice Plants
2.4. The Pentapeptide Repeats Region Encoded by bglu_2g07410 and the C-terminus of bglu_2g07420 inside the Gene Cluster of T6SS Group_5 Is Involved in Bacterial Virulence to Rice Plants
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains, Plasmids, and Growth Conditions
4.2. Construction of Markerless Deletion Mutants and Their Complemetation Strains in B. glumae BGR1
4.3. In Vivo Pathogenicity Assay at the Vegetative and Reproductive Stages of Rice Plants
4.4. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Goto, K. New bacterial diseases of rice-bacterial brown stripe and bacterial grain rot. Ann. Phytopathol. Soc. Jpn. 1956, 21, 46–47. [Google Scholar]
- Goto, T.; Nishiyama, K.; Ohata, K.-I. Bacteria causing grain rot of rice. Jpn. J. Phytopathol. 1987, 53, 141–149. [Google Scholar] [CrossRef]
- Kim, N.; Kim, J.J.; Kim, I.; Mannaa, M.; Park, J.; Kim, J.; Lee, H.; Lee, S.; Park, D.; Sul, W.J.; et al. Type VI secretion systems of plant-pathogenic Burkholderia glumae BGR1 play a functionally distinct role in interspecies interactions and virulence. Mol. Plant Pathol. 2020, 21, 1055–1069. [Google Scholar] [CrossRef] [PubMed]
- Jeong, Y.; Kim, J.; Kim, S.; Kang, Y.; Nagamatsu, T.; Hwang, I. Toxoflavin Produced by Burkholderia glumae Causing Rice Grain Rot Is Responsible for Inducing Bacterial Wilt in Many Field Crops. Plant Dis. 2003, 87, 890–895. [Google Scholar] [CrossRef]
- Iiyama, K.; Furuya, N.; Takanami, Y.; Matsuyama, N. A Role of Phytotoxin in Virulence of Pseudomonas glumae Kurita et Tabei. Jpn. J. Phytopathol. 1995, 61, 470–476. [Google Scholar] [CrossRef]
- Suzuki, F.; Sawada, H.; Azegami, K.; Tsuchiya, K. Molecular characterization of the tox operon involved in toxoflavin biosynthesis of Burkholderia glumae. J. Gen. Plant Pathol. 2004, 70, 97–107. [Google Scholar] [CrossRef]
- Kim, J.; Kang, Y.; Choi, O.; Jeong, Y.; Jeong, J.-E.; Lim, J.Y.; Kim, M.; Moon, J.S.; Suga, H.; Hwang, I. Regulation of polar flagellum genes is mediated by quorum sensing and FlhDC in Burkholderia glumae. Mol. Microbiol. 2007, 64, 165–179. [Google Scholar] [CrossRef]
- Kang, Y.; Kim, J.; Kim, S.; Kim, H.; Lim, J.Y.; Kim, M.; Kwak, J.; Moon, J.S.; Hwang, I. Proteomic analysis of the proteins regulated by HrpB from the plant pathogenic bacterium Burkholderia glumae. Proteomics 2008, 8, 106–121. [Google Scholar] [CrossRef]
- Lee, J.; Park, J.; Kim, S.; Park, I.; Seo, Y.-S. Differential regulation of toxoflavin production and its role in the enhanced virulence of Burkholderia gladioli. Mol. Plant Pathol. 2016, 17, 65–76. [Google Scholar] [CrossRef]
- Jung, B.; Park, J.; Kim, N.; Li, T.; Kim, S.; Bartley, L.E.; Kim, J.; Kim, I.; Kang, Y.; Yun, K.; et al. Cooperative interactions between seed-borne bacterial and air-borne fungal pathogens on rice. Nat. Commun. 2018, 9, 31. [Google Scholar] [CrossRef]
