Abstract
Plant viruses exploit the endomembrane system of infected cells for their replication and cell-to-cell transport. The replication of viral RNA genomes occurs in the cytoplasm in association with reorganized endomembrane compartments induced by virus-encoded proteins and is coupled with the virus intercellular transport via plasmodesmata that connect neighboring cells in plant tissues. The transport of virus genomes to and through plasmodesmata requires virus-encoded movement proteins (MPs). Distantly related plant viruses encode different MP sets, or virus transport systems, which vary in the number of MPs and their properties, suggesting their functional differences. Here, we discuss two distinct virus transport pathways based on either the modification of the endoplasmic reticulum tubules or the formation of motile vesicles detached from the endoplasmic reticulum and targeted to endosomes. The viruses with the movement proteins encoded by the triple gene block exemplify the first, and the potyviral system is the example of the second type. These transport systems use unrelated mechanisms of endomembrane reorganization. We emphasize that the mode of virus interaction with cell endomembranes determines the mechanism of plant virus cell-to-cell transport.
1. Introduction
The plant endomembrane system (EMS) is essential for biosynthetic processes and targeted distribution of macromolecules and other compounds in cells. The major compartments comprising the plant EMS include the nuclear envelope, the endoplasmic reticulum (ER), the Golgi apparatus, the trans-Golgi network (TGN), the endosomes, and the vacuole [1]. Distinct membrane compartments are connected to each other through vesicular trafficking, the process in which membrane vesicles serving as transport containers are detached from source EMS compartments and directionally delivered to target membrane compartments [1,2]. In particular, the trafficking pathways between the ER and Golgi are mediated by COPII- and COPI-coated vesicles in the case of ER-to-Golgi and retrograde Golgi-to-ER transport, respectively [3,4,5], whereas the transport between the TGN, the vacuole, the endosomes, and the plasma membrane (PM) is mediated by clathrin-coated vesicles [2,6]. The vesicular transport serves for the intracellular delivery of proteins contained within the EMS compartments (e.g., in the ER lumen, Golgi cisternae, or endosomes), as well as membrane components and membrane-embedded proteins [1]. Additionally, distinct membrane compartments are interconnected by membrane contact sites (MCSs), the sites of interactions of the ER membrane with endosomes, peroxisomes, Golgi, vacuoles, the PM, as well as organelles such as mitochondria and plastids [7]. The MCSs are proposed to constitute a non-vesicular inter-organellar communication pathway [8] required for signal transmission and exchange of lipids and possibly proteins between the ER and other organelles [9,10,11].
Plant viruses exploit the EMS for their replication and cell-to-cell transport [12]. The replication of viral (+)RNA-genomes occurs in the cytoplasm, and the virus progeny can be transported from infected cells to surrounding healthy cells through plasmodesmata (PD), plant-specific intercellular channels that traverse cell walls and provide direct contacts of the cytoplasm and the EMS of neighboring cells [13,14,15]. The PD include the PM lining the PD channel, and the desmotubule, a constricted ER tubule continuous with the ER of two adjoining cells, spanning the pore on its axis [14]. In the PD channel, the PM and the desmotubule are tethered together by protein elements characteristic of MCSs [15,16,17]. These protein tethers are believed to contribute to regulating the size of the cytoplasmic sleeve between the PM and the desmotubule that determines the PD permeability, or the PD size exclusion limit (SEL) [18]. However, the major mechanism of the regulation of the PD SEL involves the local callose deposition in the cell wall regions surrounding the PD that results in the PD ‘closure’ and degradation of PD-associated callose by β-1,3-glucanase, leading to the PD SEL increase [19]
The cell-to-cell transport of plant virus genomes requires virus-encoded movement proteins (MPs) essential for the targeting of progeny viral genomic RNA or virions to PD and their transport through the PD channels to neighboring cells [20]. The Tobacco mosaic virus (TMV) MP is the paradigm MP, the first-discovered and currently best-studied among proteins of this function [12]. Two basic properties of the TMV MP were described soon after its discovery, namely the abilities (i) to bind RNA and (ii) to accumulate in PD, increase the PD SEL, and move cell to cell [21]. Accordingly, the TMV MP has been hypothesized to bind viral RNA and form transport-competent viral ribonucleoprotein complexes (vRNPs) capable of trafficking through the PD channel dilated by the TMV MP [21]. Numerous further studies have demonstrated a tight link between the TMV replication and transport, as well as a close association of both these processes with the ER and the cytoskeleton. In particular, the TMV MP is shown to localize to the ER structures, to interact with microtubules, and to co-localize with viral RNA [12,22,23].
According to current models of TMV cell-to-cell transport, at the initial infection stages, the viral replication complexes (VRCs), which contain viral RNA, replicase, and MP, are formed on the ER membranes in association with microtubules [12,24]. These early VRCs are mobile and move in the cytoplasm along the ER in an actin-dependent manner, reaching the PD proximity [25,26,27]. The TMV MP is believed to be the protein that targets VRCs to PD, as the MP on its own is capable of PD targeting, most probably due to a specific PD localization signal that is mapped to the protein N-terminal region. This MP region is involved in the MP interaction with SYTA, a transmembrane protein of the synaptotagmin family located in the MCSs, and is involved in virus cell-to-cell transport, suggesting that SYTA may play a role in the VRC targeting to PD [28,29,30,31,32]. Further, upon replication in the PD-targeted VRCs, the virus RNA progeny is likely directed to transport through the PD channels [33].
The TMV MP has been initially predicted to be an integral membrane protein [34,35]. However, according to further experimental analysis of the MP topology in the ER membrane, the protein peripherally associates with the cytoplasmic face of the ER [36]. Additionally, the TMV RNA injected in plant cells forms granules anchored to the ER network, and the methylguanosine cap at the 5′-terminus of virus RNA is required for the anchoring [33]. Therefore, as one of possible models, the VRC formation may be initiated by virus RNA anchoring at the ER membrane and further continued upon local translation of viral membrane-interacting proteins, namely the TMV MP and the 126K protein, a TMV replicase component known to associate with the ER and co-translationally bind the 5′-terminal region of the TMV genomic RNA [37]. However, these and other available observations clarify neither the TMV VRC structure nor the mechanism of the VRC formation of the ER membranes of TMV-infected cells.
In this review, focused mainly on the intracellular transport of viral genomes to PD, we discuss two distinct TMV-unrelated plant virus transport systems, for which dissimilar mechanisms of reorganization of cell endomembranes for virus transport are currently uncovered and characterized in detail. Further, we make parallels between the mechanisms of the intracellular transport of virus-specific RNAs and cellular mRNAs.
2. TGB/BMB: The ER-Based Pathway
2.1. Properties of TGB Proteins
In plant virus-encoded multicomponent transport systems, i.e., those involving two or more MPs, multiple functions performed by the single MP of TMV are distributed among several proteins, showing a ‘division of labor’. A well-studied example of such a multicomponent transport system is a ‘triple gene block’ (TGB) [38]. TGB is an evolutionary conserved gene module found in many (+)RNA plant viruses of families Virgaviridae, Alphaflexiviridae, Betaflexiviridae, and Benyviridae. TGB consists of three partially overlapping genes encoding MPs termed TGB1, TGB2, and TGB3, which have characteristic structural features [39].
TGB1 protein contains an NTPase/helicase domain belonging to a divergent lineage of ‘accessory’ viral SFI (superfamily I) helicases that likely have evolved after a duplication of a replicative RNA helicase domain [39,40,41]. Accordingly, TGB1 proteins are experimentally shown to have the activities of NTPase [39] and RNA helicase [42]. As demonstrated by the mutagenesis of conserved motifs of the NTPase/helicase domain, the enzymatic activities of TGB1 are essential for its function in cell-to-cell transport [43,44,45]. Additionally, TGB1 protein exhibits a non-specific RNA-binding activity, which is believed to be involved in the formation of vRNPs with virus genomic RNA [45,46,47,48]. The TGB1 protein has also been shown to act as a suppressor of RNA silencing, and this activity can be linked to the efficiency of virus transport [49,50].
Based on comparative analyses, two types of TGB are distinguished, potex-like and hordei-like TGBs, termed by the names of two well-studied virus genera Potexvirus and Hordeivirus, having distinct TGBs [39]. In hordei-like TGBs, the TGB1 protein has an additional N-terminal domain, absent in potex-like TGBs, which is involved in the systemic transport of virus infection via the phloem [46,51,52]. The fundamental difference between viruses with potex-like and hordei-like TGBs is the dependence of virus cell-to-cell transport on viral capsid protein (CP) and genome encapsidation, known to be essential for transport in the case of potex-like but not hordei-like TGB [53]. As shown for Potato virus X (PVX), the type potexvirus, several TGB1 molecules can bind one end of PVX filamentous virions that renders them transport competent and enables the translation of virion-derived RNAs after the virus has moved cell to cell [54]. By contrast, encapsidation is not required for the transport of Barley stripe mosaic virus (BSMV), the type hordeivirus, and its genomic RNA is transported through the PD as a complex with TGB1, which appears to be the sole protein component of such a complex [55,56]. Therefore, the TGB1 roles in the cell-to-cell transport are not identical in potex- and hordei-like TGBs.
TGB2 and TGB3 are small integral membrane proteins, and mutations disrupting their association with membranes block virus transport [57,58]. TGB2 has two hydrophobic domains with a conserved region between them. As shown for the TGB2 of both potex- and hordei-like TGB, the N- and C-termini are exposed to the cytoplasm, while the central hydrophilic loop is exposed to the ER lumen [59,60]. However, as suggested by further analysis of TGB2 functional interactions, the protein central region can be located in the cytoplasm [61] (discussed below). The TGB3 protein of the potex-like TGBs has a single transmembrane domain, whereas the hordei-like TGB3 protein is sequence unrelated to that in the potex-like TGBs and has two transmembrane domains [39,62].
In subcellular localization experiments, the TGB2 and TGB3 proteins are found in the ER and/or ER-derived membrane structures. In particular, TGB2 fused to fluorescent proteins is found to be localized, depending on the experimental system, either in cortical ER tubules or numerous ER-related tiny vesicles, whereas TGB3 is found in both intracellular ER-associated structures and ER-derived membrane bodies located at the cell periphery in close vicinity of PD [63]. These structures are called hereafter ‘PD-associated membrane bodies’ (PAMBs). The TGB3 localization to PAMBs requires specific signals mapped in the TGB3 proteins of several viruses [64,65,66,67], and the TGB3 intracellular transport to PAMBs bypasses the conventional secretory pathway and likely occurs by a lateral translocation in the plane of the ER membrane [66]. Importantly, TGB3 is able to direct TGB2 from intracellular ER-associated sites to PAMBs [39], although in some experiments TGB2 is found in PAMBs even when expressed in the absence of TGB3 [68]. The observed TGB3 ability to change the subcellular localization of TGB2 suggests interaction between TGB2 and TGB3. Indeed, residues involved in such interactions have been mapped in both proteins of BSMV, and mutations affecting these residues inhibited virus transport [55].
Virus cell-to-cell movement is generally accepted to require a modification of the PD internal structure resulting in the PD SEL increase [14]. For potex-like TGB proteins, attempts to identify the TGB protein responsible for the PD SEL increase have resulted in contradictory data. Several experiments have shown that potex-like TGB1 introduced into cells by microinjection is able to interact with PD and increase the PD permeability [69,70,71]. In other studies, only TGB2 but not TGB1, TGB3, or CP is shown to be able to increase the PD SEL [72]. The hordei-like Potato mop-top virus (PMTV) TGB2 protein is shown to interact with cellular proteins TIP1, TIP2 и TIP3, which in turn can interact with β-1,3-glucanase and therefore influence the callose degradation at the PD neck regions, resulting in the increase of the PD SEL [73].
