To Fight or to Grow: The Balancing Role of Ethylene in Plant Abiotic Stress Responses
Abstract
:1. Introduction
2. Ethylene and JA
3. Ethylene and ABA
4. Ethylene and BR
5. Ethylene and Auxin
6. Ethylene and GA
7. Ethylene and SA
8. Ethylene and CK
9. Concluding Remarks and Future Perspectives
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, H.; Zhu, J.; Gong, Z.; Zhu, J. Abiotic stress responses in plants. Nat. Rev. Genet. 2021, 21, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Chapin, F.S. Integrated Responses of Plants to Stress. BioScience 1991, 41, 29–36. [Google Scholar] [CrossRef]
- Heil, M.; Walters, D.; Newton, A.; Lyon, G. Trade-offs associated with induced resistance. In Induced Resistance for Plant Defense; John Wiley & Sons: Chichester, UK, 2014; pp. 171–192. [Google Scholar]
- Neljubow, D. Uber die horizontale Nutation der Stengel von Pisum Sativum und einiger anderen Planzen. Bot. Centralbl. Beih. 1901, 10, 128–139. [Google Scholar]
- Merchante, C.; Stepanova, A.N. The Triple Response Assay and Its Use to Characterize Ethylene Mutants in Arabidopsis. Ethyl. Signal. 2017, 1573, 163–209. [Google Scholar] [CrossRef]
- Bleecker, A.B.; Kende, H. Ethylene: A Gaseous Signal Molecule in Plants. Annu. Rev. Cell Dev. Biol. 2000, 16, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Binder, B.M. Ethylene signaling in plants. J. Biol. Chem. 2020, 295, 7710–7725. [Google Scholar] [CrossRef] [Green Version]
- Adams, D.O.; Yang, S.F. Ethylene biosynthesis: Identification of 1-aminocyclopropane-1-carboxylic acid as an intermediate in the conversion of methionine to ethylene. Proc. Natl. Acad. Sci. USA 1979, 76, 170–174. [Google Scholar] [CrossRef] [Green Version]
- Boller, T.; Herner, R.C.; Kende, H. Assay for and enzymatic formation of an ethylene precursor, 1-aminocyclopropane-1-carboxylic acid. Planta 1979, 145, 293–303. [Google Scholar] [CrossRef]
- Hamilton, A.J.; Bouzayen, M.; Grierson, D. Identification of a tomato gene for the ethylene-forming enzyme by expression in yeast. Proc. Natl. Acad. Sci. USA 1991, 88, 7434–7437. [Google Scholar] [CrossRef] [Green Version]
- Ververidis, P.; John, P. Complete recovery in vitro of ethylene-forming enzyme activity. Phytochemistry 1991, 30, 725–727. [Google Scholar] [CrossRef]
- Kende, H. Ethylene biosynthesis. Annu. Rev. Plant Biol. 1993, 44, 283–307. [Google Scholar] [CrossRef]
- Xu, J.; Zhang, S. Ethylene Biosynthesis and Regulation in Plants. In Ethylene in Plants; Wen, C.-K., Ed.; Springer: Dordrecht, The Netherlands, 2015; pp. 1–25. ISBN 978-94-017-9483-1. [Google Scholar]
- Alonso, J.M.; Stepanova, A.N. The Ethylene Signaling Pathway. Science 2004, 306, 1513–1515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lacey, R.F.; Binder, B.M. How plants sense ethylene gas—The ethylene receptors. J. Inorg. Biochem. 2014, 133, 58–62. [Google Scholar] [CrossRef] [PubMed]
- Shakeel, S.N.; Gao, Z.; Amir, M.; Chen, Y.-F.; Rai, M.I.; Haq, N.U.; Schaller, G.E. Ethylene Regulates Levels of Ethylene Receptor/CTR1 Signaling Complexes in Arabidopsis thaliana. J. Biol. Chem. 2015, 290, 12415–12424. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Ma, M.; Feng, Y.; Li, H.; Wang, Y.; Ma, Y.; Li, M.; An, F.; Guo, H. EIN2-Directed Translational Regulation of Ethylene Signaling in Arabidopsis. Cell 2015, 163, 670–683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merchante, C.; Brumos, J.; Yun, J.; Hu, Q.; Spencer, K.R.; Enríquez, P.; Binder, B.M.; Heber, S.; Stepanova, A.N.; Alonso, J.M. Gene-Specific Translation Regulation Mediated by the Hormone-Signaling Molecule EIN2. Cell 2015, 163, 684–697. [Google Scholar] [CrossRef] [Green Version]
- Guo, H.; Ecker, J.R. Plant responses to ethylene gas are mediated by SCFEBF1/EBF2-dependent proteolysis of EIN3 transcription factor. Cell 2003, 115, 667–677. [Google Scholar] [CrossRef] [Green Version]
- Potuschak, T.; Lechner, E.; Parmentier, Y.; Yanagisawa, S.; Grava, S.; Koncz, C.; Genschik, P. EIN3-Dependent Regulation of Plant Ethylene Hormone Signaling by Two Arabidopsis F Box Proteins: EBF1 and EBF2. Cell 2003, 115, 679–689. [Google Scholar] [CrossRef] [Green Version]
- Dolgikh, V.A.; Pukhovaya, E.M.; Zemlyanskaya, E.V. Shaping ethylene response: The role of EIN3/EIL1 transcription factors. Front. Plant Sci. 2019, 10, 1030. [Google Scholar] [CrossRef] [Green Version]
- Debbarma, J.; Sarki, Y.N.; Saikia, B.; Boruah, H.P.D.; Singha, D.L.; Chikkaputtaiah, C. Ethylene Response Factor (ERF) Family Proteins in Abiotic Stresses and CRISPR-Cas9 Genome Editing of ERFs for Multiple Abiotic Stress Tolerance in Crop Plants: A Review. Mol. Biotechnol. 2019, 61, 153–172. [Google Scholar] [CrossRef]
- Dietz, K.; Vogel, M.O.; Viehhauser, A. AP2/EREBP transcription factors are part of gene regulatory networks and integrate metabolic, hormonal and environmental signals in stress acclimation and retrograde signalling. Protoplasma 2010, 245, 3–14. [Google Scholar] [CrossRef]
- Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta Gene Regul. Mech. 2012, 1819, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Chandler, J.W. Class VIIIb APETALA2 Ethylene Response Factors in Plant Development. Trends Plant. Sci. 2018, 23, 151–162. [Google Scholar] [CrossRef] [PubMed]
- Nakano, T.; Suzuki, K.; Fujimura, T.; Shinshi, H. Genome-Wide Analysis of the ERF Gene Family in Arabidopsis and Rice. Plant. Physiol. 2006, 140, 411–432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kagale, S.; Rozwadowski, K. EAR motif-mediated transcriptional repression in plants: An underlying mechanism for epigenetic regulation of gene expression. Epigenetics 2011, 6, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, S.B.; Belachew, A.; Ma, S.F.; Young, M.; Ade, J.; Shen, Y.; Marion, C.M.; Holtan, H.E.; Bailey, A.; Stone, J.K. The EDLL motif: A potent plant transcriptional activation domain from AP2/ERF transcription factors. Plant J. 2012, 70, 855–865. [Google Scholar] [CrossRef] [PubMed]
- Harkey, A.; Yoon, G.M.; Seo, D.H.; Delong, A.; Muday, G.K. Light Modulates Ethylene Synthesis, Signaling, and Downstream Transcriptional Networks to Control Plant Development. Front. Plant. Sci. 2019, 10, 1094. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pierik, R.; Sasidharan, R.; Voesenek, L.A.C.J. Growth Control by Ethylene: Adjusting Phenotypes to the Environment. J. Plant. Growth Regul. 2007, 26, 188–200. [Google Scholar] [CrossRef] [Green Version]
- Kazan, K. Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends Plant. Sci. 2015, 20, 219–229. [Google Scholar] [CrossRef]
- Kurepin, L.V.; Yeung, E.C.; Reid, D.M.; Pharis, R.P. Light Signaling Regulates Tulip Organ Growth and Ethylene Production in a Tissue-Specific Manner. Int. J. Plant. Sci. 2016, 177, 339–346. [Google Scholar] [CrossRef]
- Maksymiec, W. Signaling responses in plants to heavy metal stress. Acta Physiol. Plant. 2007, 29, 177–187. [Google Scholar] [CrossRef]