- Mougous, J.D.; Cuff, M.E.; Raunser, S.; Shen, A.; Zhou, M.; Gifford, C.A.; Goodman, A.L.; Joachimiak, G.; Ordoñez, C.L.; Lory, S.; et al. A Virulence Locus of Pseudomonas aeruginosa Encodes a Protein Secretion Apparatus. Science 2006, 312, 1526–1530. [Google Scholar] [CrossRef] [PubMed]
- Pukatzki, S.; Ma, A.; Sturtevant, D.; Krastins, B.; Sarracino, D.; Nelson, W.; Heidelberg, J.; Mekalanos, J.J. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc. Natl. Acad. Sci. USA 2006, 103, 1528–1533. [Google Scholar] [CrossRef]
- Boyer, F.; Fichant, G.; Berthod, J.; Vandenbrouck, Y.; Attree, I. Dissecting the bacterial type VI secretion system by a genome wide in silico analysis: What can be learned from available microbial genomic resources? BMC Genom. 2009, 10, 104. [Google Scholar] [CrossRef]
- Ma, L.-S.; Lin, J.-S.; Lai, E.-M. An IcmF Family Protein, ImpL M, Is an Integral Inner Membrane Protein Interacting with ImpK L, and Its Walker A Motif Is Required for Type VI Secretion System-Mediated Hcp Secretion in Agrobacterium tumefaciens. J. Bacteriol. 2009, 191, 4316–4329. [Google Scholar] [CrossRef] [PubMed]
- Filloux, A.; Freemont, P. Structural biology: Baseplates in contractile machines. Nat. Microbiol. 2016, 1, 16104. [Google Scholar] [CrossRef] [PubMed]
- Nazarov, S.; Schneider, J.P.; Brackmann, M.; Goldie, K.N.; Stahlberg, H.; Basler, M. Cryo- EM reconstruction of Type VI secretion system baseplate and sheath distal end. EMBO J. 2017, 37, e97103. [Google Scholar] [CrossRef]
- Pukatzki, S.; Ma, A.T.; Revel, A.T.; Sturtevant, D.; Mekalanos, J.J. Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc. Natl. Acad. Sci. USA 2007, 104, 15508–15513. [Google Scholar] [CrossRef]
- Pell, L.G.; Kanelis, V.; Donaldson, L.; Howell, P.L.; Davidson, A.R. The phage major tail protein structure reveals a common evolution for long-tailed phages and the type VI bacterial secretion system. Proc. Natl. Acad. Sci. USA 2009, 106, 4160–4165. [Google Scholar] [CrossRef]
- Silverman, J.M.; Brunet, Y.R.; Cascales, E.; Mougous, J.D. Structure and Regulation of the Type VI Secretion System. Annu. Rev. Microbiol. 2012, 66, 453–472. [Google Scholar] [CrossRef]
- Shneider, M.M.; Buth, S.; Ho, B.; Basler, M.; Mekalanos, J.J.; Leiman, P.G. PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature 2013, 500, 350–353. [Google Scholar] [CrossRef]
- Brackmann, M.; Nazarov, S.; Wang, J.; Basler, M. Using Force to Punch Holes: Mechanics of Contractile Nanomachines. Trends Cell Biol. 2017, 27, 623–632. [Google Scholar] [CrossRef]
- Bondage, D.D.; Lin, J.-S.; Ma, L.-S.; Kuo, C.-H.; Lai, E.-M. VgrG C terminus confers the type VI effector transport specificity and is required for binding with PAAR and adaptor–effector complex. Proc. Natl. Acad. Sci. USA 2016, 113, E3931–E3940. [Google Scholar] [CrossRef] [PubMed]