2.2. Early Model of TGB-Mediated Transport
An initial model of TGB functioning was based on experiments employing transient co-expression of individual cloned TGB proteins for analysis of their intracellular and intercellular transport. In a representative study, the PMTV TGB2 and TGB3 proteins have been shown to act in cooperation to ensure delivery of TGB1 to TGB2/TGB3-containing PAMBs at the PD entrance. This delivery does not require the TGB1 NTPase/helicase activity, which, instead, is essential for further TGB1 transport into the PD interior and to neighboring cells [43]. These and other experiments have demonstrated that TGB2 and TGB3 function in cells where these proteins have been synthesized and are not transported through the PD channels into neighboring cells. As TGB1 proteins are generally assumed to bind viral RNA or virions to produce transport-competent genome-containing vRNPs [39,63], the following model for TGB-mediated virus cell-to-cell movement has been proposed. The TGB1-containing vRNPs, which can be either TGB1:RNA complexes or TGB1-bound virions, are transported intracellularly to PD due to (i) interaction of TGB1 with TGB2/TGB3 and (ii) signals in TGB2 and/or TGB3 directing these proteins to PAMBs at the PD orifice. Further transport of viral genomes through PD depends on the enzymatic activities of TGB1, presumably required to remodel either the structure of the transported vRNP, the internal PD structure, or both [39,43]. This generalized model, although correct in describing the functions of individual proteins, appeared incomplete and has been further considerably developed in view of recently accumulated data on (i) direct and tight links between virus cell-to-cell movement and replication and (ii) the pivotal role of MP-mediated reorganization of cell membranes for virus transport.
2.3. The Role of Modified ER Membranes in TGB-Mediated Transport
Fine details of coupling virus transport to replication in ER-derived membrane compartments are being studied using potexviruses as models, and the current knowledge can be summarized as follows.
In the cytoplasm, the virus replicase molecules form high-molecular-weight complexes [74] targeted to the ER membrane by an amphipathic α-helix and form, in the absence of other virus proteins, initial viral replicative compartments (iVRCs), which are clustered in the perinuclear area into large amorphous aggregates [75] morphologically similar to structures found in potexvirus-infected cells and historically called ‘X-bodies’ [68,76]. The X-bodies contain all PVX-encoded proteins, virions, and host factors. Detailed analysis of X-bodies revealed that they represent complex layered structures. The TGB1 protein plays an important role in their organization, as TGB1 aggregates form the core of the X-body, or so-called ‘beaded sheets’, visible by electron microscopy in infected cells. Additionally, TGB1 reorganizes the actin cytoskeleton at the X-bodies [77,78]. The TGB1 aggregates are surrounded by recruited TGB2/TGB3-containing ER membranes representing a highly dense reticulated network rather than a native polygonal tubular ER network [78]. Analysis of the localization of the viral dsRNA in the X-bodies demonstrate that RdRp/dsRNA-containing bodies surround TGB1 aggregates, are covered by ‘chain mail-like’ TGB2-aggregates, and localize in the immediate vicinity to TGB3 aggregates [61]. Non-encapsidated viral RNA and progeny virions localize to the periphery of the X-bodies [61]. Therefore, the X-bodies represent highly-structured stationary virus factories associated with the ER.
At early infection stages, PVX TGB2 induces the formation of tiny mobile ER-associated membrane bodies [79]. In virus-infected cells, these bodies also contain the viral replicase [63,79]. The formation of the latter bodies likely results from the direct interaction of TGB2 with the viral replicase (Figure 1). Regions involved in this interaction have been mapped in both proteins and the TGB2 incorporation into the replicase containing structures does not require other TGB proteins [61]. Therefore, the TGB2 protein is suggested to target iVRCs [61] to convert them into movement VRCs (mVRCs) capable of intracellular transport. TGB3 also incorporates mVRCs, likely due to its interaction with TGB2 [80], whereas the TGB1 recruitment requires both TGB2 and TGB3 [68]. Thus, iVRCs can form stationary replicase-containing perinuclear X-bodies even in the absence of other viral proteins and, additionally, can develop into mobile mVRCs in the presence of the TGB proteins.
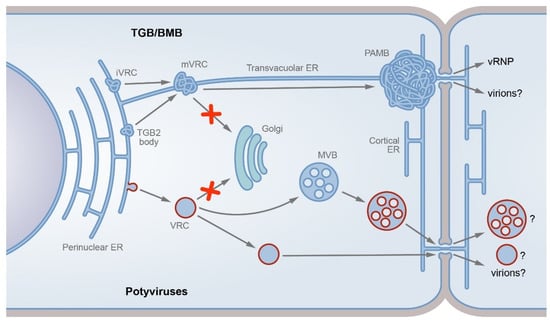
Figure 1.
TGB/BMB-specific and potyvirus-specific transport pathways. The two dissimilar pathways described are either modification of the ER tubules (TGB/BMB; upper part) or formation of motile vesicles detached from the endoplasmic reticulum and targeted to endosomes (potyviruses, bottom). In the TGB/BMB-specific pathway, the viral replicase forms initial virus replication complexes (iVRCs) on the ER membrane, whereas TGB2 can interact with the ER to form membrane TGB2 bodies. Due to TGB2 interaction with viral replicase, TGB2 targets iVRCs to convert them into movement VRCs (mVRCs). The mVRCs are capable of intracellular transport and can be transported along the cytoskeleton to the PD entrance, bypassing the Golgi-dependent trafficking pathway (shown by red cross sign). At the PD entrance, mVRCs form PD-associated membrane bodies (PAMBs), which represent ER-derived PD-anchored sites where virus replication is coupled to cell-to-cell virus transport. Virus ribonucleoprotein complexes (vRNPs) containing genomic RNA and MPs are formed in PAMBs and directed to the PD channels. Alternatively, virions, likely MP-modified, can be the virus transport form (indicated by question mark). In the potyvirus-specific pathway, the viral 6K2 protein induces the formation of VRCs on the ER membrane. The potyvirus VRCs, which carry viral proteins, virus RNA, and cellular factors, are detached from the ER membrane in a COPII-dependent manner. Further VRC intracellular trafficking bypasses the Golgi pathway (red cross sign). The VRCs target late endosomes (multivesicular bodies, MVBs), which in turn target PD. Alternatively, the VRCs can directly target PD. The potyvirus-specific entity transported through the PD channels is unknown and can be VRC, modified MVB, or virion (indicated by question marks).
The formed mVRCs are believed to be highly mobile and can be transported along the cytoskeleton to the PD entrance. In fact, TGB2-containing membrane bodies have been shown to colocalize with microfilaments (MF) and an intact actin cytoskeleton is required for the efficient cell-to-cell transport of PVX [79,81]. Thus, the signals of intracellular trafficking identified in TGB3 (see above) and presumed for TGB2 [68] can be required for the PD targeting of TGB3-containing mVRCs rather than the TGB proteins on their own. At the PD entrance, mVRCs form PAMBs, which represent ER-derived PD-anchored sites where virus replication is coupled to cell-to-cell virus transport [68] (Figure 1). Importantly, PAMBs contain reorganized densely reticulated ER tubules, which likely comprise the backbone of these structures [68]. Therefore, the ER membranes in PAMBs and X-bodies are modified in a similar manner, emphasizing the structural similarity of both types of iVRC-derived compartments.
Once vRNP complexes are synthesized in PAMBs, they are immediately directed into the PD channel by the TGB1 protein for further transport into neighboring cells [68]. Viral MPs studied so far demonstrate nonspecific RNA binding activity, suggesting that MPs are not able to distinguish between vRNAs and cellular mRNAs [82]. Therefore, besides physical concentration of necessary viral and host factors, PAMBs may possibly increase the specificity and efficiency of viral RNA transport by excluding cellular RNAs [76].
Therefore, the available data suggest that TGB2 appears to play the central role in the organization of PAMBs, as it: (1) directly interacts with the viral RdRp and therefore targets iVRCs and converts them into mVRC; (2) interacts with TGB3, which contains signals of PD targeting that facilitates the mVRC transport to the PD orifice; (3) together with TGB3 recruits TGB1 to mVRCs.
2.4. BMB2 Interaction with ER Membranes
New light is shed on the TGB2 functions in studies of MPs encoded by Hibiscus green spot virus (HGSV; family Kitaviridae), which has a transport gene module evolutionary related to TGB [40,83,84]. This transport module is called ‘binary movement block’ (BMB) as it consists of two open reading frames, accordingly called BMB1 and BMB2 genes [85]. The virus movement function of the BMB1 and BMB2 proteins has been confirmed in complementation experiments, where these two HGSV proteins complement cell-to-cell transport of the modified PVX genome, which is able to replicate and express GFP in initially infected cells, but is deficient in cell-to-cell movement because of the absence of TGB and CP genes [85].
The BMB1 protein is distantly related to the TGB1 protein in sequence and contains the NTPase/helicase domain [40,84]. Although not studied experimentally, BMB1, by analogy to TGB1, is suggested to be the RNA-binding protein that takes part in the formation of transport vRNPs. This view is indirectly confirmed in the PVX complementation experiments mentioned above, as in this system the BMB1 protein is the only viral protein able to form transport vRNPs [85].
The BMB2 protein, similar to TGB2, has two highly hydrophobic transmembrane domains, with the central region between them showing limited sequence similarity to TGB2 proteins [62,86]. BMB2 is integrated into the ER membrane and is localized in the ER-derived PAMBs formed upon BMB2 expression [85]. The intracellular BMB2 transport to PAMBs does not involve the Golgi-dependent secretory pathway, as earlier shown for TGB2, but requires the intact actin/ER network, suggesting that both BMB2 and TGB2 can employ a similar mechanism of transport to PAMBs, likely involving the lateral translocation in the plane of the ER membrane [87]. In transient co-expression experiments, BMB2 directs BMB1, which exhibits a diffuse localization when expressed alone, to PAMBs, to the PD interior, and through the PD channels to neighboring cells [85]. Therefore, as the HGSV transport system lacks a counterpart of TGB3, the functions of both TGB2 and TGB2 proteins can be apparently performed by the single BMB2 protein.
Recently, the BMB2 protein has been shown to have reticulon-like properties that provided a clue to its functions, as well as those of TGB2, in virus cell-to-cell transport [85]. Reticulons are cellular proteins essential for the generation of the ER membrane curvature and for the formation of ER tubules, serving therefore for the formation and maintenance of the ER morphology [88,89,90]. The reticulon functions are determined by a ‘reticulon homology domain’ comprising two membrane-immersed hydrophobic regions forming wedge-shaped hairpins, which bend the lipid bilayer as the reticulon oligomerizes [91,92]. These regions are connected by a loop, and therefore the reticulon molecule has a W-like topology in the ER membrane, where both ends of the protein and the central loop are located on the ER cytosolic side [91]. The BMB2 protein, similar to the reticulons, has been shown to have the W-like topology in the membrane, and the overexpression of BMB2 results in the constriction of the ER tubules [93], as previously demonstrated in similar experiments with reticulons [91]. Therefore, BMB2 is supposed to generate, either on its own or by interaction with cellular protein partner(s), the ER membrane curvature. Importantly, the PVX TGB2 protein, when transiently expressed in plant cells, is also able to induce constrictions of ER tubules, and similar constrictions have been observed in PVX-infected cells [93]. Mapping of PVX TGB2 regions interacting with virus RdRp shows that the central and C-terminal protein regions are located in the cytoplasm [61], implying that TGB2 can also adopt the W-shaped conformation in the ER membrane. Thus, the formation of aberrant densely reticulated ER tubules observed on PAMBs and X-bodies in PVX-infected cells can be attributed to the presumed TGB2 ability to modify the structure of ER tubules. As the modified ER membranes are considered a core structural element of VRCs/PAMBs/X-bodies, the modification of ER tubules by the BMB2/TGB2 proteins can be essential for the formation of initial VRCs, which are further developed into PAMBs or X-bodies upon recruitment of other viral proteins. This model is corroborated by observations that the formation of membrane replicative compartments of a variety of (+)RNA viruses replicating in the cytoplasm requires the reticulon activity, and viruses without own reticulon-like proteins recruit cellular reticulons for membrane modification [94].
Another characteristic BMB2 feature shared with reticulons is the affinity to highly curved membranes. In cells containing flat ER cisternae, both reticulons and BMB2 are targeted to cisternae rims with high membrane curvature [91,93]. These findings can explain the observed localization of BMB2 and TGB2 in the PD channel or in the PD central cavity [68,93], as these proteins may have affinity to desmotubule, the ER tubule on the PD axis that has a small diameter and, consequently, a high membrane curvature. Such BMB2/TGB2 localization makes a parallel with two plant reticulons, RTNLB3 and RTNLB6, found in the desmotubule [95,96] and, on the other hand, can be pertinent to the observed ability of BMB2 and TGB2 to increase the PD SEL [72,93]. The mechanism of BMB2/TGB2-induced SEL increase is unknown. However, taking into account the BMB2 ability to form high-molecular-weight complexes likely including BMB2 oligomers, the BMB2 protein has been hypothesized to displace, upon accumulation of such complexes in the desmotubule, native components of the PD channel and therefore increase the PD SEL [93].