- Argueso, C.T.; Hansen, M.; Kieber, J.J. Regulation of Ethylene Biosynthesis. J. Plant. Growth Regul. 2007, 26, 92–105. [Google Scholar] [CrossRef]
- Zhao, M.; Liu, W.; Xia, X.; Wang, T.; Zhang, W. Cold acclimation-induced freezing tolerance of Medicago truncatula seedlings is negatively regulated by ethylene. Physiol. Plant. 2014, 152, 115–129. [Google Scholar] [CrossRef]
- Zhang, M.; Smith, J.A.C.; Harberd, N.P.; Jiang, C. The regulatory roles of ethylene and reactive oxygen species (ROS) in plant salt stress responses. Plant Mol. Biol. 2016, 91, 651–659. [Google Scholar] [CrossRef] [PubMed]
- Dubois, M.; Claeys, H.; Broeck, L.V.D.; Inzé, D. Time of day determines Arabidopsis transcriptome and growth dynamics under mild drought. Plant Cell Environ. 2017, 40, 180–189. [Google Scholar] [CrossRef] [Green Version]
- Baerenfaller, K.; Massonnet, C.; Walsh, S.; Baginsky, S.; Bühlmann, P.; Hennig, L.; Hirsch-Hoffmann, M.; Howell, K.; Kahlau, S.; Radziejwoski, A.; et al. Systems-based analysis of Arabidopsis leaf growth reveals adaptation to water deficit. Mol. Syst. Biol. 2012, 8, 606. [Google Scholar] [CrossRef]
- Clauw, P.; Coppens, F.; De Beuf, K.; Dhondt, S.; Van Daele, T.; Maleux, K.; Storme, V.; Clement, L.; Gonzalez, N.; Inzé, D. Leaf Responses to Mild Drought Stress in Natural Variants of Arabidopsis. Plant Physiol. 2015, 167, 800–816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, H.-P.; Lin, T.-Y.; Wang, N.N.; Shih, M.-C. Differential expression of genes encoding 1-aminocyclopropane-1-carboxylate synthase in Arabidopsis during hypoxia. Plant Mol. Biol. 2005, 58, 15–25. [Google Scholar] [CrossRef]
- Eun, H.-D.; Ali, S.; Jung, H.; Kim, K.; Kim, W.-C. Profiling of ACC synthase gene (ACS11) expression in Arabidopsis induced by abiotic stresses. Appl. Biol. Chem. 2019, 62, 42. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Ma, Q.; Wang, R.; Wang, P.; Liu, Y.; Mao, T. Submergence stress-induced hypocotyl elongation through ethylene signaling-mediated regulation of cortical microtubules in Arabidopsis. J. Exp. Bot. 2019, 71, 1067–1077. [Google Scholar] [CrossRef]
- Dong, H.; Zhen, Z.; Peng, J.; Chang, L.; Gong, Q.; Wang, N.N. Loss of ACS7 confers abiotic stress tolerance by modulating ABA sensitivity and accumulation in Arabidopsis. J. Exp. Bot. 2011, 62, 4875–4887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skirycz, A.; Claeys, H.; De Bodt, S.; Oikawa, A.; Shinoda, S.; Andriankaja, M.; Maleux, K.; Eloy, N.; Coppens, F.; Yoo, S.-D.; et al. Pause-and-Stop: The Effects of Osmotic Stress on Cell Proliferation during Early Leaf Development in Arabidopsis and a Role for Ethylene Signaling in Cell Cycle Arrest. Plant Cell 2011, 23, 1876–1888. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, X.-C.; Schaller, G. Effect of salt and osmotic stress upon expression of the ethylene receptor ETR1 inArabidopsis thaliana. FEBS Lett. 2004, 562, 189–192. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Liu, C.; Li, K.; Sun, F.; Hu, H.; Li, X.; Zhao, Y.; Han, C.; Zhang, W.; Duan, Y.; et al. Arabidopsis EIN2 modulates stress response through abscisic acid response pathway. Plant Mol. Biol. 2007, 64, 633–644. [Google Scholar] [CrossRef]
- Ahmadizadeh, M.; Chen, J.-T.; Hasanzadeh, S.; Ahmar, S.; Heidari, P. Insights into the genes involved in the ethylene biosynthesis pathway in Arabidopsis thaliana and Oryza sativa. J. Genet. Eng. Biotechnol. 2020, 18, 62. [Google Scholar] [CrossRef]
- Yan, Z.; Wang, J.; Wang, F.; Xie, C.; Lv, B.; Yu, Z.; Dai, S.; Liu, X.; Xia, G.; Tian, H. MPK3/6-induced degradation of ARR1/10/12 promotes salt tolerance in Arabidopsis. EMBO Rep. 2021, 22, e52457. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Li, Z.; Wen, X.; Li, W.; Shi, H.; Yang, L.; Zhu, H.; Guo, H. Salt-induced stabilization of EIN3/EIL1 confers salinity tolerance by deterring ROS accumulation in Arabidopsis. PloS Genet. 2014, 10, e1004664. [Google Scholar] [CrossRef] [Green Version]
- Sun, L.; Jing, Y.; Liu, X.; Li, Q.; Xue, Z.; Cheng, Z.; Wang, D.; He, H.; Qian, W. Heat stress-induced transposon activation correlates with 3D chromatin organization rearrangement in Arabidopsis. Nat. Commun. 2020, 11, 1886. [Google Scholar] [CrossRef] [Green Version]
- Shi, Y.; Tian, S.; Hou, L.; Huang, X.; Zhang, X.; Guo, H.; Yang, S. Ethylene Signaling Negatively Regulates Freezing Tolerance by Repressing Expression of CBF and Type-A ARR Genes in Arabidopsis. Plant Cell 2012, 24, 2578–2595. [Google Scholar] [CrossRef] [Green Version]
- Schellingen, K.; Van Der Straeten, D.; Vandenbussche, F.; Prinsen, E.; Remans, T.; Vangronsveld, J.; Cuypers, A. Cadmium-induced ethylene production and responses in Arabidopsis thaliana rely on ACS2 and ACS6 gene expression. BMC Plant Biol. 2014, 14, 214. [Google Scholar] [CrossRef] [Green Version]
- Sun, P.; Tian, Q.-Y.; Chen, J.; Zhang, W.-H. Aluminium-induced inhibition of root elongation in Arabidopsis is mediated by ethylene and auxin. J. Exp. Bot. 2009, 61, 347–356. [Google Scholar] [CrossRef] [Green Version]
- Liang, X.; Shen, N.F.; Theologis, A. Li+-regulated 1-aminocyclopropane-1-carboxylate synthase gene expression in Arabidopsis thaliana. Plant J. 1996, 10, 1027–1036. [Google Scholar] [CrossRef]
- Cao, S.; Chen, Z.; Liu, G.; Jiang, L.; Yuan, H.; Ren, G.; Bian, X.; Jian, H.; Ma, X. The Arabidopsis Ethylene-Insensitive 2 gene is required for lead resistance. Plant Physiol. Biochem. 2009, 47, 308–312. [Google Scholar] [CrossRef]
- Husain, T.; Fatima, A.; Suhel, M.; Singh, S.; Sharma, A.; Prasad, S.M.; Singh, V.P. A brief appraisal of ethylene signaling under abiotic stress in plants. Plant Signal. Behav. 2020, 15, 1782051. [Google Scholar] [CrossRef]
- Druege, U. Ethylene and Plant Responses to Abiotic Stress. In Ethylene Action in Plants; Springer: Berlin/Heidelberg, Germany, 2006; pp. 81–118. [Google Scholar]
- Steffens, B. The role of ethylene and ROS in salinity, heavy metal, and flooding responses in rice. Front. Plant Sci. 2014, 5, 685. [Google Scholar] [CrossRef] [PubMed]
- Nazar, R.; Khan, I.R.; Iqbal, N.; Masood, A.; Khan, N.A. Involvement of ethylene in reversal of salt-inhibited photosynthesis by sulfur in mustard. Physiol. Plant. 2014, 152, 331–344. [Google Scholar] [CrossRef]
- Sharma, A.; Kumar, V.; Sidhu, G.P.S.; Kumar, R.; Kohli, S.K.; Yadav, P.; Kapoor, D.; Bali, A.S.; Shahzad, B.; Khanna, K. Abiotic stress management in plants: Role of ethylene. In Molecular Plant Abiotic Stress: Biology and Biotechnology; Wiley: Hoboken, NJ, USA, 2019; pp. 185–208. [Google Scholar]
- Ku, Y.-S.; Sintaha, M.; Cheung, M.-Y.; Lam, H.-M. Plant Hormone Signaling Crosstalks between Biotic and Abiotic Stress Responses. Int. J. Mol. Sci. 2018, 19, 3206. [Google Scholar] [CrossRef] [Green Version]