- Cianfanelli, F.R.; Alcoforado Diniz, J.; Guo, M.; De Cesare, V.; Trost, M.; Coulthurst, S.J. VgrG and PAAR Proteins Define Distinct Versions of a Functional Type VI Secretion System. PLoS Pathog. 2016, 12, e1005735. [Google Scholar] [CrossRef] [PubMed]
- Flaugnatti, N.; Le, T.T.H.; Canaan, S.; Aschtgen, M.-S.; Nguyen, V.S.; Blangy, S.; Kellenberger, C.; Roussel, A.; Cambillau, C.; Cascales, E.; et al. A phospholipase A1 antibacterial Type VI secretion effector interacts directly with the C-terminal domain of the VgrG spike protein for delivery. Mol. Microbiol. 2016, 99, 1099–1118. [Google Scholar] [CrossRef] [PubMed]
- Lien, Y.-W.; Lai, E.-M. Type VI Secretion Effectors: Methodologies and Biology. Front. Cell. Infect. Microbiol. 2017, 7, 254. [Google Scholar] [CrossRef] [PubMed]
- Unterweger, D.; Kostiuk, B.; Pukatzki, S. Adaptor Proteins of Type VI Secretion System Effectors. Trends Microbiol. 2017, 25, 8–10. [Google Scholar] [CrossRef]
- Unterweger, D.; Kostiuk, B.; Ötjengerdes, R.; Wilton, A.; Diaz-Satizabal, L.; Pukatzki, S. Chimeric adaptor proteins translocate diverse type VI secretion system effectors in Vibrio cholerae. EMBO J. 2015, 34, 2198–2210. [Google Scholar] [CrossRef]
- Liang, X.; Moore, R.; Wilton, M.; Wong, M.J.Q.; Lam, L.; Dong, T.G. Identification of divergent type VI secretion effectors using a conserved chaperone domain. Proc. Natl. Acad. Sci. USA 2015, 112, 9106–9111. [Google Scholar] [CrossRef]
- Nguyen, T.T.; Lee, H.-H.; Park, I.; Seo, Y.-S. Genome-Wide Analysis of Type VI System Clusters and Effectors in Burkholderia Species. Plant Pathol. J. 2018, 34, 11–22. [Google Scholar] [CrossRef]
- Leiman, P.; Basler, M.; Ramagopal, U.A.; Bonanno, J.B.; Sauder, J.M.; Pukatzki, S.; Burley, S.; Almo, S.C.; Mekalanos, J.J. Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc. Natl. Acad. Sci. USA 2009, 106, 4154–4159. [Google Scholar] [CrossRef]
- Felisberto-Rodrigues, C.; Durand, E.; Aschtgen, M.-S.; Blangy, S.; Ortiz-Lombardía, M.; Douzi, B.; Cambillau, C.; Cascales, E. Towards a Structural Comprehension of Bacterial Type VI Secretion Systems: Characterization of the TssJ-TssM Complex of an Escherichia coli Pathovar. PLoS Pathog. 2011, 7, e1002386. [Google Scholar] [CrossRef] [PubMed]
- Kapitein, N.; Bönemann, G.; Pietrosiuk, A.; Seyffer, F.; Hausser, I.; Locker, J.K.; Mogk, A. ClpV recycles VipA/VipB tubules and prevents non-productive tubule formation to ensure efficient type VI protein secretion. Mol. Microbiol. 2013, 87, 1013–1028. [Google Scholar] [CrossRef] [PubMed]
- Silverman, J.M.; Agnello, D.M.; Zheng, H.; Andrews, B.T.; Li, M.; Catalano, C.E.; Gonen, T.; Mougous, J.D. Haemolysin Coregulated Protein Is an Exported Receptor and Chaperone of Type VI Secretion Substrates. Mol. Cell 2013, 51, 584–593. [Google Scholar] [CrossRef] [PubMed]