Thus, two key functions of BMB2/TGB2, namely the modification of ER membranes for VRC formation and the modification of the PD internal structure resulting in the PD SEL increase, may be determined by the mode of BMB2/TGB2 interaction with the ER membrane, involving both the generation of additional membrane curvature and the targeting to membrane sites with a high curvature of the lipid bilayer.
3. Potyviruses: The Vesicles/Endosomes-Based Pathway
3.1. Potyvirus Proteins Involved in Virus Transport
Genomes of potyviruses (genus Potyvirus, family Potyviridae) encode a polyprotein precursor, which is autocatalytically processed into eleven mature proteins [97]. In addition to polyprotein-derived proteins, a 7 kDa protein named PIPO is produced in infected cells. Due to RNA polymerase slippage during the replication, an additional adenine residue is inserted in 1–2% of genomic RNA progeny that brings the PIPO-encoding sequence into the reading frame of another viral protein called P3. As a result, a fusion protein P3N-PIPO consisting of the amino-terminal half of P3 and the PIPO polypeptide is produced [98,99].
Potyviruses have a multicomponent transport system, as several viral proteins are involved in potyvirus cell-to-cell transport, including CP, P3N-PIPO, and the cylindrical inclusion protein (CI). Potyvirus mutants encoding encapsidation-deficient CP are incapable of cell-to-cell movement [100,101,102], indicating a direct link between virion formation and virus transport. Moreover, the potyvirus CP has been found in the PD interior by immunogold labeling, being detected in the form of fibrils morphologically similar to flexuous filamentous potyvirus virions [103,104]. Therefore, potyviruses are suggested to move cell-to-cell in the form of either genuine virions or virion-like complexes similar to virions in their composition but differing in their fine structure [105,106]. CI, another protein essential for potyvirus transport [103,104,107], contains a helicase domain and exhibits the NTPase and helicase activities involved in virus replication [108]. Mutations in the N-terminal protein region, which is required for the CI self-interaction and is positioned outside the helicase domain, abolish virus movement but do not block replication [107,109,110], suggesting that these two CI functions can be uncoupled. In potyvirus-infected cells, the CI protein forms pinwheel cytoplasmic inclusions, which are located at the PD entrance and often traverse the cell wall over the PD channels, and numerous virions are typically associated with the CI-formed structures [103,104]. When expressed alone in plant cells, CI is found in aggregates in the cytoplasm. However, upon co-expression with P3N-PIPO, CI forms inclusions similar to those in potyvirus-infected cells, showing that the CI transport to PD is P3N-PIPO-dependent [111]. In fact, P3N-PIPO on its own is targeted to PD and interacts with CI in plant cells [111]. Similar to the TMV MP, the P3N-PIPO protein is able to move cell-to-cell in plant tissue and increases the PD SEL [98]. P3N-PIPO binds PCaP1, a cellular cation-binding protein interacting with the PM via myristoylation. Moreover, the potyvirus transport, but not replication, is drastically reduced in PCaP1-deficient Arabidopsis plants [98]. These data demonstrate that the PCaP1-dependent targeting of P3N-PIPO to the PM is essential for virus transport.
Two additional potyvirus-encoded proteins implicated in cell-to-cell transport, VPg and HC-Pro, interact with each other [112,113]. The VPg protein is covalently linked to the 5′-terminus of potyvirus genomic RNA, and the VPg interaction with HC-Pro is suggested to be responsible for the formation of a morphologically distinct structure on one end of potyvirus virions that can be labeled with HC-Pro antibodies [114,115]. The CI protein is co-purified with a subpopulation of potyvirus particles and associated, according to immunogold labeling, with one end of the virion [116], likely due to the reported interaction of CI with HC-Pro [113].
Collectively, the available data suggest that a complex pattern of interactions between potyvirus-encoded proteins is required for virus cell-to-cell transport. According to the proposed model [53,105], which implies that the virion is the most probable transport form of the potyvirus genome, a fraction of progeny virions may have a terminal structure consisting of VPg, HC-Pro, and CI that may make these virions competent for cell-to-cell transport. The targeting of such virions to PD is possible as a result of the interaction between the virion-associated CI and P3N-PIPO, which is capable of localization to PD, and the CI self-interaction involved also in the formation of the PD-associated pinwheel inclusions. Further cell-to-cell transport of virions likely depends on the PD SEL increase induced by P3N-PIPO and, additionally, HC-Pro and CP [117]. However, this mechanistic model does not take into account the association of potyvirus replication with membranous structures and the pathways of intracellular targeting of movement-related proteins to PD.
3.2. Formation of Potyvirus VRCs
The replication of potyviruses is associated with the EMS membranes, as also documented for many other animal and plant RNA viruses replicating in the cytoplasm [118,119]. Specifically, the potyvirus replication occurs in vesicles derived from the ER membranes [120,121] (Figure 1). These vesicular VRCs carry viral proteins including CI and the RNA-dependent RNA polymerase NIb, as well as virus RNA and cellular factors [120,122,123]. However, the fine VRC structure is unstudied. In particular, it remains unclear whether the virus-specific components are located in the VRC lumen or on the cytoplasmic face of the VRC-enclosing membrane.
The biogenesis of potyvirus ER-derived VRCs depends on the viral protein termed 6K2, a small protein with a single transmembrane domain cotranslationally integrated into the ER membrane and capable of inducing the VRC formation in the absence of other viral proteins [120]. In the context of potyvirus infection, the key player in VRC formation is the 6K2-VPg-Pro precursor serving as a scaffold, the VPg moiety of which recruits several virus and host proteins involved in replication, whereas the 6K2 moiety is responsible for the membrane remodeling [124]. In fact, the 6K2-dependent reorganization of ER in potyvirus-infected cells leads to the formation of two types of structures, namely motile VRCs located at the cell periphery and a large perinuclear globular structure, which includes the ER and Golgi structures as well as chloroplasts [125]. However, the 6K2-induced structures are not functionally isolated, as the motile VRCs can derive from the perinuclear structure [125].
The 6K2-induced formation of vesicular VRCs is COPII-dependent [126], i.e., it involves the conventional cellular mechanism of COPII (coat protein complex II)-mediated formation of transport vesicles, which bud from the ER membrane and deliver cargo to the Golgi cisternae [5]. Accordingly, 6K2 is found in the ER exit sites, the discrete ER domains where secretory proteins and membrane proteins destined for the export from the ER are concentrated and packed into transport vesicles [126,127]. Furthermore, 6K2 interacts with the COPII coatomer Sec24a, the subunit interacting with cargo proteins for their packaging into COPII-vesicles [128]. The N-terminal cytoplasmic domain of 6K2 is essential for this interaction, and a point mutation of a conserved residue in this domain leads to partial retention of the protein in the ER and blocks the virus transport out of primary infected cells [128]. Therefore, the formation of potyvirus VRCs occurs upon the 6K2-induced COPII-dependent detachment of 6K2-containing vesicles from the ER. Interestingly, in cells coinfected with two fluorescently tagged versions of one potyvirus genome modified to express either GFP or mCherry fused to 6K2, a part of VRCs have been observed to carry green-only or red-only fluorescent labeling, suggesting that a particular individual VRC derives from a single genome and likely includes locally synthesized viral proteins [122].
In potyvirus-infected cells, the export of cellular proteins from the ER is inhibited, possibly due to the Sec24a mobilization by the 6K2 protein [125]. In Arabidopsis mutant lines with partially functional Sec24a, the inhibition of COPII-dependent exit from the ER is accompanied by the formation of aberrant membrane compartments somewhat similar to large perinuclear globular structures observed upon potyvirus infection [124]. These findings suggest that the formation of both discrete motile VRCs and perinuclear structures may be induced through a common mechanism involving Sec24a.
Based on available data, we conclude that the VRC nature and biogenesis differ drastically between TGB/BMB and the potyvirus transport system (Figure 1). While the TGB/BMB VRCs are structurally based on modified, likely additionally constricted, ER tubules and remain a part of the ER, the potyvirus VRCs are vesicular structures detached from the ER membrane by use of the cellular COPII machinery. Additionally, the TGB/BMB VRCs are stationarily located at the PD entrance [68], whereas the potyvirus VRCs represent motile membrane structures.
3.3. VRC Trafficking and the Endosome/Post-Golgi Pathway
The motility of potyvirus VRCs in the cytoplasm requires the actin network but not microtubules, and the VRC trafficking along actin microfilaments depends on myosins XI-2 and XI-K [122,125,126,129], suggesting a mechanism described for cellular motile endomembrane vesicles associated with F-actin [130]. In potyvirus-infected cells, the VRCs are found in association with the PM, often being located in close vicinity of PD [131], suggesting that VRCs can reach PD as a result of their intracellular movement. Moreover, using a photoactivated fluorescent protein to label 6K2, the potyvirus VRCs are shown to move cell to cell [131], demonstrating therefore the possibility of intercellular transport of virus replication membrane compartments, as it has been suggested for the TMV VRCs [132].
The VRC intracellular transport is routed by the 6K2 protein, as mutations of conserved glycine residues in the 6K2 transmembrane domain abolish the normal VRC trafficking and redirect VRCs to the Golgi structures that block the virus replication and transport [133], indicating that the normal VRC transport bypasses the Golgi structures and uses a specific nonconventional pathway (Figure 1). The RHD3 (ROOT HAIR DEFECTIVE 3) protein, a cellular dynamin-like atlastin, is mobilized to the potyvirus VRCs [134]. A potyvirus mutant carrying the substitutions of glycine residues in the 6K2 transmembrane domain is incapable of the RHD3 recruiting to VRCs and deficient in the replication and movement. Additionally, the potyvirus infection is suppressed in rhd3 mutant plants [134], suggesting that RHD3 may be required for the 6K2 function in the proper VRC routing.
The Golgi-independent VRC transport can involve post-Golgi membrane compartments. In fact, the 6K2 protein is colocalized and copurified with VTI11 [133], a Qb-SNARE protein localized to the trans-Golgi network and late endosome/multivesicular bodies (MVBs) [135]. In line with this finding, double-stranded RNA representing a replication intermediate of potyvirus genome is detected in MVBs by immunogold electron microscopy [133]. Moreover, an Arabidopsis VTI11 knock-out line is resistant to potyvirus infection [133], showing the importance of VRC targeting to MVB. This targeting depends on a specific interaction of the 6K2 protein with VSR4, an ESCRT protein, and a point mutation in VSR4 leads to resistance to potyvirus infection [136,137]. Therefore, the intracellular transport of potyvirus VRCs, which are detached from the ER and finally reach PD, can bypass the Golgi and involve post-Golgi membrane compartments (Figure 1). Interestingly, Plantago asiatica mosaic virus, a TGB-containing potexvirus, is capable of establishing systemic infection in the VTI11 knock-out plants resistant to potyvirus infection [133], emphasizing the difference between membrane-associated replication/transport systems of potyviruses and viruses with TGB/BMB.
Another role of post-Golgi membranes in the potyvirus transport is demonstrated in studies employing RabE1d(NI), a dominant negative mutant of the small GTPase RabE1d involved in the post-Golgi trafficking toward the PM. RabE1d(NI) has no effect on the virus replication and the VRC intracellular motility, but blocks the VRC cell-to-cell transport [129,138]. This RabE1d(NI) effect appeared to be indirect, as the P3N-PIPO localization to PD is suppressed in RabE1d(NI)-expressing cells that leads to blocked VRC targeting to PD [138].
The transport of 6K2-induced vesicles can be reconstructed in non-infected cells where individual potyvirus proteins are transiently co-expressed. Under these conditions, 6K2 induces the formation of vesicles morphologically indistinguishable from typical VRCs observed in infected cells, whereas the CI and P3N-PIPO proteins are necessary and sufficient for cell-to-cell transport of these vesicles [138]. The 6K2 protein interacts with CI, and such interaction is required for the intercellular movement of 6K2 vesicles [138]. These and earlier findings elucidate the specific roles played by P3N-PIPO, CI, and 6K2 in the VRC trafficking. P3N-PIPO is localized to PD, modifies the PD internal structure, and increases the PD SEL; the CI protein is targeted to PD due to its interaction with P3N-PIPO; in turn, the VRCs are targeted to the CI-formed pinwheel inclusions at the PD entrance due to the interaction of 6K2 with CI. Indeed, the 6K2-induced vesicles are observed in close association with PD and the CI-formed PD-associated inclusions [131,138].