- Devireddy, A.R.; Zandalinas, S.I.; Fichman, Y.; Mittler, R. Integration of reactive oxygen species and hormone signaling during abiotic stress. Plant J. 2020, 105, 459–476. [Google Scholar] [CrossRef]
- Knight, H.; Zarka, D.G.; Okamoto, H.; Thomashow, M.F.; Knight, M.R. Abscisic Acid Induces CBF Gene Transcription and Subsequent Induction of Cold-Regulated Genes via the CRT Promoter Element. Plant Physiol. 2004, 135, 1710–1717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Duan, G.; Li, C.; Liu, L.; Han, G.; Zhang, Y.; Wang, C. The Crosstalks Between Jasmonic Acid and Other Plant Hormone Signaling Highlight the Involvement of Jasmonic Acid as a Core Component in Plant Response to Biotic and Abiotic Stresses. Front. Plant Sci. 2019, 10, 1349. [Google Scholar] [CrossRef] [Green Version]
- Demole, E.; Lederer, E.; Mercier, D. Isolement et détermination de la structure du jasmonate de méthyle, constituant odorant caracteristique de l’essence de jasmin. Helv. Chim. Acta 1962, 45, 675–685. [Google Scholar] [CrossRef]
- Iqbal, N.; Khan, N.A.; Ferrante, A.; Trivellini, A.; Francini, A.; Khan, M.I.R. Ethylene Role in Plant Growth, Development and Senescence: Interaction with Other Phytohormones. Front. Plant Sci. 2017, 8, 475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wasternack, C.; Hause, B. Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 2013, 111, 1021–1058. [Google Scholar] [CrossRef]
- Ruan, J.; Zhou, Y.; Zhou, M.; Yan, J.; Khurshid, M.; Weng, W.; Cheng, J.; Zhang, K. Jasmonic Acid Signaling Pathway in Plants. Int. J. Mol. Sci. 2019, 20, 2479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chini, A.; Monte, I.; Zamarreño, A.M.; Hamberg, M.; Lassueur, S.; Reymond, P.; Weiss, S.; Stintzi, A.; Schaller, A.; Porzel, A.; et al. An OPR3-independent pathway uses 4,5-didehydrojasmonate for jasmonate synthesis. Nat. Chem. Biol. 2018, 14, 171–178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, Z.; Nolan, T.M.; Jiang, H.; Yin, Y. AP2/ERF transcription factor regulatory networks in hormone and abiotic stress responses in Arabidopsis. Front. Plant Sci. 2019, 10, 228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riyazuddin, R.; Verma, R.; Singh, K.; Nisha, N.; Keisham, M.; Bhati, K.K.; Kim, S.T.; Gupta, R. Ethylene: A Master Regulator of Salinity Stress Tolerance in Plants. Biomolecules 2020, 10, 959. [Google Scholar] [CrossRef]
- Voesenek, L.A.C.J.; Sasidharan, R.; Visser, E.J.; Bailey-Serres, J. Flooding stress signaling through perturbations in oxygen, ethylene, nitric oxide and light. New Phytol. 2015, 209, 39–43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Ou, B.; Zhang, J.; Si, W.; Gu, H.; Qin, G.; Qu, L. The Arabidopsis Mediator subunit MED16 regulates iron homeostasis by associating with EIN 3/EIL1 through subunit MED25. Plant J. 2014, 77, 838–851. [Google Scholar] [CrossRef]
- Huang, J.; Zhao, X.; Bürger, M.; Wang, Y.; Chory, J. Two interacting ethylene response factors regulate heat stress response. Plant Cell 2020, 33, 338–357. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.-C.; Liao, P.-M.; Kuo, W.-W.; Lin, T.-P. The Arabidopsis ETHYLENE RESPONSE FACTOR1 Regulates Abiotic Stress-Responsive Gene Expression by Binding to Different cis-Acting Elements in Response to Different Stress Signals. Plant Physiol. 2013, 162, 1566–1582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chini, A.; Gimenez-Ibanez, S.; Goossens, A.; Solano, R. Redundancy and specificity in jasmonate signalling. Curr. Opin. Plant Biol. 2016, 33, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Earley, K.; Lawrence, R.J.; Pontes, O.; Reuther, R.; Enciso, A.J.; Silva, M.; Neves, N.; Gross, M.; Viegas, W.; Pikaard, C.S. Erasure of histone acetylation by Arabidopsis I6 mediates large-scale gene silencing in nucleolar dominance. Genes Dev. 2006, 20, 1283–1293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Z.; An, F.; Feng, Y.; Li, P.; Xue, L.A.M.; Jiang, Z.; Kim, J.M.; To, T.K.; Li, W.; Zhang, X.; et al. Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 12539–12544. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Z.; Lee, B. Friends or Foes: New Insights in Jasmonate and Ethylene Co-Actions. Plant Cell Physiol. 2014, 56, 414–420. [Google Scholar] [CrossRef]
- Cui, M.; Lin, Y.; Zu, Y.; Efferth, T.; Li, D.; Tang, Z. Ethylene increases accumulation of compatible solutes and decreases oxidative stress to improve plant tolerance to water stress in Arabidopsis. J. Plant Biol. 2015, 58, 193–201. [Google Scholar] [CrossRef]
- Kim, J.; To, T.K.; Matsui, A.; Tanoi, K.; Kobayashi, N.I.; Matsuda, F.; Habu, Y.; Ogawa, D.; Sakamoto, T.; Matsunaga, S. Acetate-mediated novel survival strategy against drought in plants. Nat. Plants 2017, 3, 17097. [Google Scholar] [CrossRef]
- Fu, J.; Wu, H.; Ma, S.; Xiang, D.; Liu, R.; Xiong, L. OsJAZ1 Attenuates Drought Resistance by Regulating JA and ABA Signaling in Rice. Front. Plant Sci. 2017, 8, 2108. [Google Scholar] [CrossRef] [Green Version]
- Jin, J.; Duan, J.; Shan, C.; Mei, Z.; Chen, H.; Feng, H.; Zhu, J.; Cai, W. Ethylene insensitive3-like2 (OsEIL2) confers stress sensitivity by regulating OsBURP16, the β subunit of polygalacturonase (PG1β-like) subfamily gene in rice. Plant Sci. 2019, 292, 110353. [Google Scholar] [CrossRef]
- Riemann, M.; Dhakarey, R.; Hazman, M.; Miro, B.; Kohli, A.; Nick, P. Exploring jasmonates in the hormonal network of drought and salinity responses. Front. Plant Sci. 2015, 6, 1077. [Google Scholar] [CrossRef] [Green Version]
- Zhu, C.; Gan, L.; Shen, Z.; Xia, K. Interactions between jasmonates and ethylene in the regulation of root hair development in Arabidopsis. J. Exp. Bot. 2006, 57, 1299–1308. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Zhu, Z.; An, F.; Hao, D.; Li, P.; Song, J.; Yi, C.; Guo, H. Jasmonate-Activated MYC2 Represses ETHYLENE INSENSITIVE3 Activity to Antagonize Ethylene-Promoted Apical Hook Formation in Arabidopsis. Plant Cell 2014, 26, 1105–1117. [Google Scholar] [CrossRef] [Green Version]
- Song, S.; Huang, H.; Gao, H.; Wang, J.; Wu, D.; Liu, X.; Yang, S.; Zhai, Q.; Li, C.; Qi, T.; et al. Interaction between MYC2 and ETHYLENE INSENSITIVE3 Modulates Antagonism between Jasmonate and Ethylene Signaling in Arabidopsis. Plant Cell 2014, 26, 263–279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Moerkercke, A.; Duncan, O.; Zander, M.; Simura, J.; Broda, M.; Vanden Bossche, R.; Lewsey, M.G.; Lama, S.; Singh, K.B.; Ljung, K.; et al. A MYC2/MYC3/MYC4-dependent transcription factor network regulates water spray-responsive gene expression and jasmonate levels. Proc. Natl. Acad. Sci. USA 2019, 116, 23345–23356. [Google Scholar] [CrossRef]
- Lorenzo, O.; Piqueras, R.; Sánchez-serrano, J.J.; Solano, R. ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 2003, 15, 165–178. [Google Scholar] [CrossRef] [Green Version]