- Taylor, N.M.I.; Prokhorov, N.; Guerrero-Ferreira, R.; Shneider, M.M.; Browning, C.; Goldie, K.N.; Stahlberg, H.; Leiman, P. Structure of the T4 baseplate and its function in triggering sheath contraction. Nature 2016, 533, 346–352. [Google Scholar] [CrossRef]
- Diniz, J.A.; Coulthurst, S.J. Intraspecies Competition in Serratia marcescens Is Mediated by Type VI-Secreted Rhs Effectors and a Conserved Effector-Associated Accessory Protein. J. Bacteriol. 2015, 197, 2350–2360. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, Z.; Wang, F.; Li, D.-D.; Li, Y.-Z. Identification of type VI secretion system toxic effectors using adaptors as markers. Comput. Struct. Biotechnol. J. 2020, 18, 3723–3733. [Google Scholar] [CrossRef] [PubMed]
- Zong, B.; Zhang, Y.; Wang, X.; Liu, M.; Zhang, T.; Zhu, Y.; Zheng, Y.; Hu, L.; Li, P.; Chen, H.; et al. Characterization of multiple type-VI secretion system (T6SS) VgrG proteins in the pathogenicity and antibacterial activity of porcine extra-intestinal pathogenic Escherichia coli. Virulence 2019, 10, 118–132. [Google Scholar] [CrossRef]
- Shalom, G.; Shaw, J.G.; Thomas, M.S. In vivo expression technology identifies a type VI secretion system locus in Burkholderia pseudomallei that is induced upon invasion of macrophages. Microbiology 2007, 153, 2689–2699. [Google Scholar] [CrossRef]
- Hopf, V.; Göhler, A.; Eske-Pogodda, K.; Bast, A.; Steinmetz, I.; Breitbach, K. BPSS1504, a Cluster 1 Type VI Secretion Gene, Is Involved in Intracellular Survival and Virulence of Burkholderia pseudomallei. Infect. Immun. 2014, 82, 2006–2015. [Google Scholar] [CrossRef]
- Bateman, A.; Murzin, A.G.; Teichmann, S.A. Structure and distribution of pentapeptide repeats in bacteria. Protein Sci. 1998, 7, 1477–1480. [Google Scholar] [CrossRef]
- Tran, J.H.; Jacoby, G.A.; Hooper, D.C. Interaction of the Plasmid-Encoded Quinolone Resistance Protein QnrA with Escherichia coli Topoisomerase IV. Antimicrob. Agents Chemother. 2005, 49, 3050–3052. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Henry, T.; Couillault, C.; Rockenfeller, P.; Boucrot, E.; Dumont, A.; Schroeder, N.; Hermant, A.; Knodler, L.; Lécine, P.; Steele-Mortimer, O.; et al. The Salmonella effector protein PipB2 is a linker for kinesin-1. Proc. Natl. Acad. Sci. USA 2006, 103, 13497–13502. [Google Scholar] [CrossRef] [PubMed]
- Vetting, M.W.; Hegde, S.S.; Fajardo, J.E.; Fiser, A.; Roderick, S.L.; Takiff, H.E.; Blanchard, J.S. Pentapeptide Repeat Proteins. Biochemistry 2003, 45, 1–10. [Google Scholar] [CrossRef]
- Zhang, R.; Kennedy, M. Current Understanding of the Structure and Function of Pentapeptide Repeat Proteins. Biomolecules 2021, 11, 638. [Google Scholar] [CrossRef] [PubMed]
- Murtza, T.; You, M.P.; Barbetti, M.J. Canola Growth Stage at Time of Infection Determines Magnitude of White Leaf Spot (Neopseudocercosporella capsellae) Impact. Plant Dis. 2021, 105, 1515–1521. [Google Scholar] [CrossRef] [PubMed]