Therefore, two models, both supported by experimental data, are currently suggested for the potyvirus cell-to-cell movement: the model of virion transport outlined above and the more recent model of VRC cell-to-cell transport. Two key protein components of the potyvirus transport system, as well as their proposed functions, are common for both models, i.e., (i) P3N-PIPO, which targets PD, and (ii) the CI protein, which is directed to PD due to its interaction with P3N-PIPO and forms characteristic PD-associated inclusions. The genome-containing infectious entities transported through the PD channels are, however, different in the two models, being either modified virions carrying CI at one end and therefore capable of interaction with P3N-PIPO and/or CI molecules forming inclusions, or VRCs carrying 6K2 and therefore interacting with CI and the pinwheel inclusions. At the moment, it is not clear how these two models can converge, as the VRC model does not explain why the potyvirus CP is essential for virus cell-to-cell movement, while the virion model does not consider the observed intercellular transport of 6K2-containing vesicles.
Irrespective of the nature of a potyvirus infectious entity transported through the PD channels, recent findings showing that entering the endosome/post-Golgi pathway is essential for a potyvirus infection making parallels with the mechanism of intracellular transport discovered for cellular mRNAs.
4. Endosome Structures in mRNA Transport
Generally, the intracellular transport of mRNA is required for specific localization of particular mRNA in eukaryotic cells that ensures the localized protein synthesis essential for establishing cell polarity, patterning, and cell fate determination [139]. Additionally, the mRNA transport is indispensable in polarized cells, such as mammalian neurons, in which distal dendrites and axons spatially separated from the nucleus-containing cell body are provided with transcripts by means of the mRNA transport [140] or cells of distal fast-growing tips of fungal hyphae, in which the mRNA delivery from the nucleus is required for the localized synthesis of protein components essential for the growth [141].
Although the mRNA transport has been well characterized in several non-plant systems (see below), the studies of mRNA transport mechanisms in plants are in their very beginning. Currently, the main model for such studies is the mRNA transport in endosperm cells of developing rice seeds, where mRNAs encoding the storage proteins glutelin and prolamine are transported to two distinct ER-associated endomembrane compartments [142]. For their differential localization, the two mRNAs contain cis-acting signals, which are termed zip codes. In the prolamine mRNA, two zip-code elements with a common sequence motif are located in the protein coding region and the 3′-UTR, whereas the glutelin RNA contains six zip-code elements of two types found in the coding region (five such elements) and the 3′-UTR [143]. Several RNA-binding proteins (RBPs) interact with the zip codes to form transport RNPs. The key role is played by the RBP-P/RBP-L complex, which specifically binds zip codes and serves as a scaffold for interaction with other RNP components [144]. In endosperm cells, the glutelin and prolamine RNAs are detected as particles moving in the cytoplasm [145,146]. However, the nature of these particles remained unclear until recently, when other proteins interacting with the RNPs have been discovered. In particular, the RBP-P/RBP-L complex has been shown to interact with an N-ethylmaleimide-sensitive factor (NSF) and the small GTPase Rab5a, the two proteins participating in endosomal membrane trafficking; moreover, the RBP-P/RBP-L proteins colocalize with NSF and Rab5a on endosomes carrying the glutelin mRNA [147]. In Rab5-deficient plants, the glutelin mRNA-containing RNPs are mistargeted to extracellular structures as a result of corrupted endosome trafficking [147]. Collectively, these findings demonstrate that mRNAs can be transported in plant cells in the form of RNPs anchored to the surface of endosomes, making evident parallels with the potyvirus-specific transport pathway involving endosomes. This mechanism of mRNA transport is not unique for plants and the role of endosomes in mRNA transport is well documented for other systems, including fungal hyphae and animal axons.
Similar to rice endosperm cells, the endosome-dependent intracellular mRNA trafficking in the filamentous forms of the fungus Ustilago maydis requires a number of proteins involved in transport RNP formation and its anchoring to the endosome surface [141,148]. In particular, the RNPs are formed with the participation of Rrm4 and Grp1, two RNA-binding proteins interacting predominantly with the 3′-UTRs of shared cargo mRNAs, often in close proximity, and Pab1, a poly(A)-binding protein [148,149]. The transport RNPs are anchored to endosomes by UPA1, a protein that interacts with Pab1 and, on the other hand, contains the FYVE zinc finger domain enabling specific interaction with lipids on the endosome surface [150]. The transport RNP-carrying endosomes move along the microtubule by the use of kinesin and dynein motors [151]. Importantly, translationally active polyribosomes are efficiently co-transported with endosomes in U. maydis [152,153], suggesting that the endosomal transport system enables translocation of functional translation apparatus that ensures the immediate local protein synthesis upon mRNA delivery to growing distal hyphae ends.
Neuronal axons are often extremely long and can extend for millimeters. To avoid the prolonged time of response to external stimuli that would drastically slow down the axon functioning, neurons employ rapid mRNA trafficking from the nucleus to distal cell parts and localized protein synthesis as the ways to overcome the distance constraints [154,155]. In the axons of Xenopus laevis retinal ganglion cells, 20–30% of mRNAs colocalize with endosomes displaying bidirectional intracellular transport, and a number of ribosomal proteins and RBPs, such as Fragile X mental retardation protein, Staufen2, and Pumilio2, are associated with both mRNAs and endosomes, suggesting that these organelles could act as mRNA transport platforms [156]. In mice models, the localized translation of β-actin in axons is critical to the process of growth cone pathfinding [157]. For selective intracellular transport and localized translation, the neuron β-actin mRNA is targeted to late endosomes together with ribosomes and the RNA-binding protein ZBP1 that binds to a 54-nucleotide zip code sequence in the 3′-UTR of β-actin mRNA [158,159]. Similar to the β-actin mRNA, mRNA of GAP-43, another protein selectively localized to axonal growth cones during development, localizes in axons in a ZBP1-dependent manner [158,160]. Recently, FERRY, a novel early endosome-transported RNP complex composed of five subunits called Fy-1 to Fy-5, has been identified in human neurons. The small GTPase Rab5 binds both early endosomes and Fy-2, therefore tethering the FERRY RNP on moving membrane organelles [161].
The above examples demonstrate that the endosome-dependent mRNA trafficking is observed in plant, fungal, and animal cells, therefore the involvement of endosomes in the potyvirus cell-to-cell transport does not appear somewhat uncommon. However, the TGB/BMB-specific transport pathway has no parallels among currently known cellular mechanisms of RNA transport.
5. Conclusions
Here, using the TGB/BMB-mediated virus transport and the potyvirus-specific transport as examples, we substantiate the idea that plant viruses can use dissimilar, unrelated mechanisms of modification of cell endomembranes for their movement in plants. In particular, the proteins of the TGB/BMB transport system modify the ER tubules, thereby generating movement-related replication compartments that remain a part of the ER and are stationarily located at the PD entrance, so that the genomic RNA progeny is directed into the PD microchannels for cell-to-cell transport. By contrast, the potyvirus-specific replication/movement membrane compartments represent motile vesicular structures detached from the ER membrane, and the trafficking of these structures to PD involves targeting to post-Golgi compartments, including endosomes, which appear to be an essential hub for the potyvirus infection. Moreover, vesicular membrane structures carrying virus proteins and genomic RNA are suggested to be transported through PD to enable virus cell-to-cell transport. Therefore, according to the different ways of interaction with endomembranes, the mechanisms of transport, both intracellular and intercellular, appear to be fundamentally different between potyviruses and TGB/BMB-encoding viruses. Undoubtedly, detailed analyses of host membrane modifications observed for plant virus transport systems of other types may either allow their assignment to one of the transport mechanisms discussed in this review, or lead to the discovery of yet unknown mechanisms of endomembrane targeting and cell-to-cell transport. For example, studies of ‘double gene block’ (DGB), the transport system of carmoviruses, demonstrate that the intracellular trafficking of a DGB-encoded protein involves Golgi [162], showing a marked difference from both transport systems discussed in this review and suggesting that DGB may use a distinct specific transport pathway. In conclusion, we emphasize that studies of plant virus interaction with endomembranes are of key importance for understanding the mechanisms of virus cell-to-cell transport.
Author Contributions