- Zander, M.; Lewsey, M.G.; Clark, N.M.; Yin, L.; Bartlett, A.; Guzmán, J.P.S.; Hann, E.; Langford, A.E.; Jow, B.; Wise, A.; et al. Integrated multi-omics framework of the plant response to jasmonic acid. Nat. Plants 2020, 6, 290–302. [Google Scholar] [CrossRef]
- Hao, D.; Jin, L.; Wen, X.; Yu, F.; Xie, Q.; Guo, H. The RING E3 ligase SDIR1 destabilizes EBF1/EBF2 and modulates the ethylene response to ambient temperature fluctuations in Arabidopsis. Proc. Natl. Acad. Sci. USA 2021, 118, e2024592118. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, C.; Li, Y.; Zheng, N.; Chen, H.; Zhao, Q.; Gao, T.; Guo, H.; Xie, Q. SDIR1 Is a RING Finger E3 Ligase That Positively Regulates Stress-Responsive Abscisic Acid Signaling in Arabidopsis. Plant Cell 2007, 19, 1912–1929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dar, N.A.; Amin, I.; Wani, W.; Wani, S.A.; Shikari, A.B.; Wani, S.H.; Masoodi, K.Z. Abscisic acid: A key regulator of abiotic stress tolerance in plants. Plant Gene 2017, 11, 106–111. [Google Scholar] [CrossRef]
- Emenecker, R.J.; Strader, L.C. Auxin-Abscisic Acid Interactions in Plant Growth and Development. Biomolecules 2020, 10, 281. [Google Scholar] [CrossRef] [Green Version]
- Sah, S.K.; Reddy, K.R.; Li, J. Abscisic acid and abiotic stress tolerance in crop plants. Front. Plant Sci. 2016, 7, 571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Müller, M. Foes or Friends: ABA and Ethylene Interaction under Abiotic Stress. Plants 2021, 10, 448. [Google Scholar] [CrossRef] [PubMed]
- Kumar, D.; Hazra, S.; Datta, R.; Chattopadhyay, S. Transcriptome analysis of Arabidopsis mutants suggests a crosstalk between ABA, ethylene and GSH against combined cold and osmotic stress. Sci. Rep. 2016, 6, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Marczak, M.; Ciesla, A.; Janicki, M.; Kasprowicz-Maluski, A.; Kubiak, P.; Ludwikow, A. Protein Phosphatases Type 2C Group A Interact with and Regulate the Stability of ACC Synthase 7 in Arabidopsis. Cells 2020, 9, 978. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ueda, H.; Ito, T.; Inoue, R.; Masuda, Y.; Nagashima, Y.; Kozuka, T.; Kusaba, M. Genetic Interaction Among Phytochrome, Ethylene and Abscisic Acid Signaling During Dark-Induced Senescence in Arabidopsis Thaliana. Front. Plant Sci. 2020, 11, 564. [Google Scholar] [CrossRef]
- Lei, G.; Shen, M.; Li, Z.-G.; Zhang, B.; Duan, K.-X.; Wang, N.; Cao, Y.-R.; Zhang, W.-K.; Ma, B.; Ling, H.-Q.; et al. EIN2 regulates salt stress response and interacts with a MA3 domain-containing protein ECIP1 in Arabidopsis. Plant Cell Environ. 2011, 34, 1678–1692. [Google Scholar] [CrossRef]
- Cao, W.-H.; Liu, J.; He, X.-J.; Mu, R.-L.; Zhou, H.-L.; Chen, S.-Y.; Zhang, J.-S. Modulation of Ethylene Responses Affects Plant Salt-Stress Responses. Plant Physiol. 2006, 143, 707–719. [Google Scholar] [CrossRef] [Green Version]
- Achard, P.; Chen, D.; De Grauwe, L.; Decat, J.; Schoutteten, H.; Moritz, T.; Van Der Straeten, D.; Peng, J.; Harberd, N.P. Integration of Plant Responses to Environmentally Activated Phytohormonal Signals. Science 2006, 311, 91–94. [Google Scholar] [CrossRef]
- Munemasa, S.; Hauser, F.; Park, J.; Waadt, R.; Brandt, B.; Schroeder, J.I. Mechanisms of abscisic acid-mediated control of stomatal aperture. Curr. Opin. Plant Biol. 2015, 28, 154–162. [Google Scholar] [CrossRef] [Green Version]
- Madhavan, S.; Chrominiski, A.; Smith, B.N. Effect of Ethylene on Stomatal Opening in Tomato and Carnation Leaves. Plant Cell Physiol. 1983, 24. [Google Scholar] [CrossRef]
- Tanaka, Y.; Sano, T.; Tamaoki, M.; Nakajima, N.; Kondo, N.; Hasezawa, S. Ethylene Inhibits Abscisic Acid-Induced Stomatal Closure in Arabidopsis. Plant Physiol. 2005, 138, 2337–2343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benlloch-González, M.; Arquero, O.; Fournier, J.M.; Barranco, D.; Benlloch, M. K+ starvation inhibits water-stress-induced stomatal closure. J. Plant Physiol. 2008, 165, 623–630. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, N.; Nazar, R.; Syeed, S.; Masood, A.; Khan, N.A. Exogenously-sourced ethylene increases stomatal conductance, photosynthesis, and growth under optimal and deficient nitrogen fertilization in mustard. J. Exp. Bot. 2011, 62, 4955–4963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watkins, J.M.; Hechler, P.J.; Muday, G.K. Ethylene-Induced Flavonol Accumulation in Guard Cells Suppresses Reactive Oxygen Species and Moderates Stomatal Aperture. Plant Physiol. 2014, 164, 1707–1717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Desikan, R.; Last, K.; Harrett-Williams, R.; Tagliavia, C.; Harter, K.; Hooley, R.; Hancock, J.T.; Neill, S.J. Ethylene-induced stomatal closure in Arabidopsis occurs via AtrbohF-mediated hydrogen peroxide synthesis. Plant J. 2006, 47, 907–916. [Google Scholar] [CrossRef]
- Shi, C.; Qi, C.; Ren, H.; Huang, A.; Hei, S.; She, X. Ethylene mediates brassinosteroid-induced stomatal closure via Gα protein-activated hydrogen peroxide and nitric oxide production in Arabidopsis. Plant J. 2015, 82, 280–301. [Google Scholar] [CrossRef] [PubMed]
- Nazareno, A.L.; Hernandez, B. A mathematical model of the interaction of abscisic acid, ethylene and methyl jasmonate on stomatal closure in plants. PLoS ONE 2017, 12, e0171065. [Google Scholar] [CrossRef]
- Xu, Z.-S.; Xia, L.-Q.; Chen, M.; Cheng, X.-G.; Zhang, R.-Y.; Li, L.-C.; Zhao, Y.-X.; Lu, Y.; Ni, Z.-Y.; Liu, L.; et al. Isolation and molecular characterization of the Triticum aestivum L. ethylene-responsive factor 1 (TaERF1) that increases multiple stress tolerance. Plant Mol. Biol. 2007, 65, 719–732. [Google Scholar] [CrossRef]
- Xing, L.; Di, Z.; Yang, W.; Liu, J.; Li, M.; Wang, X.; Cui, C.; Wang, X.; Zhang, R.; Xiao, J. Overexpression of ERF1-V from Haynaldia villosa can enhance the resistance of wheat to powdery mildew and increase the tolerance to salt and drought stresses. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef] [Green Version]
- Wu, L.; Chen, X.; Ren, H.; Zhang, Z.; Zhang, H.; Wang, J.; Wang, X.-C.; Huang, R. ERF protein JERF1 that transcriptionally modulates the expression of abscisic acid biosynthesis-related gene enhances the tolerance under salinity and cold in tobacco. Planta 2007, 226, 815–825. [Google Scholar] [CrossRef] [PubMed]
- Quan, R.; Hu, S.; Zhang, Z.; Zhang, H.; Zhang, Z.; Huang, R. Overexpression of an ERF transcription factorTSRF1improves rice drought tolerance. Plant Biotechnol. J. 2010, 8, 476–488. [Google Scholar] [CrossRef]
- Dong, Z.; Yu, Y.; Li, S.; Wang, J.; Tang, S.; Huang, R. Abscisic Acid Antagonizes Ethylene Production through the ABI4-Mediated Transcriptional Repression of ACS4 and ACS8 in Arabidopsis. Mol. Plant 2015, 9, 126–135. [Google Scholar] [CrossRef] [Green Version]
- Agurla, S.; Gahir, S.; Munemasa, S.; Murata, Y.; Raghavendra, A.S. Mechanism of stomatal closure in plants exposed to drought and cold stress. In Survival Strategies in Extreme Cold and Desiccation; Springer: Singapore, 2018; pp. 215–232. [Google Scholar]