- Kalogeraki, V.S.; Winans, S.C. Suicide plasmids containing promoterless reporter genes can simultaneously disrupt and create fusions to target genes of diverse bacteria. Gene 1997, 188, 69–75. [Google Scholar] [CrossRef]
- Schäfer, A.; Tauch, A.; Jsger, W.; Kalinowski, J.; Thierbachb, G.; Piihler, A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: Selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 1994, 145, 49–5201. [Google Scholar] [CrossRef]
- Kovach, M.E.; Elzer, P.H.; Hill, D.S.; Robertson, G.T.; Farris, M.A.; Roop, R.M., II; Peterson, K.M. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 1995, 166, 175–176. [Google Scholar] [CrossRef]
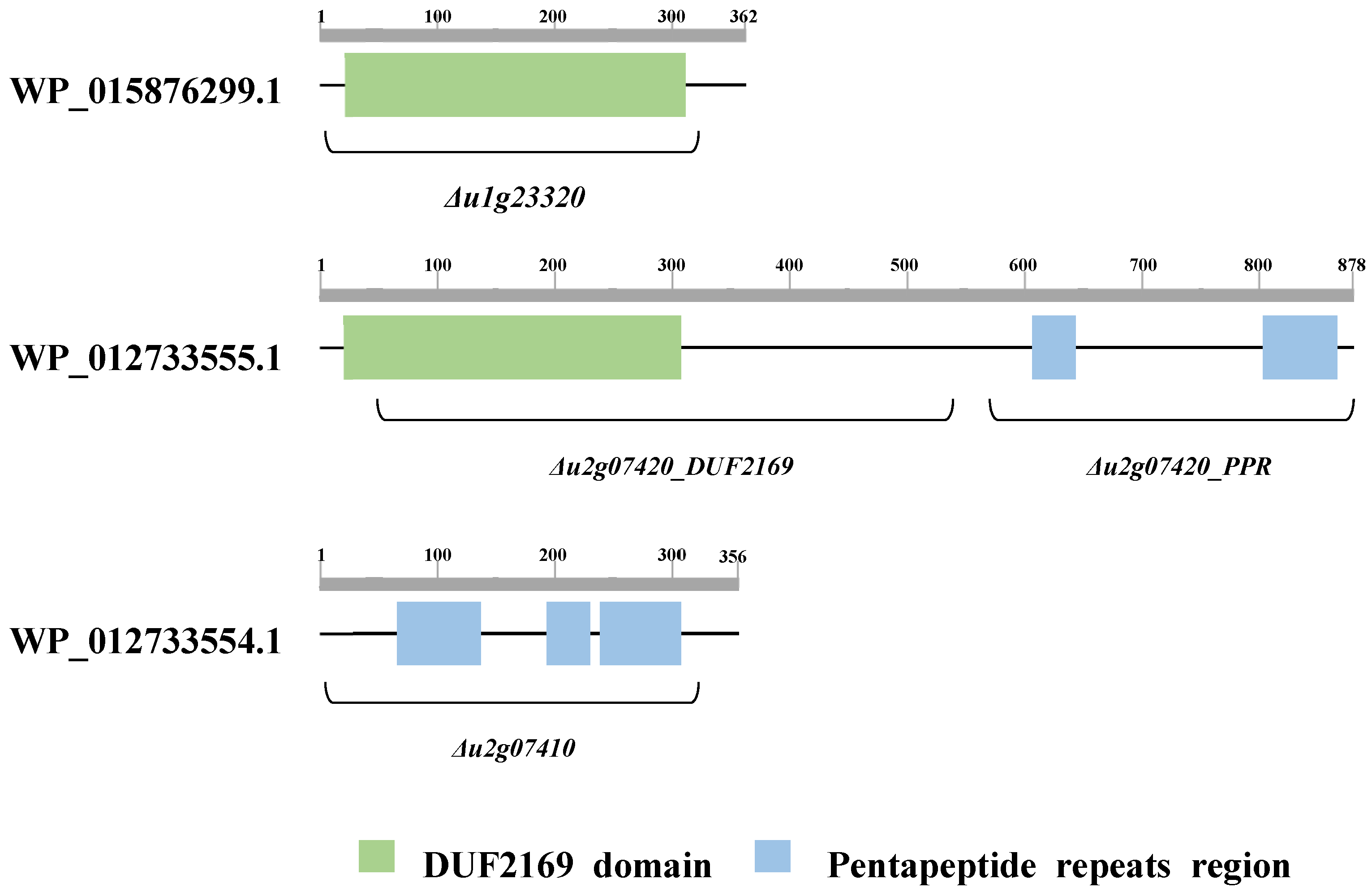



| Analysis | Accession | Description | InterPro Accession | InterPro Description | Interval | E-Value |
|---|---|---|---|---|---|---|
| WP_015876299.1 | ||||||
| Pfam | PF09937 | Uncharacterized protein conserved in bacteria (DUF2169) | IPR018683 | DUF2169 | 21~311 | 5.6 × 10−91 |
| WP_012733554.1 | ||||||
| Pfam | PF13599 | Pentapeptide repeats (9 copies) | IPR001646 | Pentapeptide repeat | 66~137 | 2.5 × 10−10 |
| Pfam | PF00805 | Pentapeptide repeats (8 copies) | IPR001646 | Pentapeptide repeat | 193~230 | 3.2 × 10−9 |
| Pfam | PF13599 | Pentapeptide repeats (9 copies) | IPR001646 | Pentapeptide repeat | 238~308 | 1.7 × 10−7 |
| WP_012733555.1 | ||||||
| Pfam | PF09937 | Uncharacterized protein conserved in bacteria (DUF2169) | IPR018683 | DUF2169 | 20~307 | 6.4 × 10−73 |
| Pfam | PF00808 | Pentapeptide repeats (8 copies) | IPR001646 | Pentapeptide repeat | 605~642 | 2.0 × 10−11 |
| Pfam | PF13599 | Pentapeptide repeats (9 copies) | IPR001646 | Pentapeptide repeat | 801~864 | 3.1 × 10−6 |
| Name | Characteristics | Source |
|---|---|---|
| Bacterial strains | ||
| B. glumae | ||
| BGR1 | Burkholderia glumae isolate from rice, wild-type, Rifr * | [4] |
| Δu2g07410 | BGR1 derivative, deletion of 824 bp within bglu_2g07410 | This study |
| Δu2g07420_DUF2169 | BGR1 derivative, deletion of 1466 bp of the DUF2169 domain region within bglu_2g07420 | This study |
| Δu2g07420_PPR | BGR1 derivative, deletion of 927 bp of the pentapeptide repeats region within bglu_2g0720 | This study |
| Δu2g07410-20 | bglu_2g07410 and pentapeptide repeats region of bglu_2g07420 double deletion mutant derived from BGR1 | This study |
| Δu1g23320 | BGR1 derivative, deletion of 951 bp within bglu_1g23320 | This study |
| Δu2g07410-C | BGR1 bglu_2g07410 deletion mutant containing pBu2g07410 | This study |
| Δu2g07420_DUF2169-C | BGR1 bglu_2g07420 deletion mutant (Δu2g07420_DUF2169) containing pBu2g07420 | This study |
| Δu2g07420_PPR-C | BGR1 bglu_2g07420 deletion mutant (Δu2g07420_PPR) containing pBu2g07420 | This study |