All authors, original draft preparation, review, and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Russian Science Foundation, grant number 22-14-00063.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Aniento, F.; de Medina Hernández, V.S.; Dagdas, Y.; Rojas-Pierce, M.; Russinova, E. Molecular Mechanisms of Endomembrane Trafficking in Plants. Plant Cell 2022, 34, 146–173. [Google Scholar] [CrossRef] [PubMed]
- Arora, D.; Damme, D. Van Motif-Based Endomembrane Trafficking. Plant Physiol. 2021, 186, 221–238. [Google Scholar] [CrossRef] [PubMed]
- Brandizzi, F. Transport from the Endoplasmic Reticulum to the Golgi in Plants: Where Are We Now? Semin. Cell Dev. Biol. 2018, 80, 94–105. [Google Scholar] [CrossRef] [PubMed]
- Weigel, A.V.; Chang, C.L.; Shtengel, G.; Xu, C.S.; Hoffman, D.P.; Freeman, M.; Iyer, N.; Aaron, J.; Khuon, S.; Bogovic, J.; et al. ER-to-Golgi Protein Delivery through an Interwoven, Tubular Network Extending from ER. Cell 2021, 184, 2412–2429.e16. [Google Scholar] [CrossRef]
- Brandizzi, F.; Barlowe, C. Organization of the ER–Golgi Interface for Membrane Traffic Control. Nat. Rev. Mol. Cell Biol. 2013, 14, 382–392. [Google Scholar] [CrossRef]
- González Solís, A.; Berryman, E.; Otegui, M.S. Plant Endosomes as Protein Sorting Hubs. FEBS Lett. 2022, 17, 2288–2304. [Google Scholar] [CrossRef]
- Pérez-Sancho, J.; Tilsner, J.; Samuels, A.L.; Botella, M.A.; Bayer, E.M.; Rosado, A. Stitching Organelles: Organization and Function of Specialized Membrane Contact Sites in Plants. Trends Cell Biol. 2016, 26, 705–717. [Google Scholar] [CrossRef]
- Cohen, S.; Valm, A.M.; Lippincott-Schwartz, J. Interacting Organelles. Curr. Opin. Cell Biol. 2018, 53, 84–91. [Google Scholar] [CrossRef]
- Shai, N.; Yifrach, E.; Van Roermund, C.W.T.; Cohen, N.; Bibi, C.; Ijlst, L.; Cavellini, L.; Meurisse, J.; Schuster, R.; Zada, L.; et al. Systematic Mapping of Contact Sites Reveals Tethers and a Function for the Peroxisome-Mitochondria Contact. Nat. Commun. 2018, 9, 1761. [Google Scholar] [CrossRef]
- Valm, A.M.; Cohen, S.; Legant, W.R.; Melunis, J.; Hershberg, U.; Wait, E.; Cohen, A.R.; Davidson, M.W.; Betzig, E.; Lippincott-Schwartz, J. Applying Systems-Level Spectral Imaging and Analysis to Reveal the Organelle Interactome. Nature 2017, 546, 162–167. [Google Scholar] [CrossRef]
- Pankratenko, A.V.; Atabekova, A.K.; Morozov, S.Y.; Solovyev, A.G. Membrane Contacts in Plasmodesmata: Structural Components and Their Functions. Biochemistry 2020, 85, 531–544. [Google Scholar] [CrossRef] [PubMed]
- Heinlein, M. Plant Virus Replication and Movement. Virology 2015, 479–480, 657–671. [Google Scholar] [CrossRef]
- Nicolas, W.J.; Grison, M.S.; Bayer, E.M. Shaping Intercellular Channels of Plasmodesmata: The Structure-to-Function Missing Link. J. Exp. Bot. 2017, 69, 91–103. [Google Scholar] [CrossRef]
- Heinlein, M. Plasmodesmata: Channels for Viruses on the Move. Methods Mol. Biol. 2015, 1217, 25–52. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.P.; Paterlini, A.; Glavier, M.; Bayer, E.M. Intercellular Trafficking via Plasmodesmata: Molecular Layers of Complexity. Cell. Mol. Life Sci. 2021, 78, 799–816. [Google Scholar] [CrossRef]
- Brault, M.L.; Petit, J.D.; Immel, F.; Nicolas, W.J.; Glavier, M.; Brocard, L.; Gaston, A.; Fouché, M.; Hawkins, T.J.; Crowet, J.; et al. Multiple C2 Domains and Transmembrane Region Proteins (MCTPs) Tether Membranes at Plasmodesmata. EMBO Rep. 2019, 20, e47182. [Google Scholar] [CrossRef] [PubMed]
- Tilsner, J.; Nicolas, W.; Rosado, A.; Bayer, E.M. Staying Tight: Plasmodesmal Membrane Contact Sites and the Control of Cell-to-Cell Connectivity in Plants. Annu. Rev. Plant Biol. 2016, 67, 337–364. [Google Scholar] [CrossRef]
- Huang, C.; Heinlein, M. Function of Plasmodesmata in the Interaction of Plants with Microbes and Viruses. Methods Mol. Biol. 2022, 2457, 23–54. [Google Scholar] [CrossRef]
- Wu, S.-W.; Kumar, R.; Iswanto, A.B.B.; Kim, J.-Y. Callose Balancing at Plasmodesmata. J. Exp. Bot. 2018, 69, 5325–5339. [Google Scholar] [CrossRef]
- Lucas, W.J. Plant Viral Movement Proteins: Agents for Cell-to-Cell Trafficking of Viral Genomes. Virology 2006, 344, 169–184. [Google Scholar] [CrossRef]
- Citovsky, V. Tobacco Mosaic Virus: A Pioneer to Cell–to–Cell Movement. Philos. Trans. R. Soc. London. 1999, 354, 637–643. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Boyko, V.; Ferralli, J.; Ashby, J.; Schellenbaum, P.; Heinlein, M. Function of Microtubules in Intercellular Transport of Plant Virus RNA. Nat. Cell Biol. 2000, 2, 826–832. [Google Scholar] [CrossRef] [PubMed]
- Heinlein, M.; Padgett, H.S.; Gens, J.S.; Pickard, B.G.; Casper, S.J.; Epel, B.L.; Beachy, R.N. Changing Patterns of Localization of the Tobacco Mosaic Virus Movement Protein and Replicase to the Endoplasmic Reticulum and Microtubules during Infection. Plant Cell 1998, 10, 1107–1120. [Google Scholar] [CrossRef]
- Sambade, A.; Brandner, K.; Hofmann, C.; Seemanpillai, M.; Mutterer, J.; Heinlein, M. Transport of TMV Movement Protein Particles Associated with the Targeting of RNA to Plasmodesmata. Traffic 2008, 9, 2073–2088. [Google Scholar] [CrossRef] [PubMed]
- Wright, K.M.; Wood, N.T.; Roberts, A.G.; Chapman, S.; Boevink, P.; Mackenzie, K.M.; Oparka, K.J. Targeting of TMV Movement Protein to Plasmodesmata Requires the Actin/ER Network: Evidence from FRAP. Traffic 2007, 8, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.-Z.; Blancaflor, E.B.; Nelson, R.S. The Tobacco Mosaic Virus 126-Kilodalton Protein, a Constituent of the Virus Replication Complex, Alone or within the Complex Aligns with and Traffics along Microfilaments. Plant Physiol. 2005, 138, 1853–1865. [Google Scholar] [CrossRef]
- Amari, K.; Di Donato, M.; Dolja, V.V.; Heinlein, M. Myosins VIII and XI Play Distinct Roles in Reproduction and Transport of Tobacco Mosaic Virus. PLoS Pathog. 2014, 10, e1004448. [Google Scholar] [CrossRef]
- Lewis, J.D.; Lazarowitz, S.G. Arabidopsis Synaptotagmin SYTA Regulates Endocytosis and Virus Movement Protein Cell-to-Cell Transport. Proc. Natl. Acad. Sci. USA 2010, 107, 2491–2496. [Google Scholar] [CrossRef]
- Uchiyama, A.; Shimada-Beltran, H.; Levy, A.; Zheng, J.Y.; Javia, P.A.; Lazarowitz, S.G. The Arabidopsis Synaptotagmin SYTA Regulates the Cell-to-Cell Movement of Diverse Plant Viruses. Front. Plant Sci. 2014, 5, 584. [Google Scholar] [CrossRef]
- Levy, A.; Zheng, J.Y.; Lazarowitz, S.G. Synaptotagmin SYTA Forms ER-Plasma Membrane Junctions That Are Recruited to Plasmodesmata for Plant Virus Movement. Curr. Biol. 2015, 25, 2018–2025. [Google Scholar] [CrossRef]
- Yuan, C.; Lazarowitz, S.G.; Citovsky, V. Identification of a Functional Plasmodesmal Localization Signal in a Plant Viral Cell-To-Cell-Movement Protein. mBio 2016, 7, e02052-15. [Google Scholar] [CrossRef] [PubMed]
- Yuan, C.; Lazarowitz, S.G.; Citovsky, V. The Plasmodesmal Localization Signal of TMV MP Is Recognized by Plant Synaptotagmin SYTA. mBio 2018, 9, e01314-18. [Google Scholar] [CrossRef]
- Christensen, N.; Tilsner, J.; Bell, K.; Hammann, P.; Parton, R.; Lacomme, C.; Oparka, K. The 5′ Cap of Tobacco Mosaic Virus (TMV) Is Required for Virion Attachment to the Actin/Endoplasmic Reticulum Network during Early Infection. Traffic 2009, 10, 536–551. [Google Scholar] [CrossRef] [PubMed]
- Brill, L.M.; Nunn, R.S.; Kahn, T.W.; Yeager, M.; Beachy, R.N. Recombinant Tobacco Mosaic Virus Movement Protein Is an RNA-Binding, α-Helical Membrane Protein. Proc. Natl. Acad. Sci. USA 2000, 97, 7112–7117. [Google Scholar] [CrossRef] [PubMed]
- Fujiki, M.; Kawakami, S.; Kim, R.W.; Beachy, R.N. Domains of Tobacco Mosaic Virus Movement Protein Essential for Its Membrane Association. J. Gen. Virol. 2006, 87, 2699–2707. [Google Scholar] [CrossRef]
- Peiró, A.; Martínez-Gil, L.; Tamborero, S.; Pallás, V.; Sánchez-Navarro, J.A.; Mingarro, I. The Tobacco Mosaic Virus Movement Protein Associates with but Does Not Integrate into Biological Membranes. J. Virol. 2014, 88, 3016–3026. [Google Scholar] [CrossRef]
- Kawamura-Nagaya, K.; Ishibashi, K.; Huang, Y.-P.; Miyashita, S.; Ishikawa, M. Replication Protein of Tobacco Mosaic Virus Cotranslationally Binds the 5′ Untranslated Region of Genomic RNA to Enable Viral Replication. Proc. Natl. Acad. Sci. USA 2014, 111, E1620-8. [Google Scholar] [CrossRef]
- Morozov, S.Y.; Dolja, V.V.; Atabekov, J.G. Probable Reassortment of Genomic Elements among Elongated RNA-Containing Plant Viruses. J. Mol. Evol. 1989, 29, 52–62. [Google Scholar] [CrossRef]
- Morozov, S.Y.; Solovyev, A.G. Triple Gene Block: Modular Design of a Multifunctional Machine for Plant Virus Movement. J. Gen. Virol. 2003, 84, 1351–1366. [Google Scholar] [CrossRef]
- Morozov, S.Y.; Solovyev, A.G. Did Silencing Suppression Counter-Defensive Strategy Contribute to Origin and Evolution of the Triple Gene Block Coding for Plant Virus Movement Proteins? Front. Plant Sci. 2012, 3, 136. [Google Scholar] [CrossRef]
- Koonin, E.V.; Dolja, V.V. Evolution and Taxonomy of Positive-Strand RNA Viruses: Implications of Comparative Analysis of Amino Acid Sequences. Crit. Rev. Biochem. Mol. Biol. 1993, 28, 375–430. [Google Scholar] [CrossRef]
- Kalinina, N.O.; Rakitina, D.V.; Solovyev, A.G.; Schiemann, J.; Morozov, S.Y. RNA Helicase Activity of the Plant Virus Movement Proteins Encoded by the First Gene of the Triple Gene Block. Virology 2002, 296, 321–329. [Google Scholar] [CrossRef] [PubMed]
- Zamyatnin, A.A.; Solovyev, A.G.; Savenkov, E.I.; Germundsson, A.; Sandgren, M.; Valkonen, J.P.T.; Morozov, S.Y. Transient Coexpression of Individual Genes Encoded by the Triple Gene Block of Potato Mop-Top Virus Reveals Requirements for TGBp1 Trafficking. Mol. Plant. Microbe. Interact. 2004, 17, 921–930. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Zhang, K.; Li, Z.; Li, Z.; Yang, M.; Jin, X.; Cao, Q.; Wang, X.; Yue, N.; Li, D.; et al. The Barley Stripe Mosaic Virus Γb Protein Promotes Viral Cell-to-Cell Movement by Enhancing ATPase-Mediated Assembly of Ribonucleoprotein Movement Complexes. PLoS Pathog. 2020, 16, e1008709. [Google Scholar] [CrossRef] [PubMed]