- Khan, M.I.R.; Trivellini, A.; Chhillar, H.; Chopra, P.; Ferrante, A.; Khan, N.A.; Ismail, A.M. The significance and functions of ethylene in flooding stress tolerance in plants. Environ. Exp. Bot. 2020, 179, 104188. [Google Scholar] [CrossRef]
- Benschop, J.J.; Jackson, M.B.; Gühl, K.; Vreeburg, R.A.M.; Croker, S.J.; Peeters, A.J.M.; Voesenek, L.A.C.J. Contrasting interactions between ethylene and abscisic acid in Rumex species differing in submergence tolerance. Plant J. 2005, 44, 756–768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benschop, J.J.; Bou, J.; Peeters, A.J.; Wagemaker, N.; Gühl, K.; Ward, D.; Hedden, P.; Moritz, T.; Voesenek, L.A. Long-Term Submergence-Induced Elongation in Rumex palustris Requires Abscisic Acid-Dependent Biosynthesis of Gibberellin1. Plant Physiol. 2006, 141, 1644–1652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saika, H.; Okamoto, M.; Miyoshi, K.; Kushiro, T.; Shinoda, S.; Jikumaru, Y.; Fujimoto, M.; Arikawa, T.; Takahashi, H.; Ando, M.; et al. Ethylene Promotes Submergence-Induced Expression of OsABA8ox1, a Gene that Encodes ABA 8’-Hydroxylase in Rice. Plant Cell Physiol. 2006, 48, 287–298. [Google Scholar] [CrossRef] [Green Version]
- Arroyo, A.; Bossi, F.; Finkelstein, R.R.; León, P. Three Genes That Affect Sugar Sensing (Abscisic Acid Insensitive 4, Abscisic Acid Insensitive 5, and Constitutive Triple Response 1) Are Differentially Regulated by Glucose in Arabidopsis. Plant Physiol. 2003, 133, 231–242. [Google Scholar] [CrossRef] [Green Version]
- Benschop, J.J.; Millenaar, F.F.; Smeets, M.E.; Van Zanten, M.; Voesenek, L.; Peeters, A.J. Abscisic Acid Antagonizes Ethylene-Induced Hyponastic Growth in Arabidopsis. Plant Physiol. 2006, 143, 1013–1023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ortega-Martínez, O.; Pernas, M.; Carol, R.J.; Dolan, L. Ethylene Modulates Stem Cell Division in the Arabidopsis thaliana Root. Science 2007, 317, 507–510. [Google Scholar] [CrossRef]
- Linkies, A.; Müller, K.; Morris, K.; Turečková, V.; Wenk, M.; Cadman, C.S.; Corbineau, F.; Strnad, M.; Lynn, J.R.; Finch-Savage, W.E.; et al. Ethylene Interacts with Abscisic Acid to Regulate Endosperm Rupture during Germination: A Comparative Approach Using Lepidium sativum and Arabidopsis thaliana. Plant Cell 2009, 21, 3803–3822. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Han, W.; De Smet, I.; Talboys, P.; Loya, R.; Hassan, A.; Rong, H.; Jürgens, G.; Knox, J.P.; Wang, M.-H. ABA promotes quiescence of the quiescent centre and suppresses stem cell differentiation in the Arabidopsis primary root meristem. Plant J. 2010, 64, 764–774. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.K.H.M.; McCouch, S.R.M. Getting to the roots of it: Genetic and hormonal control of root architecture. Front. Plant Sci. 2013, 4, 186. [Google Scholar] [CrossRef] [Green Version]
- Chang, K.N.; Zhong, S.; Weirauch, M.T.; Hon, G.; Pelizzola, M.; Li, H.; Huang, S.-S.C.; Schmitz, R.; Urich, M.A.; Kuo, D.; et al. Temporal transcriptional response to ethylene gas drives growth hormone cross-regulation in Arabidopsis. eLife 2013, 2, e00675. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Huang, S.-S.C.; Wise, A.; Castanon, R.; Nery, J.R.; Chen, H.; Watanabe, M.; Thomas, J.; Bar-Joseph, Z.; Ecker, J.R. A transcription factor hierarchy defines an environmental stress response network. Science 2016, 354, aag1550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Planas-Riverola, A.; Gupta, A.; Betegón-Putze, I.; Bosch, N.; Ibañes, M.; Caño-Delgado, A.I. Brassinosteroid signaling in plant development and adaptation to stress. Development 2019, 146, dev151894. [Google Scholar] [CrossRef] [Green Version]
- Nolan, T.M.; Vukašinović, N.; Liu, D.; Russinova, E.; Yin, Y. Brassinosteroids: Multidimensional Regulators of Plant Growth, Development, and Stress Responses. Plant Cell 2019, 32, 295–318. [Google Scholar] [CrossRef] [Green Version]
- Jiroutova, P.; Oklestkova, J.; Strnad, M. Crosstalk between Brassinosteroids and Ethylene during Plant Growth and under Abiotic Stress Conditions. Int. J. Mol. Sci. 2018, 19, 3283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, B.; Zhang, J.; Xia, X.; Zhang, W.-H. Ameliorative effect of brassinosteroid and ethylene on germination of cucumber seeds in the presence of sodium chloride. Plant Growth Regul. 2011, 65, 407–413. [Google Scholar] [CrossRef]
- Zhu, T.; Deng, X.; Zhou, X.; Zhu, L.; Zou, L.; Li, P.; Zhang, D.; Lin, H. Ethylene and hydrogen peroxide are involved in brassinosteroid-induced salt tolerance in tomato. Sci. Rep. 2016, 6, 35392. [Google Scholar] [CrossRef] [Green Version]
- Wei, L.-J.; Deng, X.-G.; Zhu, T.; Zheng, T.; Li, P.-X.; Wu, J.-Q.; Zhang, D.-W.; Lin, H.-H. Ethylene is Involved in Brassinosteroids Induced Alternative Respiratory Pathway in Cucumber (Cucumis sativus L.) Seedlings Response to Abiotic Stress. Front. Plant Sci. 2015, 6, 982. [Google Scholar] [CrossRef] [Green Version]
- Schmitz, A.; Folsom, J.J.; Jikamaru, Y.; Ronald, P.; Walia, H. SUB 1 A-mediated submergence tolerance response in rice involves differential regulation of the brassinosteroid pathway. New Phytol. 2013, 198, 1060–1070. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lavy, M.; Estelle, M. Mechanisms of auxin signaling. Development 2016, 143, 3226–3229. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y. Auxin biosynthesis. Arab. Book 2014, 12, e0173. [Google Scholar] [CrossRef] [Green Version]
- Enders, T.A.; Strader, L.C. Auxin activity: Past, present, and future. Am. J. Bot. 2015, 102, 180–196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muday, G.K.; Rahman, A.; Binder, B.M. Auxin and ethylene: Collaborators or competitors? Trends Plant Sci. 2012, 17, 181–195. [Google Scholar] [CrossRef] [PubMed]
- Stepanova, A.N.; Alonso, J.M. From Ethylene-Auxin Interactions to Auxin Biosynthesis and Signal Integration. Plant Cell 2019, 31, 1393–1394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mazzoni-Putman, S.M.; Brumos, J.; Zhao, C.; Alonso, J.M.; Stepanova, A.N. Auxin Interactions with Other Hormones in Plant Development. Cold Spring Harb. Perspect. Biol. 2021, 13, a039990. [Google Scholar] [CrossRef]
- Koevoets, I.T.; Venema, J.H.; Elzenga, J.T.M.; Testerink, C. Roots Withstanding their Environment: Exploiting Root System Architecture Responses to Abiotic Stress to Improve Crop Tolerance. Front. Plant Sci. 2016, 7, 1335. [Google Scholar] [CrossRef] [Green Version]
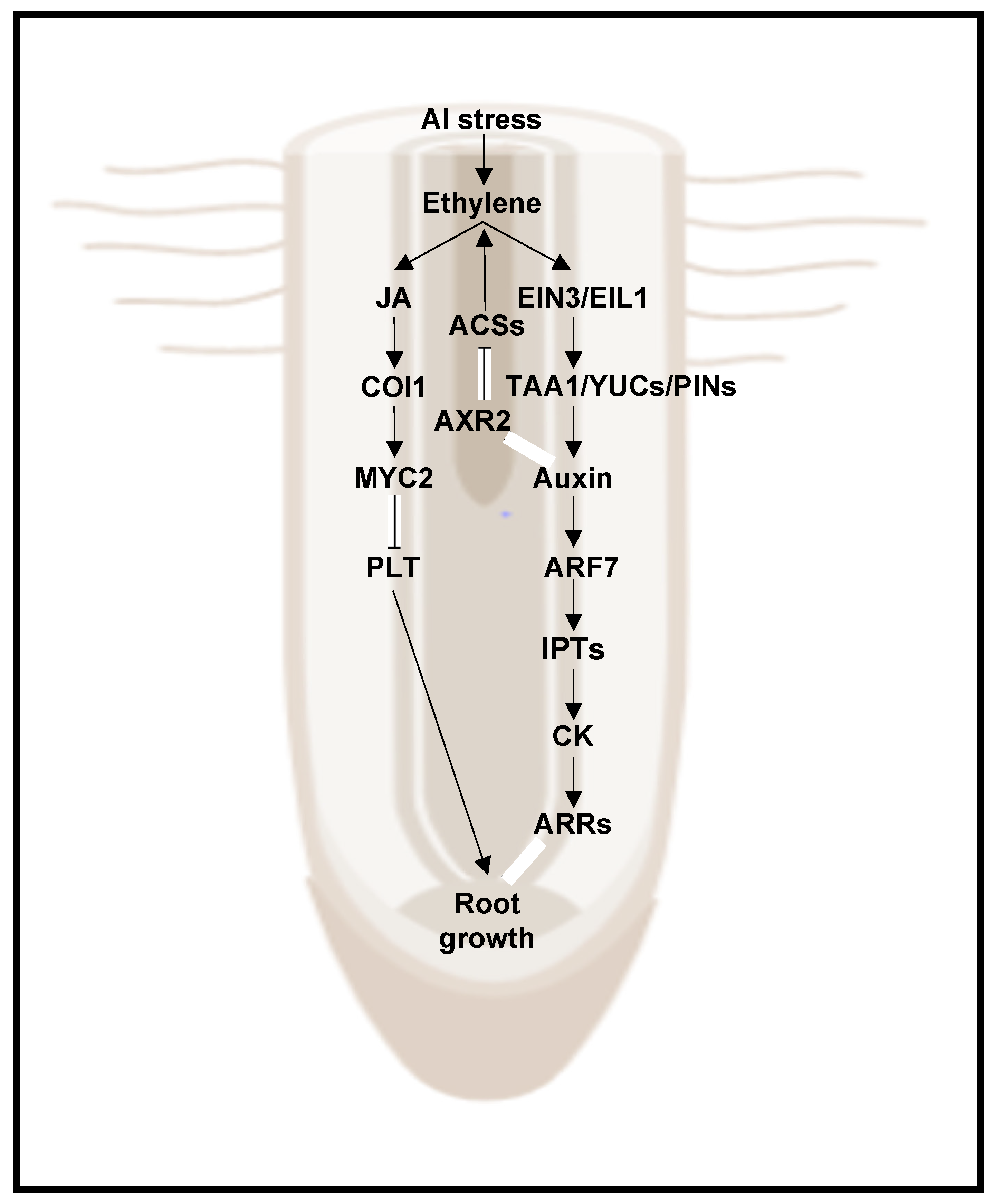
- Bojórquez-Quintal, E.; Escalante-Magaña, C.; Echevarría-Machado, I.; Martínez-Estévez, M. Aluminum, a Friend or Foe of Higher Plants in Acid Soils. Front. Plant Sci. 2017, 8, 1767. [Google Scholar] [CrossRef]
