Rapid Generation and Analysis of a Barley Doubled Haploid Line with Higher Nitrogen Use Efficiency Than Parental Lines by F1 Microspore Embryogenesis
Abstract
1. Introduction
2. Results
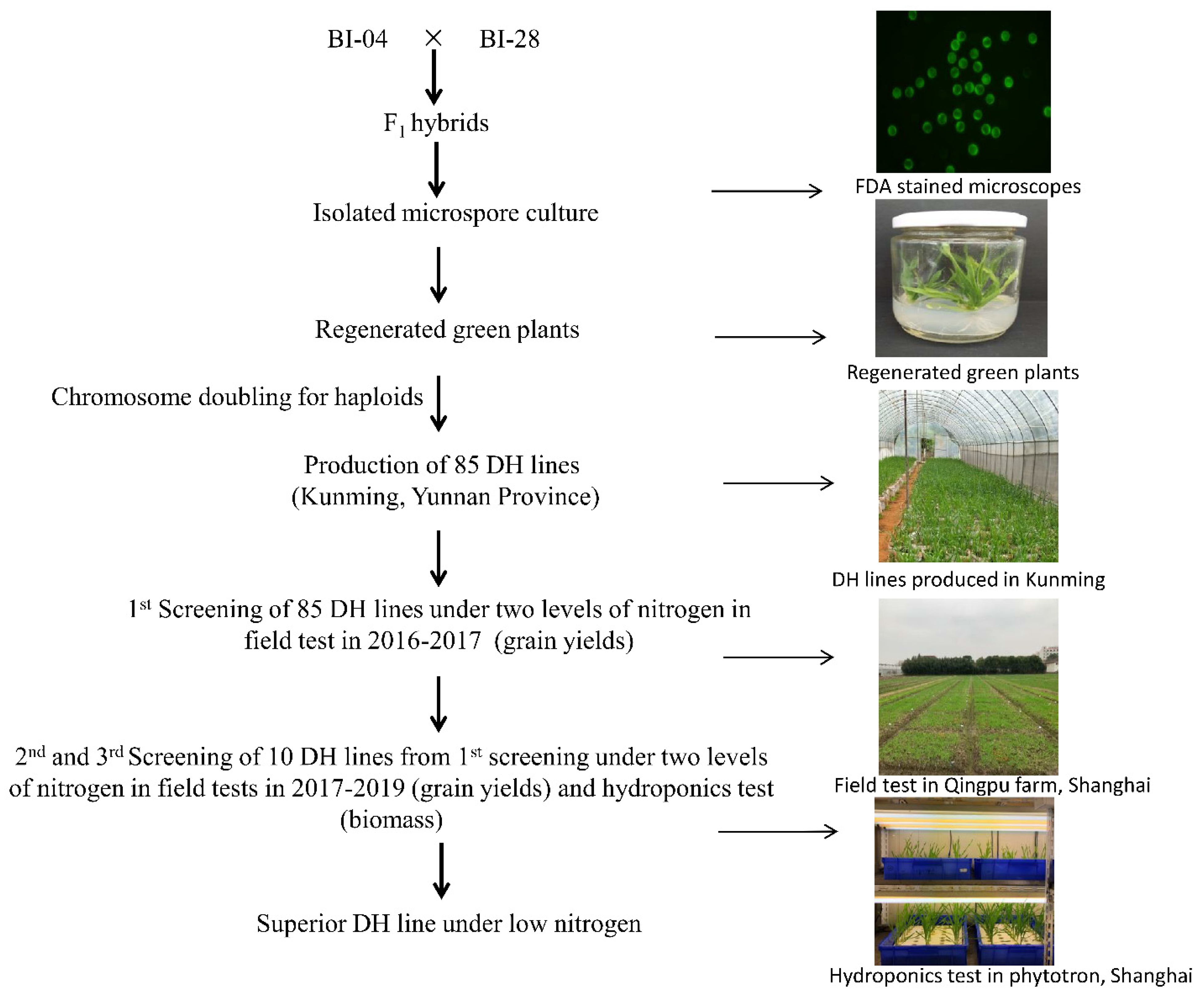
2.1. DH Lines Produced by a Cross Combined with F1 Isolated Microspore Culture