- Donald, R.G.; Lawrence, D.M.; Jackson, A.O. The Barley Stripe Mosaic Virus 58-Kilodalton Beta(b) Protein Is a Multifunctional RNA Binding Protein. J. Virol. 1997, 71, 1538–1546. [Google Scholar] [CrossRef] [PubMed]
- Kalinina, N.O.; Rakitina, D.A.; Yelina, N.E.; Zamyatnin, A.A.; Stroganova, T.A.; Klinov, D.V.; Prokhorov, V.V.; Ustinova, S.V.; Chernov, B.K.; Schiemann, J.; et al. RNA-Binding Properties of the 63 KDa Protein Encoded by the Triple Gene Block of Poa Semilatent Hordeivirus. J. Gen. Virol. 2001, 82, 2569–2578. [Google Scholar] [CrossRef]
- Bleykasten, C.; Gilmer, D.; Guilley, H.; Richards, K.E.; Jonard, G. Beet Necrotic Yellow Vein Virus 42 KDa Triple Gene Block Protein Binds Nucleic Acid in Vitro. J. Gen. Virol. 1996, 77, 889–897. [Google Scholar] [CrossRef] [PubMed]
- Cowan, G.H.; Lioliopoulou, F.; Ziegler, A.; Torrance, L. Subcellular Localisation, Protein Interactions, and RNA Binding of Potato Mop-Top Virus Triple Gene Block Proteins. Virology 2002, 298, 106–115. [Google Scholar] [CrossRef][Green Version]
- Bayne, E.H.; Rakitina, D.V.; Morozov, S.Y.; Baulcombe, D.C. Cell-to-Cell Movement of Potato Potexvirus X Is Dependent on Suppression of RNA Silencing. Plant J. 2005, 44, 471–482. [Google Scholar] [CrossRef]
- Lim, H.-S.; Vaira, A.M.; Reinsel, M.D.; Bae, H.; Bailey, B.; Domier, L.L.; Hammond, J. Pathogenicity of Alternanthera Mosaic Virus Is Affected by Determinants in RNA-Dependent RNA Polymerase and by Reduced Efficacy of Silencing Suppression in a Movement-Competent TGB1. J. Gen. Virol. 2010, 91, 277–287. [Google Scholar] [CrossRef]
- Solovyev, A.G.; Savenkov, E.I.; Grdzelishvili, V.Z.; Kalinina, N.O.; Morozov, S.Y.; Schiemann, J.; Atabekov, J.G. Movement of Hordeivirus Hybrids with Exchanges in the Triple Gene Block. Virology 1999, 253, 278–287. [Google Scholar] [CrossRef] [PubMed]
- Wright, K.M.; Cowan, G.H.; Lukhovitskaya, N.I.; Tilsner, J.; Roberts, A.G.; Savenkov, E.I.; Torrance, L. The N-Terminal Domain of PMTV TGB1 Movement Protein Is Required for Nucleolar Localization, Microtubule Association, and Long-Distance Movement. Mol. Plant-Microbe Interact. 2010, 23, 1486–1497. [Google Scholar] [CrossRef] [PubMed]
- Solovyev, A.G.; Makarov, V.V. Helical Capsids of Plant Viruses: Architecture with Structural Lability. J. Gen. Virol. 2016, 97, 1739–1754. [Google Scholar] [CrossRef] [PubMed]
- Atabekov, J.G.; Rodionova, N.P.; Karpova, O.V.; Kozlovsky, S.V.; Poljakov, V.Y. The Movement Protein-Triggered in Situ Conversion of Potato Virus X Virion RNA from a Nontranslatable into a Translatable Form. Virology 2000, 271, 259–263. [Google Scholar] [CrossRef]
- Lim, H.-S.; Bragg, J.N.; Ganesan, U.; Lawrence, D.M.; Yu, J.; Isogai, M.; Hammond, J.; Jackson, A.O. Triple Gene Block Protein Interactions Involved in Movement of Barley Stripe Mosaic Virus. J. Virol. 2008, 82, 4991–5006. [Google Scholar] [CrossRef] [PubMed]
- Jackson, A.O.; Lim, H.-S.; Bragg, J.; Ganesan, U.; Lee, M.Y. Hordeivirus Replication, Movement, and Pathogenesis. Annu. Rev. Phytopathol. 2009, 47, 385–422. [Google Scholar] [CrossRef]
- Mitra, R.; Krishnamurthy, K.; Blancaflor, E.; Payton, M.; Nelson, R.S.; Verchot-Lubicz, J. The Potato Virus x TGBp2 Protein Association with the Endoplasmic Reticulum Plays a Role in but Is Not Sufficient for Viral Cell-to-Cell Movement. Virology 2003, 312, 35–48. [Google Scholar] [CrossRef]
- Krishnamurthy, K.; Heppler, M.; Mitra, R.; Blancaflor, E.; Payton, M.; Nelson, R.S.; Verchot-Lubicz, J. The Potato Virus X TGBp3 Protein Associates with the ER Network for Virus Cell-to-Cell Movement. Virology 2003, 309, 135–151. [Google Scholar] [CrossRef]
- Zamyatnin, A.A., Jr.; Solovyev, A.G.; Bozhkov, P.V.; Valkonen, J.P.T.; Morozov, S.Y.; Savenkov, E.I. Assessment of the Integral Membrane Protein Topology in Living Cells. Plant J. 2006, 46, 145–154. [Google Scholar] [CrossRef] [PubMed]
- Hsu, H.-T.; Chou, Y.-L.; Tseng, Y.-H.; Lin, Y.-H.; Lin, T.-M.; Lin, N.-S.; Hsu, Y.-H.; Chang, B.-Y. Topological Properties of the Triple Gene Block Protein 2 of Bamboo Mosaic Virus. Virology 2008, 379, 1–9. [Google Scholar] [CrossRef]
- Wu, X.; Liu, J.; Chai, M.; Wang, J.; Li, D.; Wang, A.; Cheng, X. The Potato Virus X TGBp2 Protein Plays Dual Functional Roles. J. Virol. 2019, 93, e01635-18. [Google Scholar] [CrossRef] [PubMed]
- Morozov, S.; Solovyev, A. Small Hydrophobic Viral Proteins Involved in Intercellular Movement of Diverse Plant Virus Genomes. AIMS Microbiol. 2020, 6, 305–329. [Google Scholar] [CrossRef] [PubMed]
- Verchot-Lubicz, J.; Torrance, L.; Solovyev, A.G.; Morozov, S.Y.; Jackson, A.O.; Gilmer, D. Varied Movement Strategies Employed by Triple Gene Block-Encoding Viruses. Mol. Plant-Microbe Interact. 2010, 23, 1231–1247. [Google Scholar] [CrossRef] [PubMed]
- Tilsner, J.; Cowan, G.H.; Roberts, A.G.; Chapman, S.N.; Ziegler, A.; Savenkov, E.; Torrance, L. Plasmodesmal Targeting and Intercellular Movement of Potato Mop-Top Pomovirus Is Mediated by a Membrane Anchored Tyrosine-Based Motif on the Lumenal Side of the Endoplasmic Reticulum and the C-Terminal Transmembrane Domain in the TGB3 Movement Protein. Virology 2010, 402, 41–51. [Google Scholar] [CrossRef]
- Shemyakina, E.A.; Erokhina, T.N.; Gorshkova, E.N.; Schiemann, J.; Solovyev, A.G.; Morozov, S.Y. Formation of Protein Complexes Containing Plant Virus Movement Protein TGBp3 Is Necessary for Its Intracellular Trafficking. Biochimie 2011, 93, 742–748. [Google Scholar] [CrossRef]
- Schepetilnikov, M.V.; Solovyev, A.G.; Gorshkova, E.N.; Schiemann, J.; Prokhnevsky, A.I.; Dolja, V.V.; Morozov, S.Y. Intracellular Targeting of a Hordeiviral Membrane-Spanning Movement Protein: Sequence Requirements and Involvement of an Unconventional Mechanism. J. Virol. 2008, 82, 1284–1293. [Google Scholar] [CrossRef]
- Lim, H.-S.; Bragg, J.N.; Ganesan, U.; Ruzin, S.; Schichnes, D.; Lee, M.Y.; Vaira, A.M.; Ryu, K.H.; Hammond, J.; Jackson, A.O. Subcellular Localization of the Barley Stripe Mosaic Virus Triple Gene Block Proteins. J. Virol. 2009, 83, 9432–9448. [Google Scholar] [CrossRef]
- Tilsner, J.; Linnik, O.; Louveaux, M.; Roberts, I.M.; Chapman, S.N.; Oparka, K.J. Replication and Trafficking of a Plant Virus Are Coupled at the Entrances of Plasmodesmata. J. Cell Biol. 2013, 201, 981–995. [Google Scholar] [CrossRef]
- Howard, A.R.; Heppler, M.L.; Ju, H.-J.; Krishnamurthy, K.; Payton, M.E.; Verchot-Lubicz, J. Potato Virus X TGBp1 Induces Plasmodesmata Gating and Moves between Cells in Several Host Species Whereas CP Moves Only in N. Benthamiana Leaves. Virology 2004, 328, 185–197. [Google Scholar] [CrossRef]
- Angell, S.M.; Davies, C.; Baulcombe, D.C. Cell-to-Cell Movement of Potato Virus X Is Associated with a Change in the Size-Exclusion Limit of Plasmodesmata in Trichome Cells of Nicotiana Clevelandii. Virology 1996, 216, 197–201. [Google Scholar] [CrossRef]
- Lough, T.J.; Shash, K.; Xoconostle-Cázares, B.; Hofstra, K.R.; Beck, D.L.; Balmori, E.; Forster, R.L.S.; Lucas, W.J. Molecular Dissection of the Mechanism by Which Potexvirus Triple Gene Block Proteins Mediate Cell-to-Cell Transport of Infectious RNA. Mol. Plant-Microbe Interact. 1998, 11, 801–814. [Google Scholar] [CrossRef]
- Tamai, A.; Meshi, T. Cell-to-Cell Movement of Potato Virus X: The Role of P12 and P8 Encoded by the Second and Third Open Reading Frames of the Triple Gene Block. Mol. Plant-Microbe Interact. 2001, 14, 1158–1167. [Google Scholar] [CrossRef] [PubMed]
- Fridborg, I.; Grainger, J.; Page, A.; Coleman, M.; Findlay, K.; Angell, S. TIP, a Novel Host Factor Linking Callose Degradation with the Cell-to-Cell Movement of Potato Virus X. Mol. Plant. Microbe. Interact. 2003, 16, 132–140. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Shiraishi, T.; Hagiwara-Komoda, Y.; Komatsu, K.; Maejima, K.; Okano, Y.; Fujimoto, Y.; Yusa, A.; Yamaji, Y.; Namba, S. The Plant Noncanonical Antiviral Resistance Protein JAX1 Inhibits Potexviral Replication by Targeting the Viral RNA-Dependent RNA Polymerase. J. Virol. 2019, 93, e01506-18. [Google Scholar] [CrossRef]
- Komatsu, K.; Sasaki, N.; Yoshida, T.; Suzuki, K.; Masujima, Y.; Hashimoto, M.; Watanabe, S.; Tochio, N.; Kigawa, T.; Yamaji, Y.; et al. Identification of a Proline-Kinked Amphipathic α-Helix Downstream from the Methyltransferase Domain of a Potexvirus Replicase and Its Role in Virus Replication and Perinuclear Complex Formation. J. Virol. 2021, 95, e0190620. [Google Scholar] [CrossRef]
- Wu, X.; Cheng, X. Intercellular Movement of Plant RNA Viruses: Targeting Replication Complexes to the Plasmodesma for Both Accuracy and Efficiency. Traffic 2020, 21, 725–736. [Google Scholar] [CrossRef]
- Tilsner, J.; Linnik, O.; Wright, K.M.; Bell, K.; Roberts, A.G.; Lacomme, C.; Santa Cruz, S.; Oparka, K.J. The TGB1 Movement Protein of Potato Virus X Reorganizes Actin and Endomembranes into the X-Body, a Viral Replication Factory. Plant Physiol. 2012, 158, 1359–1370. [Google Scholar] [CrossRef]
- Linnik, O.; Liesche, J.; Tilsner, J.; Oparka, K.J. Unraveling the Structure of Viral Replication Complexes at Super-Resolution. Front. Plant Sci. 2013, 4, 6. [Google Scholar] [CrossRef]
- Ju, H.-J.; Samuels, T.D.; Wang, Y.-S.; Blancaflor, E.; Payton, M.; Mitra, R.; Krishnamurthy, K.; Nelson, R.S.; Verchot-Lubicz, J. The Potato Virus X TGBp2 Movement Protein Associates with Endoplasmic Reticulum-Derived Vesicles during Virus Infection. Plant Physiol. 2005, 138, 1877–1895. [Google Scholar] [CrossRef]