- Shen, H.; Hou, N.; Schlicht, M.; Wan, Y.; Mancuso, S.; Baluska, F. Aluminium toxicity targets PIN2 in Arabidopsis root apices: Effects on PIN2 endocytosis, vesicular recycling, and polar auxin transport. Sci. Bull. 2008, 53, 2480–2487. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.B.; Geng, X.Y.; He, C.M.; Zhang, F.; Wang, R.; Horst, W.J.; Ding, Z.J. TAA1-Regulated Local Auxin Biosynthesis in the Root-Apex Transition Zone Mediates the Aluminum-Induced Inhibition of Root Growth in Arabidopsis. Plant Cell 2014, 26, 2889–2904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, G.; Gao, S.; Tian, H.; Wu, W.; Robert, H.S.; Ding, Z. Local Transcriptional Control of YUCCA Regulates Auxin Promoted Root-Growth Inhibition in Response to Aluminium Stress in Arabidopsis. PLoS Genet. 2016, 12, e1006360. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Liu, G.; Liu, J.; Zhang, B.; Meng, W.; Müller, B.; Hayashi, K.; Zhang, X.; Zhao, Z.; De Smet, I.; et al. Synergistic action of auxin and cytokinin mediates aluminum—Induced root growth inhibition in Arabidopsis. EMBO Rep. 2017, 18, 1213–1230. [Google Scholar] [CrossRef]
- Wakeel, A.; Ali, I.; Upreti, S.; Azizullah, A.; Liu, B.; Khan, A.R.; Huang, L.; Wu, M.; Gan, Y. Ethylene mediates dichromate-induced inhibition of primary root growth by altering AUX1 expression and auxin accumulation in Arabidopsis thaliana. Plant Cell Environ. 2018, 41, 1453–1467. [Google Scholar] [CrossRef]
- Wakeel, A.; Gan, Y. A model for the ethylene-mediated auxin distribution under Cr(VI) stress in Arabidopsis thaliana. Plant Signal. Behav. 2018, 13, e1473685. [Google Scholar] [CrossRef]
- Li, G.; Song, H.; Li, B.; Kronzucker, H.J.; Shi, W. Auxin resistant1 and PIN-FORMED2 protect lateral root formation in Arabidopsis under iron stress. Plant Physiol. 2015, 169, 2608–2623. [Google Scholar]
- Li, J.; Xu, H.-H.; Liu, W.-C.; Zhang, X.-W.; Lu, Y.-T. Ethylene Inhibits Root Elongation during Alkaline Stress through AUXIN1 and Associated Changes in Auxin Accumulation. Plant Physiol. 2015, 168, 1777–1791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gratão, P.; Monteiro, C.C.; Carvalho, R.F.; Tezotto, T.; Piotto, F.; Peres, L.; Azevedo, R.A. Biochemical dissection of diageotropica and Never ripe tomato mutants to Cd-stressful conditions. Plant Physiol. Biochem. 2012, 56, 79–96. [Google Scholar] [CrossRef]
- Gruber, B.D.; Giehl, R.F.; Friedel, S.; von Wirén, N. Plasticity of the Arabidopsis root system under nutrient deficiencies. Plant Physiol. 2013, 163, 161–179. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Zhang, H.; Fang, X.; Zhang, Y.; Jin, C. Auxin acts downstream of ethylene and nitric oxide to regulate magnesium deficiency-induced root hair development in Arabidopsis thaliana. Plant Cell Physiol. 2018, 59, 1452–1465. [Google Scholar] [CrossRef]
- Martín-Rejano, E.M.; Camacho-Cristóbal, J.J.; Herrera-Rodríguez, M.B.; Rexach, J.; Navarro-Gochicoa, M.T.; Gonzalez-Fontes, A. Auxin and ethylene are involved in the responses of root system architecture to low boron supply in Arabidopsis seedlings. Physiol. Plant. 2011, 142, 170–178. [Google Scholar] [CrossRef] [PubMed]
- Camacho-Cristóbal, J.J.; Martín-Rejano, E.M.; Herrera-Rodríguez, M.B.; Navarro-Gochicoa, M.T.; Rexach, J.; González-Fontes, A. Boron deficiency inhibits root cell elongation via an ethylene/auxin/ROS-dependent pathway in Arabidopsis seedlings. J. Exp. Bot. 2015, 66, 3831–3840. [Google Scholar] [CrossRef] [PubMed]
- Jacobsen, A.G.R.; Jervis, G.; Xu, J.; Topping, J.F.; Lindsey, K. Root growth responses to mechanical impedance are regulated by a network of ROS, ethylene and auxin signalling in Arabidopsis. New Phytol. 2021, 231, 225–242. [Google Scholar] [CrossRef] [PubMed]
- Sasidharan, R.; Voesenek, L.A.C.J. Ethylene-Mediated Acclimations to Flooding Stress. Plant Physiol. 2015, 169, 3–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Visser, E.; Cohen, J.; Barendse, G.; Blom, C.; Voesenek, L. An Ethylene-Mediated Increase in Sensitivity to Auxin Induces Adventitious Root Formation in Flooded Rumex palustris Sm. Plant Physiol. 1996, 112, 1687–1692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vidoz, M.L.; Loreti, E.; Mensuali, A.; Alpi, A.; Perata, P. Hormonal interplay during adventitious root formation in flooded tomato plants. Plant J. 2010, 63, 551–562. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.; Li, Q.; Ma, X.; Qian, C.; Wang, H.; Ren, N.; Shen, C.; Huang, S.; Xu, X.; Xu, Q.; et al. Waterlogging—Induced adventitious root formation in cucumber is regulated by ethylene and auxin through reactive oxygen species signalling. Plant Cell Environ. 2018, 42, 1458–1470. [Google Scholar] [CrossRef]
- Yamauchi, T.; Shimamura, S.; Nakazono, M.; Mochizuki, T. Aerenchyma formation in crop species: A review. Field Crop. Res. 2013, 152, 8–16. [Google Scholar] [CrossRef]
- Yamauchi, T.; Tanaka, A.; Tsutsumi, N.; Inukai, Y.; Nakazono, M. A Role for Auxin in Ethylene-Dependent Inducible Aerenchyma Formation in Rice Roots. Plants 2020, 9, 610. [Google Scholar] [CrossRef]
- Zhong, S.; Shi, H.; Xue, C.; Wang, L.; Xi, Y.; Li, J.; Quail, P.H.; Deng, X.W.; Guo, H. A Molecular Framework of Light-Controlled Phytohormone Action in Arabidopsis. Curr. Biol. 2012, 22, 1530–1535. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.Y.; Park, Y.-J.; Lee, J.-H.; Kim, Z.H.; Park, C.-M. EIN3-Mediated Ethylene Signaling Attenuates Auxin Response during Hypocotyl Thermomorphogenesis. Plant Cell Physiol. 2021, 62, 708–720. [Google Scholar] [CrossRef]
- Fei, Q.; Wei, S.; Zhou, Z.; Gao, H.; Li, X. Adaptation of root growth to increased ambient temperature requires auxin and ethylene coordination in Arabidopsis. Plant Cell Rep. 2017, 36, 1507–1518. [Google Scholar] [CrossRef] [PubMed]
- Binenbaum, J.; Weinstain, R.; Shani, E. Gibberellin Localization and Transport in Plants. Trends Plant Sci. 2018, 23, 410–421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hernández-García, J.; Briones-Moreno, A.; Blázquez, M.A. Origin and evolution of gibberellin signaling and metabolism in plants. Semin. Cell Dev. Biol. 2020, 109, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Hattori, Y.; Nagai, K.; Furukawa, S.; Song, X.-J.; Kawano, R.; Sakakibara, H.; Wu, J.; Matsumoto, T.; Yoshimura, A.; Kitano, H.; et al. The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature 2009, 460, 1026–1030. [Google Scholar] [CrossRef] [PubMed]
- Bailey-Serres, J.; Fukao, T.; Gibbs, D.; Holdsworth, M.; Lee, S.C.; Licausi, F.; Perata, P.; Voesenek, L.; van Dongen, J. Making sense of low oxygen sensing. Trends Plant Sci. 2012, 17, 129–138. [Google Scholar] [CrossRef]
- Kuroha, T.; Nagai, K.; Gamuyao, R.; Wang, D.R.; Furuta, T.; Nakamori, M.; Kitaoka, T.; Adachi, K.; Minami, A.; Mori, Y.; et al. Ethylene-gibberellin signaling underlies adaptation of rice to periodic flooding. Science 2018, 361, 181–186. [Google Scholar] [CrossRef] [Green Version]