| Δu2g07410-20-C | BGR1 bglu_2g07410-20 double deletion mutant (Δu2g07410-20) containing pBu2g07410 and pBu2g07420 | This study |
| E. coli | ||
| E. coli DH5α λpir | F- 80dlacZΔM15 (lacZYA-argF) U169 recA1 endA1hsdR17 (rk-, mk+) phoAsupE44 -thi-1 gyrA96 relA1 | Lab collection |
| E. coli S17-1 λpir | hsdR recA pro RP4-2 (Tc::Mu; Km::Tn7) (λ pir) | [46] |
| Plasmids | ||
| pK18mobsacB | Allelic exchange suicide vector, sacB Kmr * | [47] |
| pK18u2g07410 | For constructing bglu_2g07410 KO mutant, pK18mobsacB:: LR fragment of bglu_2g07410 region restricted by EcoRI-HindIII | This study |
| pK18u2g07420DUF2169 | For constructing the DUF2169 domain in the bglu_2g07420 KO mutant, pK18mobsacB:: LR fragment of the DUF2169 domain region in bglu_2g07420 restricted by EcoRI-HindIII | This study |
| pK18u2g07420PPR | For constructing pentapeptide repeats region in the bglu_2g07420 KO mutant, pK18mobsacB::LR fragment of pentapeptide repeats region in bglu_2g07420 restricted by BamHI-HindIII | This study |
| pK18u1g23320 | For constructing bglu_1g23320 KO mutant, pK18mobsacB:: LR fragment of bglu_1g23320 region restricted by BamHI-HindIII | This study |
| pBBR1MCS2 | Broad-host-range plasmid, Kmr *, used to construct complementation strains. | [48] |
| pBu2g07410 | For constructing the Δu2g07410 complementation strain, pBBR1MCS2::CDS of bglu_2g07410 | This study |
| pBu2g07420 | For constructing the Δu2g07420_DUF2169 and Δu2g07420_PPR complementation strain, pBBR1MCS2::CDS of bglu_2g07420 | This study |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, N.; Han, G.; Jung, H.; Lee, H.-H.; Park, J.; Seo, Y.-S. T6SS Accessory Proteins, Including DUF2169 Domain-Containing Protein and Pentapeptide Repeats Protein, Contribute to Bacterial Virulence in T6SS Group_5 of Burkholderia glumae BGR1. Plants 2022, 11, 34. https://doi.org/10.3390/plants11010034
Kim N, Han G, Jung H, Lee H-H, Park J, Seo Y-S. T6SS Accessory Proteins, Including DUF2169 Domain-Containing Protein and Pentapeptide Repeats Protein, Contribute to Bacterial Virulence in T6SS Group_5 of Burkholderia glumae BGR1. Plants. 2022; 11(1):34. https://doi.org/10.3390/plants11010034
Chicago/Turabian StyleKim, Namgyu, Gil Han, Hyejung Jung, Hyun-Hee Lee, Jungwook Park, and Young-Su Seo. 2022. "T6SS Accessory Proteins, Including DUF2169 Domain-Containing Protein and Pentapeptide Repeats Protein, Contribute to Bacterial Virulence in T6SS Group_5 of Burkholderia glumae BGR1" Plants 11, no. 1: 34. https://doi.org/10.3390/plants11010034
APA StyleKim, N., Han, G., Jung, H., Lee, H.-H., Park, J., & Seo, Y.-S. (2022). T6SS Accessory Proteins, Including DUF2169 Domain-Containing Protein and Pentapeptide Repeats Protein, Contribute to Bacterial Virulence in T6SS Group_5 of Burkholderia glumae BGR1. Plants, 11(1), 34. https://doi.org/10.3390/plants11010034