- Samuels, T.D.; Ju, H.-J.; Ye, C.-M.; Motes, C.M.; Blancaflor, E.B.; Verchot-Lubicz, J. Subcellular Targeting and Interactions among the Potato Virus X TGB Proteins. Virology 2007, 367, 375–389. [Google Scholar] [CrossRef]
- Harries, P.; Park, J.-W.; Sasaki, N.; Ballard, K.D.; Maule, A.J.; Nelson, R.S. Differing Requirements for Actin and Myosin by Plant Viruses for Sustained Intercellular Movement. Proc. Natl. Acad. Sci. USA 2009, 106, 17594–17599. [Google Scholar] [CrossRef] [PubMed]
- Waigmann, E.; Ueki, S.; Trutnyeva, K.; Citovsky, V. The Ins and Outs of Nondestructive Cell-to-Cell and Systemic Movement of Plant Viruses. CRC. Crit. Rev. Plant Sci. 2004, 23, 195–250. [Google Scholar] [CrossRef]
- Melzer, M.J.; Sether, D.M.; Borth, W.B.; Hu, J.S. Characterization of a Virus Infecting Citrus Volkameriana with Citrus Leprosis-like Symptoms. Phytopathology 2012, 102, 122–127. [Google Scholar] [CrossRef] [PubMed]
- Morozov, S.Y.; Solovyev, A.G. Phylogenetic Relationship of Some “Accessory” Helicases of Plant Positive-Stranded RNA Viruses: Toward Understanding the Evolution of Triple Gene Block. Front. Microbiol. 2015, 6, 508. [Google Scholar] [CrossRef]
- Lazareva, E.A.; Lezzhov, A.A.; Komarova, T.V.; Morozov, S.Y.; Heinlein, M.; Solovyev, A.G. A Novel Block of Plant Virus Movement Genes. Mol. Plant Pathol. 2017, 18, 611–624. [Google Scholar] [CrossRef]
- Solovyev, A.G.; Morozov, S.Y. Non-Replicative Integral Membrane Proteins Encoded by Plant Alpha-like Viruses: Emergence of Diverse Orphan ORFs and Movement Protein Genes. Front. Plant Sci. 2017, 8, 1820. [Google Scholar] [CrossRef]
- Lazareva, E.A.; Lezzhov, A.A.; Golyshev, S.A.; Morozov, S.Y.; Heinlein, M.; Solovyev, A.G. Similarities in Intracellular Transport of Plant Viral Movement Proteins BMB2 and TGB3. J. Gen. Virol. 2017, 98, 2379–2391. [Google Scholar] [CrossRef]
- Hawes, C.; Kiviniemi, P.; Kriechbaumer, V. The Endoplasmic Reticulum: A Dynamic and Well-Connected Organelle. J. Integr. Plant Biol. 2015, 57, 50–62. [Google Scholar] [CrossRef]
- Kriechbaumer, V.; Brandizzi, F. The Plant Endoplasmic Reticulum: An Organized Chaos of Tubules and Sheets with Multiple Functions. J. Microsc. 2020, 280, 122–133. [Google Scholar] [CrossRef]
- Hu, J.; Shibata, Y.; Voss, C.; Shemesh, T.; Li, Z.; Coughlin, M.; Kozlov, M.M.; Rapoport, T.; Prinz, W. a Membrane Proteins of the Endoplasmic Reticulum Induce High-Curvature Tubules. Science 2008, 319, 1247–1250. [Google Scholar] [CrossRef]
- Sparkes, I.; Tolley, N.; Aller, I.; Svozil, J.; Osterrieder, A.; Botchway, S.; Mueller, C.; Frigerio, L.; Hawes, C. Five Arabidopsis Reticulon Isoforms Share Endoplasmic Reticulum Location, Topology, and Membrane-Shaping Properties. Plant Cell 2010, 22, 1333–1343. [Google Scholar] [CrossRef] [PubMed]
- Shibata, Y.; Voss, C.; Rist, J.M.; Hu, J.; Rapoport, T.A.; Prinz, W.A.; Voeltz, G.K. The Reticulon and DP1/Yop1p Proteins Form Immobile Oligomers in the Tubular Endoplasmic Reticulum. J. Biol. Chem. 2008, 283, 18892–18904. [Google Scholar] [CrossRef] [PubMed]
- Lazareva, E.A.; Lezzhov, A.A.; Chergintsev, D.A.; Golyshev, S.A.; Dolja, V.V.; Morozov, S.Y.; Heinlein, M.; Solovyev, A.G. Reticulon-like Properties of a Plant Virus-Encoded Movement Protein. New Phytol. 2021, 229, 1052–1066. [Google Scholar] [CrossRef] [PubMed]
- Diaz, A.; Ahlquist, P. Role of Host Reticulon Proteins in Rearranging Membranes for Positive-Strand RNA Virus Replication. Curr. Opin. Microbiol. 2012, 15, 519–524. [Google Scholar] [CrossRef] [PubMed]
- Kriechbaumer, V.; Botchway, S.W.; Slade, S.E.; Knox, K.; Frigerio, L.; Oparka, K.; Hawes, C. Reticulomics: Protein-Protein Interaction Studies with Two Plasmodesmata-Localized Reticulon Family Proteins Identify Binding Partners Enriched at Plasmodesmata, Endoplasmic Reticulum, and the Plasma Membrane. Plant Physiol. 2015, 169, 1933–1945. [Google Scholar] [CrossRef]
- Knox, K.; Wang, P.; Kriechbaumer, V.; Tilsner, J.; Frigerio, L.; Sparkes, I.; Hawes, C.; Oparka, K. Putting the Squeeze on Plasmodesmata: A Role for Reticulons in Primary Plasmodesmata Formation. Plant Physiol. 2015, 168, 1563–1572. [Google Scholar] [CrossRef]
- Revers, F.; García, J.A. Molecular Biology of Potyviruses. Adv. Virus Res. 2015, 92, 101–199. [Google Scholar] [CrossRef]
- Vijayapalani, P.; Maeshima, M.; Nagasaki-Takekuchi, N.; Miller, W.A. Interaction of the Trans-Frame Potyvirus Protein P3N-PIPO with Host Protein PCaP1 Facilitates Potyvirus Movement. PLoS Pathog. 2012, 8, e1002639. [Google Scholar] [CrossRef]
- Chung, B.Y.-W.; Miller, W.A.; Atkins, J.F.; Firth, A.E. An Overlapping Essential Gene in the Potyviridae. Proc. Natl. Acad. Sci. USA 2008, 105, 5897–5902. [Google Scholar] [CrossRef]
- Seo, J.-K.; Vo Phan, M.S.; Kang, S.-H.; Choi, H.-S.; Kim, K.-H. The Charged Residues in the Surface-Exposed C-Terminus of the Soybean Mosaic Virus Coat Protein Are Critical for Cell-to-Cell Movement. Virology 2013, 446, 95–101. [Google Scholar] [CrossRef]
- Dolja, V.V.; Haldeman, R.; Robertson, N.L.; Dougherty, W.G.; Carrington, J.C. Distinct Functions of Capsid Protein in Assembly and Movement of Tobacco Etch Potyvirus in Plants. EMBO J. 1994, 13, 1482–1491. [Google Scholar] [CrossRef]
- Dolja, V.V.; Haldeman-Cahill, R.; Montgomery, A.E.; Vandenbosch, K.A.; Carrington, J.C. Capsid Protein Determinants Involved in Cell-to-Cell and Long Distance Movement of Tobacco Etch Potyvirus. Virology 1995, 206, 1007–1016. [Google Scholar] [CrossRef] [PubMed]
- Roberts, I.M.; Wang, D.; Findlay, K.; Maule, A.J. Ultrastructural and Temporal Observations of the Potyvirus Cylindrical Inclusions (CIs) Show That the CI Protein Acts Transiently in Aiding Virus Movement. Virology 1998, 245, 173–181. [Google Scholar] [CrossRef]
- Rodríguez-Cerezo, E.; Findlay, K.; Shaw, J.G.; Lomonossoff, G.P.; Qiu, S.G.; Linstead, P.; Shanks, M.; Risco, C. The Coat and Cylindrical Inclusion Proteins of a Potyvirus Are Associated with Connections between Plant Cells. Virology 1997, 236, 296–306. [Google Scholar] [CrossRef]
- Wang, A. Cell-to-Cell Movement of Plant Viruses via Plasmodesmata: A Current Perspective on Potyviruses. Curr. Opin. Virol. 2021, 48, 10–16. [Google Scholar] [CrossRef]
- Taliansky, M.; Torrance, L.; Kalinina, N.O. Role of Plant Virus Movement Proteins. Methods Mol. Biol. 2008, 451, 33–54. [Google Scholar] [CrossRef] [PubMed]
- Carrington, J.C.; Jensen, P.E.; Schaad, M.C. Genetic Evidence for an Essential Role for Potyvirus CI Protein in Cell-to-Cell Movement. Plant J. 1998, 14, 393–400. [Google Scholar] [CrossRef] [PubMed]
- Fernández, A.; Guo, H.S.; Sáenz, P.; Simón-Buela, L.; Gómez de Cedrón, M.; García, J.A. The Motif V of Plum Pox Potyvirus CI RNA Helicase Is Involved in NTP Hydrolysis and Is Essential for Virus RNA Replication. Nucleic Acids Res. 1997, 25, 4474–4480. [Google Scholar] [CrossRef]
- Gómez de Cedrón, M.; Osaba, L.; López, L.; García, J.A. Genetic Analysis of the Function of the Plum Pox Virus CI RNA Helicase in Virus Movement. Virus Res. 2006, 116, 136–145. [Google Scholar] [CrossRef]
- López, L.; Urzainqui, A.; Domínguez, E.; García, J.A. Identification of an N-Terminal Domain of the Plum Pox Potyvirus CI RNA Helicase Involved in Self-Interaction in a Yeast Two-Hybrid System. J. Gen. Virol. 2001, 82, 677–686. [Google Scholar] [CrossRef]
- Wei, T.; Zhang, C.; Hong, J.; Xiong, R.; Kasschau, K.D.; Zhou, X.; Carrington, J.C.; Wang, A. Formation of Complexes at Plasmodesmata for Potyvirus Intercellular Movement Is Mediated by the Viral Protein P3N-PIPO. PLoS Pathog. 2010, 6, e1000962. [Google Scholar] [CrossRef] [PubMed]
- Yambao, M.L.M.; Masuta, C.; Nakahara, K.; Uyeda, I. The Central and C-Terminal Domains of VPg of Clover Yellow Vein Virus Are Important for VPg–HCPro and VPg–VPg Interactions. J. Gen. Virol. 2003, 84, 2861–2869. [Google Scholar] [CrossRef] [PubMed]
- Guo, D.; Rajamäki, M.-L.; Saarma, M.; Valkonen, J.P.T. Towards a Protein Interaction Map of Potyviruses: Protein Interaction Matrixes of Two Potyviruses Based on the Yeast Two-Hybrid System. J. Gen. Virol. 2001, 82, 935–939. [Google Scholar] [CrossRef]
- Dunoyer, P.; Thomas, C.; Harrison, S.; Revers, F.; Maule, A. A Cysteine-Rich Plant Protein Potentiates Potyvirus Movement through an Interaction with the Virus Genome-Linked Protein VPg. J. Virol. 2004, 78, 2301–2309. [Google Scholar] [CrossRef] [PubMed]
- Torrance, L.; Andreev, I.A.; Gabrenaite-Verhovskaya, R.; Cowan, G.; Mäkinen, K.; Taliansky, M.E. An Unusual Structure at One End of Potato Potyvirus Particles. J. Mol. Biol. 2006, 357, 1–8. [Google Scholar] [CrossRef]
- Gabrenaite-Verkhovskaya, R.; Andreev, I.A.; Kalinina, N.O.; Torrance, L.; Taliansky, M.E.; Mäkinen, K. Cylindrical Inclusion Protein of Potato Virus A Is Associated with a Subpopulation of Particles Isolated from Infected Plants. J. Gen. Virol. 2008, 89, 829–838. [Google Scholar] [CrossRef]
- Rojas, M.R.; Zerbini, F.M.; Allison, R.F.; Gilbertson, R.L.; Lucas, W.J. Capsid Protein and Helper Component-Proteinase Function as Potyvirus Cell-to-Cell Movement Proteins. Virology 1997, 237, 283–295. [Google Scholar] [CrossRef]
- Salonen, A.; Ahola, T.; Kääriäinen, L. Viral RNA Replication in Association with Cellular Membranes. Curr. Top. Microbiol. Immunol. 2004, 285, 139–173. [Google Scholar] [CrossRef]
- Gushchin, V.A.; Solovyev, A.G.; Erokhina, T.N.; Morozov, S.Y.; Agranovsky, A.A. Beet Yellows Virus Replicase and Replicative Compartments: Parallels with Other RNA Viruses. Front. Microbiol. 2013, 4, 38. [Google Scholar] [CrossRef]
- Schaad, M.C.; Jensen, P.E.; Carrington, J.C. Formation of Plant RNA Virus Replication Complexes on Membranes: Role of an Endoplasmic Reticulum-Targeted Viral Protein. EMBO J. 1997, 16, 4049–4059. [Google Scholar] [CrossRef]