- Fukao, T.; Xu, K.; Ronald, P.C.; Bailey-Serres, J. A Variable Cluster of Ethylene Response Factor–Like Genes Regulates Metabolic and Developmental Acclimation Responses to Submergence in Rice. Plant Cell 2006, 18, 2021–2034. [Google Scholar] [CrossRef] [Green Version]
- Xu, K.; Xu, X.; Fukao, T.; Canlas, P.; Maghirang-Rodriguez, R.; Heuer, S.; Ismail, A.M.; Bailey-Serres, J.; Ronald, P.C.; Mackill, D.J. Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nat. Cell Biol. 2006, 442, 705–708. [Google Scholar] [CrossRef] [Green Version]
- Fukao, T.; Bailey-Serres, J. Submergence tolerance conferred by Sub1A is mediated by SLR1 and SLRL1 restriction of gibberellin responses in rice. Proc. Natl. Acad. Sci. USA 2008, 105, 16814–16819. [Google Scholar] [CrossRef] [Green Version]
- Lin, C.-C.; Chao, Y.-T.; Chen, W.-C.; Ho, H.-Y.; Chou, M.-Y.; Li, Y.-R.; Wu, Y.-L.; Yang, H.-A.; Hsieh, H.; Lin, C.-S.; et al. Regulatory cascade involving transcriptional and N-end rule pathways in rice under submergence. Proc. Natl. Acad. Sci. USA 2019, 116, 3300–3309. [Google Scholar] [CrossRef] [Green Version]
- Dubois, M.; Skirycz, A.; Claeys, H.; Maleux, K.; Dhondt, S.; De Bodt, S.; Bossche, R.V.; De Milde, L.; Yoshizumi, T.; Matsui, M.; et al. Ethylene response factor6 Acts as a Central Regulator of Leaf Growth under Water-Limiting Conditions in Arabidopsis. Plant Physiol. 2013, 162, 319–332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dubois, M.; Broeck, L.V.D.; Claeys, H.; Van Vlierberghe, K.; Matsui, M.; Inzé, D. The Ethylene Response factors ERF6 and ERF11 Antagonistically Regulate Mannitol-Induced Growth Inhibition in Arabidopsis. Plant Physiol. 2015, 169, 166–179. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Zhang, Z.-L.; Park, J.; Tyler, L.; Yusuke, J.; Qiu, K.; Nam, E.A.; Lumba, S.; Desveaux, D.; McCourt, P.; et al. The ERF11 Transcription Factor Promotes Internode Elongation by Activating Gibberellin Biosynthesis and Signaling. Plant Physiol. 2016, 171, 2760–2770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Broeck, L.V.D.; Dubois, M.; Vermeersch, M.; Storme, V.; Matsui, M.; Inzé, D. From network to phenotype: The dynamic wiring of an Arabidopsis transcriptional network induced by osmotic stress. Mol. Syst. Biol. 2017, 13, 961. [Google Scholar] [CrossRef]
- An, F.; Zhang, X.; Zhu, Z.; Ji, Y.; He, W.; Jiang, Z.; Li, M.; Guo, H. Coordinated regulation of apical hook development by gibberellins and ethylene in etiolated Arabidopsis seedlings. Cell Res. 2012, 22, 915–927. [Google Scholar] [CrossRef] [Green Version]
- Lefevere, H.; Bauters, L.; Gheysen, G. Salicylic Acid Biosynthesis in Plants. Front. Plant Sci. 2020, 11, 338. [Google Scholar] [CrossRef]
- Fu, Z.Q.; Yan, S.; Saleh, A.; Wang, W.; Ruble, J.; Oka, N.; Mohan, R.; Spoel, S.; Tada, Y.; Zheng, N.; et al. NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 2012, 486, 228–232. [Google Scholar] [CrossRef] [Green Version]
- Iqbal, N.; Masood, A.; Khan, N.A. Phytohormones in salinity tolerance: Ethylene and gibberellins cross talk. In Phytohormones and Abiotic Stress Tolerance in Plants; Springer: Berlin/Heidelberg, Germany, 2012; pp. 77–98. [Google Scholar]
- Khan, M.I.R.; Fatma, M.; Per, T.S.; Anjum, N.A.; Khan, N.A. Salicylic acid-induced abiotic stress tolerance and underlying mechanisms in plants. Front. Plant Sci. 2015, 6, 462. [Google Scholar] [CrossRef] [Green Version]
- Peng, Y.; Yang, J.; Li, X.; Zhang, Y. Salicylic Acid: Biosynthesis and Signaling. Annu. Rev. Plant Biol. 2021, 72, 761–791. [Google Scholar] [CrossRef]
- Khan, M.I.R.; Iqbal, N.; Masood, A.; Per, T.S.; Khan, N.A. Salicylic acid alleviates adverse effects of heat stress on photosynthesis through changes in proline production and ethylene formation. Plant Signal. Behav. 2013, 8, e26374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, M.I.R.; Asgher, M.; Khan, N.A. Alleviation of salt-induced photosynthesis and growth inhibition by salicylic acid involves glycinebetaine and ethylene in mungbean (Vigna radiata L.). Plant Physiol. Biochem. 2014, 80, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, W.; Imran, M.; Yaseen, M.; Haq, T.U.; Jamshaid, M.U.; Rukh, S.; Ikram, R.M.; Ali, M.; Ali, A.; Maqbool, M.; et al. Role of salicylic acid in regulating ethylene and physiological characteristics for alleviating salinity stress on germination, growth and yield of sweet pepper. PeerJ 2020, 8, e8475. [Google Scholar] [CrossRef] [PubMed]
- Nazar, R.; Umar, S.; Khan, N.; Sareer, O. Salicylic acid supplementation improves photosynthesis and growth in mustard through changes in proline accumulation and ethylene formation under drought stress. S. Afr. J. Bot. 2015, 98, 84–94. [Google Scholar] [CrossRef]
- Leslie, C.A.; Romani, R.J. Salicylic acid: A new inhibitor of ethylene biosynthesis. Plant Cell Rep. 1986, 5, 144–146. [Google Scholar] [CrossRef]
- Rao, M.V.; Lee, H.-I.; Davis, K.R. Ozone-induced ethylene production is dependent on salicylic acid, and both salicylic acid and ethylene act in concert to regulate ozone-induced cell death. Plant J. 2002, 32, 447–456. [Google Scholar] [CrossRef] [Green Version]
- Ogawa, D.; Nakajima, N.; Sano, T.; Tamaoki, M.; Aono, M.; Kubo, A.; Kanna, M.; Ioki, M.; Kamada, H.; Saji, H. Salicylic Acid Accumulation Under O3 Exposure is Regulated by Ethylene in Tobacco Plants. Plant Cell Physiol. 2005, 46, 1062–1072. [Google Scholar] [CrossRef]
- Thaler, J.S.; Humphrey, P.T.; Whiteman, N.K. Evolution of jasmonate and salicylate signal crosstalk. Trends Plant Sci. 2012, 17, 260–270. [Google Scholar] [CrossRef]
- Kieber, J.J.; Schaller, G.E. Cytokinin signaling in plant development. Development 2018, 145, dev149344. [Google Scholar] [CrossRef] [Green Version]
- Hutchison, C.E.; Li, J.; Argueso, C.; Gonzalez, M.; Lee, E.; Lewis, M.W.; Maxwell, B.B.; Perdue, T.D.; Schaller, G.E.; Alonso, J.M.; et al. The Arabidopsis Histidine Phosphotransfer Proteins Are Redundant Positive Regulators of Cytokinin Signaling. Plant Cell 2006, 18, 3073–3087. [Google Scholar] [CrossRef] [Green Version]
- Zwack, P.J.; Rashotte, A.M. Interactions between cytokinin signalling and abiotic stress responses. J. Exp. Bot. 2015, 66, 4863–4871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vogel, J.; Woeste, K.E.; Theologis, A.; Kieber, J.J. Recessive and dominant mutations in the ethylene biosynthetic gene ACS5 of Arabidopsis confer cytokinin insensitivity and ethylene overproduction, respectively. Proc. Natl. Acad. Sci. USA 1998, 95, 4766–4771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chae, H.S.; Faure, F.; Kieber, J.J. The eto1, eto2, and eto3 Mutations and Cytokinin Treatment Increase Ethylene Biosynthesis in Arabidopsis by Increasing the Stability of ACS Protein. Plant Cell 2003, 15, 545–559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hansen, M.; Chae, H.S.; Kieber, J.J. Regulation of ACS protein stability by cytokinin and brassinosteroid. Plant J. 2009, 57, 606–614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jalmi, S.K.; Bhagat, P.K.; Verma, D.; Noryang, S.; Tayyeba, S.; Singh, K.; Sharma, D.; Sinha, A.K. Traversing the Links between Heavy Metal Stress and Plant Signaling. Front. Plant Sci. 2018, 9, 12. [Google Scholar] [CrossRef] [PubMed]
- Artner, C.; Benkova, E. Ethylene and Cytokinin: Partners in Root Growth Regulation. Mol. Plant 2019, 12, 1312–1314. [Google Scholar] [CrossRef]
- Skalak, J.; Nicolas, K.L.; Vankova, R.; Hejatko, J. Signal Integration in Plant Abiotic Stress Responses via Multistep Phosphorelay Signaling. Front. Plant Sci. 2021, 12. [Google Scholar] [CrossRef]
- Yamauchi, T.; Yoshioka, M.; Fukazawa, A.; Mori, H.; Nishizawa, N.K.; Tsutsumi, N.; Yoshioka, H.; Nakazono, M. An NADPH Oxidase RBOH Functions in Rice Roots during Lysigenous Aerenchyma Formation under Oxygen-Deficient Conditions. Plant Cell 2017, 29, 775–790. [Google Scholar] [CrossRef] [Green Version]
- Yamauchi, T.; Tanaka, A.; Inahashi, H.; Nishizawa, N.K.; Tsutsumi, N.; Inukai, Y.; Nakazono, M. Fine control of aerenchyma and lateral root development through AUX/IAA- and ARF-dependent auxin signaling. Proc. Natl. Acad. Sci. USA 2019, 116, 20770–20775. [Google Scholar] [CrossRef] [Green Version]
- Tavares, E.Q.P.; Grandis, A.; Lembke, C.G.; Souza, G.M.; Purgatto, E.; De Souza, A.P.; Buckeridge, M.S. Roles of auxin and ethylene in aerenchyma formation in sugarcane roots. Plant Signal. Behav. 2018, 13, e1422464. [Google Scholar] [CrossRef]
- Ponce, G.; Barlow, P.W.; Feldman, L.J.; Cassab, G.I. Auxin and ethylene interactions control mitotic activity of the quiescent centre, root cap size, and pattern of cap cell differentiation in maize. Plant Cell Environ. 2005, 28, 719–732. [Google Scholar] [CrossRef]
- Habben, J.E.; Bao, X.; Bate, N.J.; DeBruin, J.L.; Dolan, D.; Hasegawa, D.; Helentjaris, T.G.; Lafitte, R.H.; Lovan, N.; Mo, H.; et al. Transgenic alteration of ethylene biosynthesis increases grain yield in maize under field drought-stress conditions. Plant Biotechnol. J. 2014, 12, 685–693. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, X.; Zhao, Y.; Li, Y.; Zhang, G.; Peng, Z.; Zhang, J. Enhancing auxin accumulation in maize root tips improves root growth and dwarfs plant height. Plant Biotechnol. J. 2017, 16, 86–99. [Google Scholar] [CrossRef] [PubMed]
- Emerick, K.; Ronald, P.C. Sub1 Rice: Engineering Rice for Climate Change. Cold Spring Harb. Perspect. Biol. 2019, 11, a034637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Licausi, F.; Kosmacz, M.; Weits, D.A.; Giuntoli, B.; Giorgi, F.M.; Voesenek, L.A.C.J.; Perata, P.; van Dongen, J.T. Oxygen sensing in plants is mediated by an N-end rule pathway for protein destabilization. Nature 2011, 479, 419–422. [Google Scholar] [CrossRef]
- Gasch, P.; Fundinger, M.; Müller, J.T.; Lee, T.; Bailey-Serres, J.; Mustroph, A. Redundant ERF-VII Transcription Factors Bind to an Evolutionarily Conserved cis-Motif to Regulate Hypoxia-Responsive Gene Expression in Arabidopsis. Plant Cell 2015, 28, 160–180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perata, P. Ethylene Signaling Controls Fast Oxygen Sensing in Plants. Trends Plant Sci. 2019, 25, 3–6. [Google Scholar] [CrossRef] [PubMed]
- Lamers, J.; van der Meer, T.; Testerink, C. How Plants Sense and Respond to Stressful Environments. Plant Physiol. 2020, 182, 1624–1635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, J.-H.; Domijan, M.; Klose, C.; Biswas, S.; Ezer, D.; Gao, M.; Khattak, A.K.; Box, M.S.; Charoensawan, V.; Cortijo, S.; et al. Phytochromes function as thermosensors in Arabidopsis. Science 2016, 354, 886–889. [Google Scholar] [CrossRef] [Green Version]
- Legris, M.; Klose, C.; Burgie, E.S.; Rojas, C.C.R.; Neme, M.; Hiltbrunner, A.; Wigge, P.A.; Schäfer, E.; Vierstra, R.D.; Casal, J.J. Phytochrome B integrates light and temperature signals in Arabidopsis. Science 2016, 354, 897–900. [Google Scholar] [CrossRef] [Green Version]
- Yuan, F.; Yang, H.; Xue, Y.; Kong, D.; Ye, R.; Li, C.; Zhang, J.; Theprungsirikul, J.; Shrift, T.; Krichilsky, B.; et al. OSCA1 mediates osmotic-stress-evoked Ca2+ increases vital for osmosensing in Arabidopsis. Nature 2014, 514, 367–371. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Lv, J.; Shi, Y.; Gao, J.; Hua, J.; Song, C.; Gong, Z.; Yang, S. EGR 2 phosphatase regulates OST 1 kinase activity and freezing tolerance in Arabidopsis. EMBO J. 2018, 38, e99819. [Google Scholar] [CrossRef]
- Jiang, Z.; Zhou, X.; Tao, M.; Yuan, F.; Liu, L.; Wu, F.; Wu, X.; Xiang, Y.; Niu, Y.; Liu, F.; et al. Plant cell-surface GIPC sphingolipids sense salt to trigger Ca2+ influx. Nature 2019, 572, 341–346. [Google Scholar] [CrossRef] [PubMed]
- Huot, B.; Yao, J.; Montgomery, B.L.; He, S.Y. Growth–Defense Tradeoffs in Plants: A Balancing Act to Optimize Fitness. Mol. Plant 2014, 7, 1267–1287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
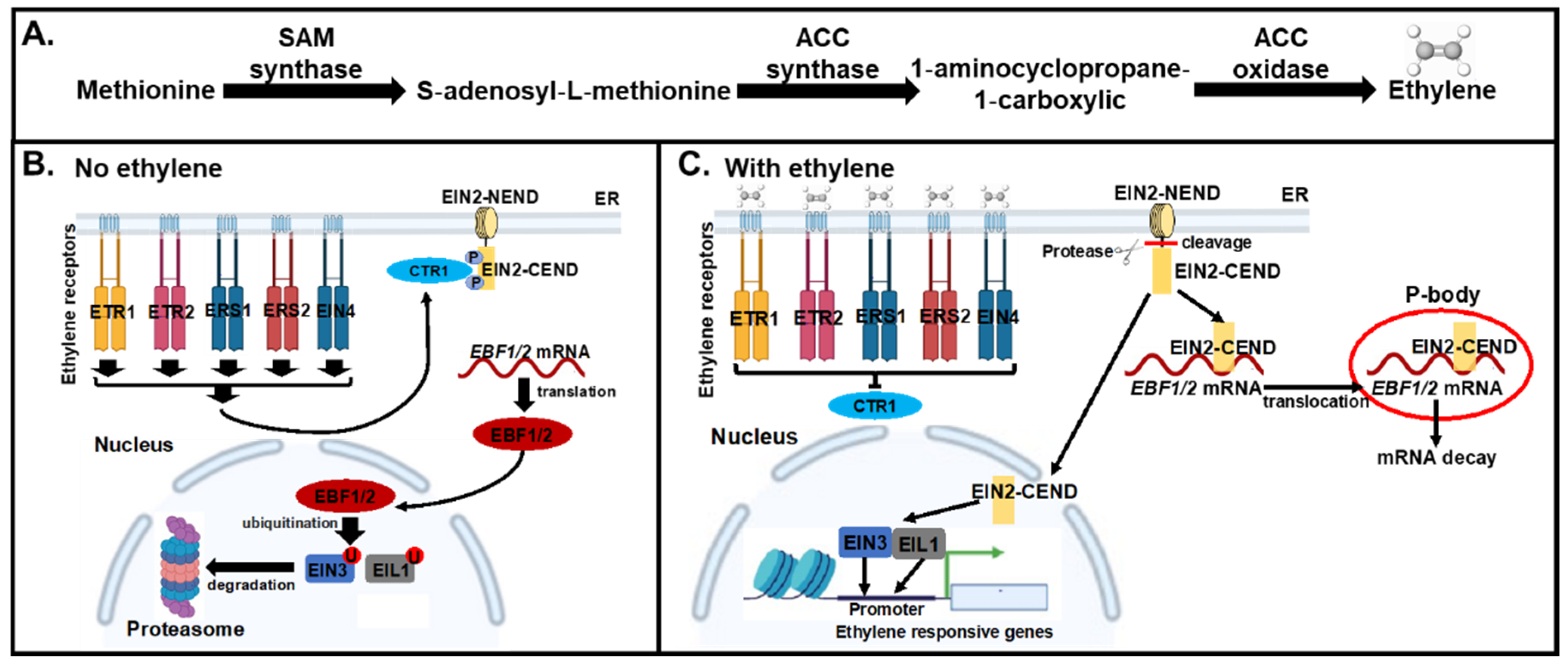
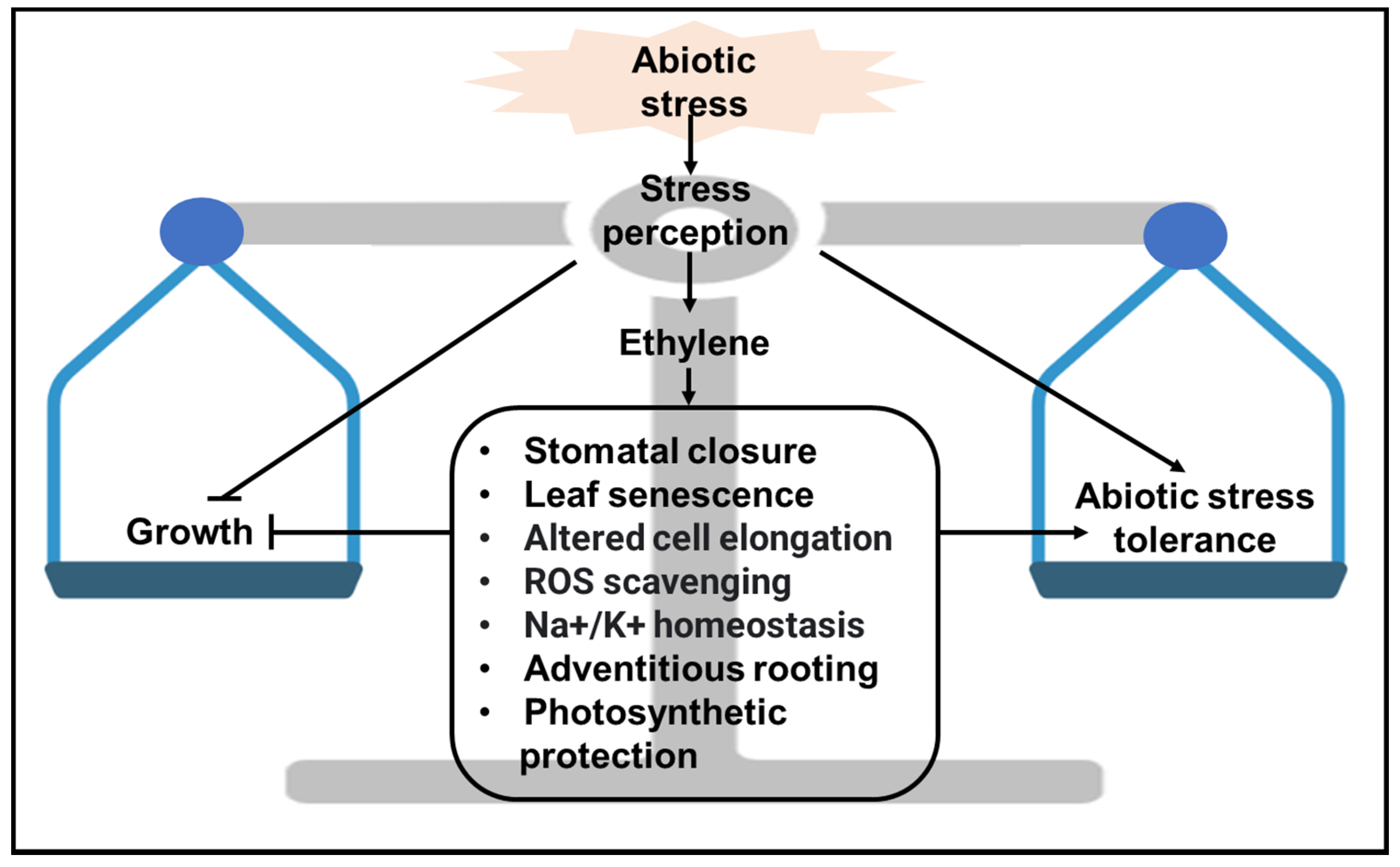
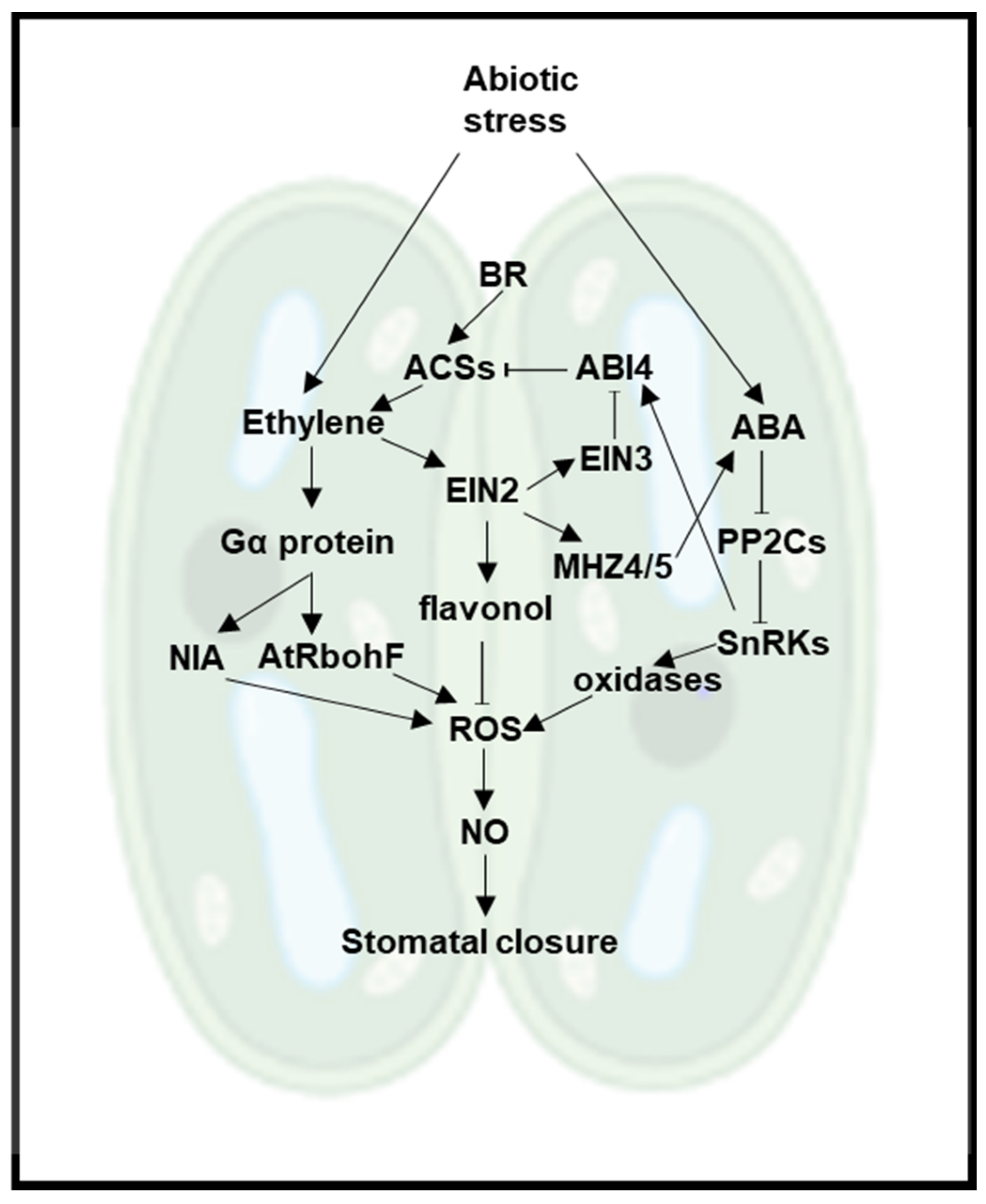
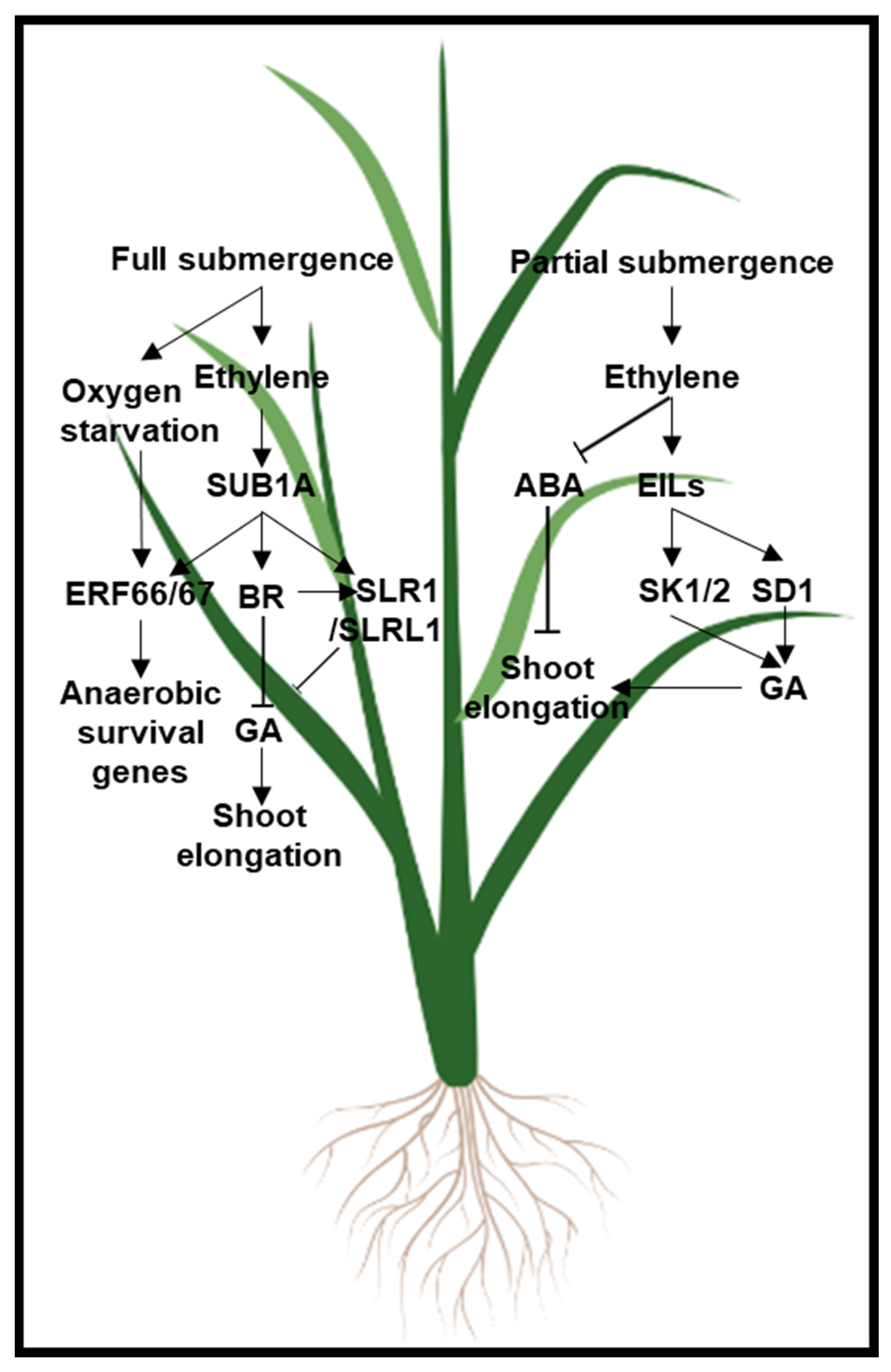
| Stress Type | Regulation | Ethylene Biosynthetic Genes | Ethylene Signaling Genes |
|---|---|---|---|
| Drought stress | Up | ACS8, 12 [37,38] | |
| Down | ACO2, 4, 5, ACS10, 11 [38,39] | ||
| Flooding/hypoxia stress | Up | ACS2, 6, 7, 9, 11 [40,41] | EIN3 [42] |
| Down | |||
| Osmotic stress | Up | ACS7, ACO2 [43,44] | ETR2, ERS1, CTR1, EIN3/EIL1, EBF1/2 [44,45] |
| Down | ETR1, EIN2 [46,47] | ||
| Salt stress | Up | ACS2, 6, 7, 11, ACO2, 4 [47,48] | EIN3 [49] |
| Down | ETR1, EIN2 [45,46] | ||
| Heat stress | Up | ACS6, 7, 8, 10, 11, 12, ACO2, 4 [43,50] | ERS2, ETR2, EIN3 [50] |
| Down | ACS2, 4, 5, ACO1, 3 [50] | ERS1, CTR1, EIN2 [50] | |
| Cold stress | Up | ACS2, 11, ACO2 [41,47] | EIN3 [51] |
| Down | ETR1, EIN4, EIN2, EBF1/2 [51] | ||
| Heavy metal stress | Up | ACS2, 5, 6, ACO1, 2 [52,53,54] | EIN2 [55] |
| Down |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, H.; Bullock, D.A., Jr.; Alonso, J.M.; Stepanova, A.N. To Fight or to Grow: The Balancing Role of Ethylene in Plant Abiotic Stress Responses. Plants 2022, 11, 33. https://doi.org/10.3390/plants11010033
Chen H, Bullock DA Jr., Alonso JM, Stepanova AN. To Fight or to Grow: The Balancing Role of Ethylene in Plant Abiotic Stress Responses. Plants. 2022; 11(1):33. https://doi.org/10.3390/plants11010033
Chicago/Turabian StyleChen, Hao, David A. Bullock, Jr., Jose M. Alonso, and Anna N. Stepanova. 2022. "To Fight or to Grow: The Balancing Role of Ethylene in Plant Abiotic Stress Responses" Plants 11, no. 1: 33. https://doi.org/10.3390/plants11010033
APA StyleChen, H., Bullock, D. A., Jr., Alonso, J. M., & Stepanova, A. N. (2022). To Fight or to Grow: The Balancing Role of Ethylene in Plant Abiotic Stress Responses. Plants, 11(1), 33. https://doi.org/10.3390/plants11010033