- Wan, J.; Basu, K.; Mui, J.; Vali, H.; Zheng, H.; Laliberté, J.-F. Ultrastructural Characterization of Turnip Mosaic Virus-Induced Cellular Rearrangements Reveals Membrane-Bound Viral Particles Accumulating in Vacuoles. J. Virol. 2015, 89, 12441–12456. [Google Scholar] [CrossRef]
- Cotton, S.; Grangeon, R.; Thivierge, K.; Mathieu, I.; Ide, C.; Wei, T.; Wang, A.; Laliberté, J.-F. Turnip Mosaic Virus RNA Replication Complex Vesicles Are Mobile, Align with Microfilaments, and Are Each Derived from a Single Viral Genome. J. Virol. 2009, 83, 10460–10471. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; Shi, Y.; Dai, Z.; Wang, A. The RNA-Dependent RNA Polymerase NIb of Potyviruses Plays Multifunctional, Contrasting Roles during Viral Infection. Viruses 2020, 12, 77. [Google Scholar] [CrossRef] [PubMed]
- Grangeon, R.; Jiang, J.; Laliberté, J.-F. Host Endomembrane Recruitment for Plant RNA Virus Replication. Curr. Opin. Virol. 2012, 2, 683–690. [Google Scholar] [CrossRef] [PubMed]
- Grangeon, R.; Agbeci, M.; Chen, J.; Grondin, G.; Zheng, H.; Laliberté, J.-F. Impact on the Endoplasmic Reticulum and Golgi Apparatus of Turnip Mosaic Virus Infection. J. Virol. 2012, 86, 9255–9265. [Google Scholar] [CrossRef]
- Wei, T.; Wang, A. Biogenesis of Cytoplasmic Membranous Vesicles for Plant Potyvirus Replication Occurs at Endoplasmic Reticulum Exit Sites in a COPI- and COPII-Dependent Manner. J. Virol. 2008, 82, 12252–12264. [Google Scholar] [CrossRef]
- Lerich, A.; Langhans, M.; Sturm, S.; Robinson, D.G. Is the 6 KDa Tobacco Etch Viral Protein a Bona Fide ERES Marker? J. Exp. Bot. 2011, 62, 5013–5023. [Google Scholar] [CrossRef]
- Jiang, J.; Patarroyo, C.; Garcia Cabanillas, D.; Zheng, H.; Laliberté, J.-F. The Vesicle-Forming 6K2 Protein of Turnip Mosaic Virus Interacts with the COPII Coatomer Sec24a for Viral Systemic Infection. J. Virol. 2015, 89, 6695–6710. [Google Scholar] [CrossRef]
- Agbeci, M.; Grangeon, R.; Nelson, R.S.; Zheng, H.; Laliberté, J.-F. Contribution of Host Intracellular Transport Machineries to Intercellular Movement of Turnip Mosaic Virus. PLoS Pathog. 2013, 9, e1003683. [Google Scholar] [CrossRef]
- Peremyslov, V.V.; Klocko, A.L.; Fowler, J.E.; Dolja, V.V. Arabidopsis Myosin XI-K Localizes to the Motile Endomembrane Vesicles Associated with F-Actin. Front. Plant Sci. 2012, 3, 184. [Google Scholar] [CrossRef]
- Grangeon, R.; Jiang, J.; Wan, J.; Agbeci, M.; Zheng, H.; Laliberté, J.-F. 6K2-Induced Vesicles Can Move Cell to Cell during Turnip Mosaic Virus Infection. Front. Microbiol. 2013, 4, 351. [Google Scholar] [CrossRef] [PubMed]
- Kawakami, S.; Watanabe, Y.; Beachy, R.N. Tobacco Mosaic Virus Infection Spreads Cell to Cell as Intact Replication Complexes. Proc. Natl. Acad. Sci. USA 2004, 101, 6291–6296. [Google Scholar] [CrossRef]
- Cabanillas, D.G.; Jiang, J.; Movahed, N.; Germain, H.; Yamaji, Y.; Zheng, H.; Laliberté, J.-F. Turnip Mosaic Virus Uses the SNARE Protein VTI11 in an Unconventional Route for Replication Vesicle Trafficking. Plant Cell 2018, 30, 2594–2615. [Google Scholar] [CrossRef] [PubMed]
- Movahed, N.; Sun, J.; Vali, H.; Laliberté, J.-F.; Zheng, H. A Host ER Fusogen Is Recruited by Turnip Mosaic Virus for Maturation of Viral Replication Vesicles. Plant Physiol. 2019, 179, 507–518. [Google Scholar] [CrossRef] [PubMed]
- Ito, E.; Uemura, T. RAB GTPases and SNAREs at the Trans-Golgi Network in Plants. J. Plant Res. 2022, 135, 389–403. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Jia, Z.; Ding, K.; Zheng, H.; Lu, Y.; Lin, L.; Peng, J.; Rao, S.; Wang, A.; Chen, J.; et al. Turnip Mosaic Virus Co-Opts the Vacuolar Sorting Receptor VSR4 to Promote Viral Genome Replication in Plants by Targeting Viral Replication Vesicles to the Endosome. PLOS Pathog. 2022, 18, e1010257. [Google Scholar] [CrossRef]
- Agaoua, A.; Rittener, V.; Troadec, C.; Desbiez, C.; Bendahmane, A.; Moquet, F.; Dogimont, C. A Single Substitution in Vacuolar Protein Sorting 4 Is Responsible for Resistance to Watermelon Mosaic Virus in Melon. J. Exp. Bot. 2022, 73, 4008–4021. [Google Scholar] [CrossRef]
- Movahed, N.; Patarroyo, C.; Sun, J.; Vali, H.; Laliberté, J.-F.; Zheng, H. Cylindrical Inclusion Protein of Turnip Mosaic Virus Serves as a Docking Point for the Intercellular Movement of Viral Replication Vesicles. Plant Physiol. 2017, 175, 1732–1744. [Google Scholar] [CrossRef]
- Chin, A.; Lécuyer, E. RNA Localization: Making Its Way to the Center Stage. Biochim. Biophys. Acta—Gen. Subj. 2017, 1861, 2956–2970. [Google Scholar] [CrossRef] [PubMed]
- Zappulo, A.; van den Bruck, D.; Ciolli Mattioli, C.; Franke, V.; Imami, K.; McShane, E.; Moreno-Estelles, M.; Calviello, L.; Filipchyk, A.; Peguero-Sanchez, E.; et al. RNA Localization Is a Key Determinant of Neurite-Enriched Proteome. Nat. Commun. 2017, 8, 583. [Google Scholar] [CrossRef]
- Haag, C.; Steuten, B.; Feldbrügge, M. Membrane-Coupled MRNA Trafficking in Fungi. Annu. Rev. Microbiol. 2015, 69, 265–281. [Google Scholar] [CrossRef] [PubMed]
- Chou, H.-L.; Tian, L.; Washida, H.; Fukuda, M.; Kumamaru, T.; Okita, T.W. The Rice Storage Protein MRNAs as a Model System for RNA Localization in Higher Plants. Plant Sci. 2019, 284, 203–211. [Google Scholar] [CrossRef]
- Tian, L.; Chou, H.-L.; Zhang, L.; Hwang, S.-K.; Starkenburg, S.R.; Doroshenk, K.A.; Kumamaru, T.; Okita, T.W. RNA-Binding Protein RBP-P Is Required for Glutelin and Prolamine MRNA Localization in Rice Endosperm Cells. Plant Cell 2018, 30, 2529–2552. [Google Scholar] [CrossRef]
- Tian, L.; Chou, H.-L.; Fukuda, M.; Kumamaru, T.; Okita, T.W. MRNA Localization in Plant Cells. Plant Physiol. 2020, 182, 97–109. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.-B.; Wang, C.; Muench, D.G.; Ozawa, K.; Franceschi, V.R.; Wu, Y.; Okita, T.W. Messenger RNA Targeting of Rice Seed Storage Proteins to Specific ER Subdomains. Nature 2000, 407, 765–767. [Google Scholar] [CrossRef] [PubMed]
- Hamada, S.; Ishiyama, K.; Choi, S.-B.; Wang, C.; Singh, S.; Kawai, N.; Franceschi, V.R.; Okita, T.W. The Transport of Prolamine RNAs to Prolamine Protein Bodies in Living Rice Endosperm Cells. Plant Cell 2003, 15, 2253–2264. [Google Scholar] [CrossRef] [PubMed]
- Tian, L.; Doroshenk, K.A.; Zhang, L.; Fukuda, M.; Washida, H.; Kumamaru, T.; Okita, T. Zipcode RNA-Binding Proteins and Membrane Trafficking Proteins Cooperate to Transport Glutelin MRNAs in Rice Endosperm. Plant Cell 2020, 32, 2566–2581. [Google Scholar] [CrossRef]
- Müntjes, K.; Devan, S.K.; Reichert, A.S.; Feldbrügge, M. Linking Transport and Translation of MRNAs with Endosomes and Mitochondria. EMBO Rep. 2021, 22, e52445. [Google Scholar] [CrossRef]
- Olgeiser, L.; Haag, C.; Boerner, S.; Ule, J.; Busch, A.; Koepke, J.; König, J.; Feldbrügge, M.; Zarnack, K. The Key Protein of Endosomal MRNP Transport Rrm4 Binds Translational Landmark Sites of Cargo MRNAs. EMBO Rep. 2019, 20, e46588. [Google Scholar] [CrossRef]
- Pohlmann, T.; Baumann, S.; Haag, C.; Albrecht, M.; Feldbrügge, M. A FYVE Zinc Finger Domain Protein Specifically Links MRNA Transport to Endosome Trafficking. Elife 2015, 4, e06041. [Google Scholar] [CrossRef]
- Baumann, S.; Pohlmann, T.; Jungbluth, M.; Brachmann, A.; Feldbrügge, M. Kinesin-3 and Dynein Mediate Microtubule-Dependent Co-Transport of MRNPs and Endosomes. J. Cell Sci. 2012, 125, 2740–2752. [Google Scholar] [CrossRef]
- Baumann, S.; König, J.; Koepke, J.; Feldbrügge, M. Endosomal Transport of Septin mRNA and Protein Indicates Local Translation on Endosomes and Is Required for Correct Septin Filamentation. EMBO Rep. 2014, 15, 94–102. [Google Scholar] [CrossRef] [PubMed]
- Higuchi, Y.; Ashwin, P.; Roger, Y.; Steinberg, G. Early Endosome Motility Spatially Organizes Polysome Distribution. J. Cell Biol. 2014, 204, 343–357. [Google Scholar] [CrossRef] [PubMed]
- Sahoo, P.K.; Smith, D.S.; Perrone-Bizzozero, N.; Twiss, J.L. Axonal MRNA Transport and Translation at a Glance. J. Cell Sci. 2018, 131, jcs196808. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Yu, J.; Ji, S.-J. Axonal MRNA Localization and Translation: Local Events with Broad Roles. Cell. Mol. Life Sci. 2021, 78, 7379–7395. [Google Scholar] [CrossRef]
- Cioni, J.-M.; Lin, J.Q.; Holtermann, A.V.; Koppers, M.; Jakobs, M.A.H.; Azizi, A.; Turner-Bridger, B.; Shigeoka, T.; Franze, K.; Harris, W.A.; et al. Late Endosomes Act as MRNA Translation Platforms and Sustain Mitochondria in Axons. Cell 2019, 176, 56–72.e15. [Google Scholar] [CrossRef]
- Bassell, G.J.; Zhang, H.; Byrd, A.L.; Femino, A.M.; Singer, R.H.; Taneja, K.L.; Lifshitz, L.M.; Herman, I.M.; Kosik, K.S. Sorting of β-Actin MRNA and Protein to Neurites and Growth Cones in Culture. J. Neurosci. 1998, 18, 251–265. [Google Scholar] [CrossRef]
- Agrawal, M.; Welshhans, K. Local Translation Across Neural Development: A Focus on Radial Glial Cells, Axons, and Synaptogenesis. Front. Mol. Neurosci. 2021, 14, 717170. [Google Scholar] [CrossRef]
- Zhang, H.L.; Eom, T.; Oleynikov, Y.; Shenoy, S.M.; Liebelt, D.A.; Dictenberg, J.B.; Singer, R.H.; Bassell, G.J. Neurotrophin-Induced Transport of a β-Actin MRNP Complex Increases β-Actin Levels and Stimulates Growth Cone Motility. Neuron 2001, 31, 261–275. [Google Scholar] [CrossRef]
- Donnelly, C.J.; Park, M.; Spillane, M.; Yoo, S.; Pacheco, A.; Gomes, C.; Vuppalanchi, D.; McDonald, M.; Kim, H.H.; Merianda, T.T.; et al. Axonally Synthesized -Actin and GAP-43 Proteins Support Distinct Modes of Axonal Growth. J. Neurosci. 2013, 33, 3311–3322. [Google Scholar] [CrossRef]
- Quentin, D.; Schuhmacher, J.; Klink, B.; Lauer, J.; Shaikh, T.; Huis in t’t Veld, P.; Welp, L.; Urlaub, H.; Zerial, M.; Raunser, S. Structure of the Human FERRY Rab5 Effector Complex. bioRxiv 2021. [Google Scholar] [CrossRef]
- Navarro, J.A.; Pallás, V. An Update on the Intracellular and Intercellular Trafficking of Carmoviruses. Front. Plant Sci. 2017, 8, 1801. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).