2.2. Identification of DH45 by Screening the DH Population for Lines with Higher NUE Than the Parents in Field Experiments
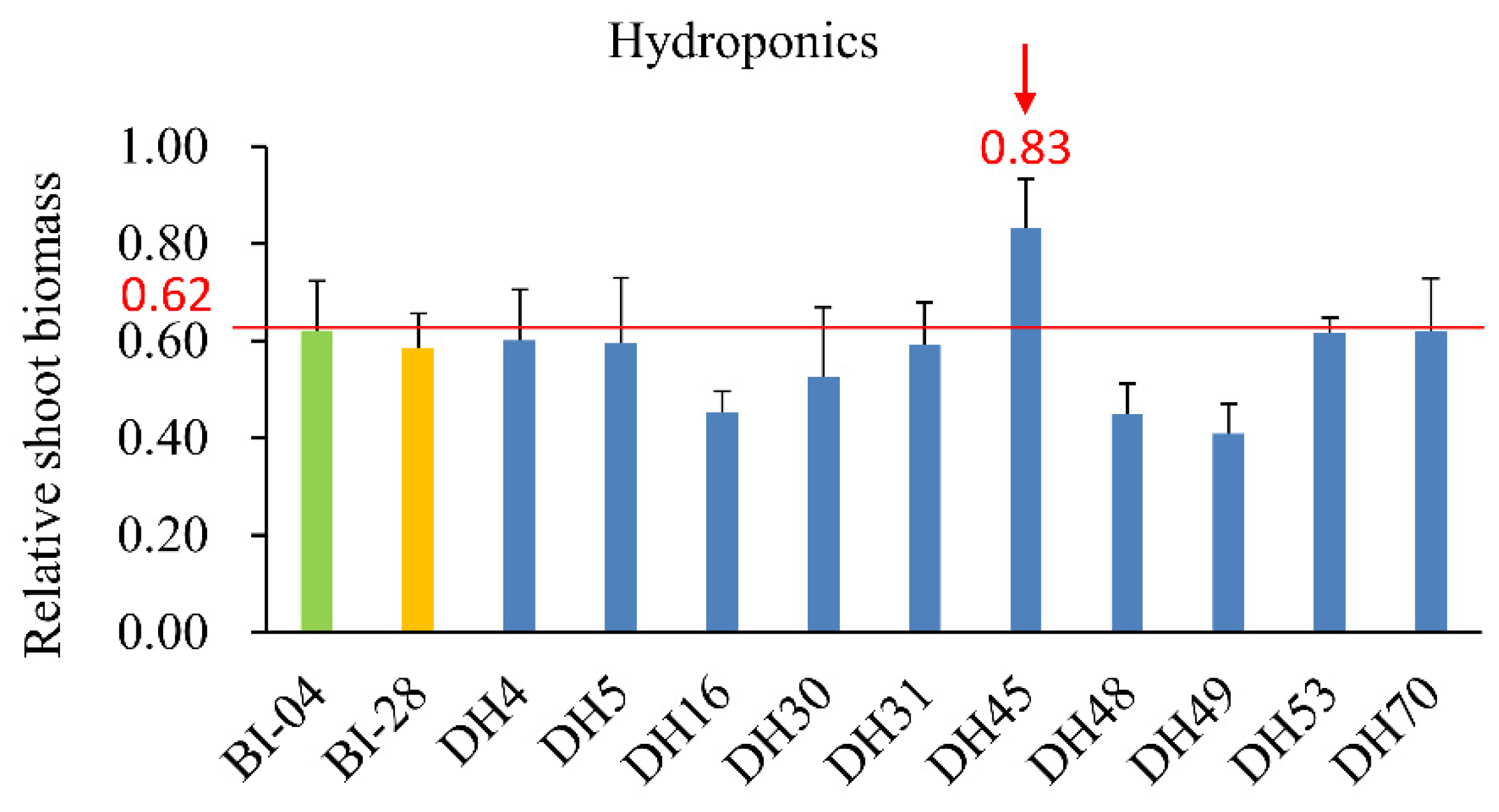
2.3. Comparing the NUE of DH45 and Its Parents in Hydroponics Experiments
2.4. Transcriptomic Profiling of DH45 and its Parents
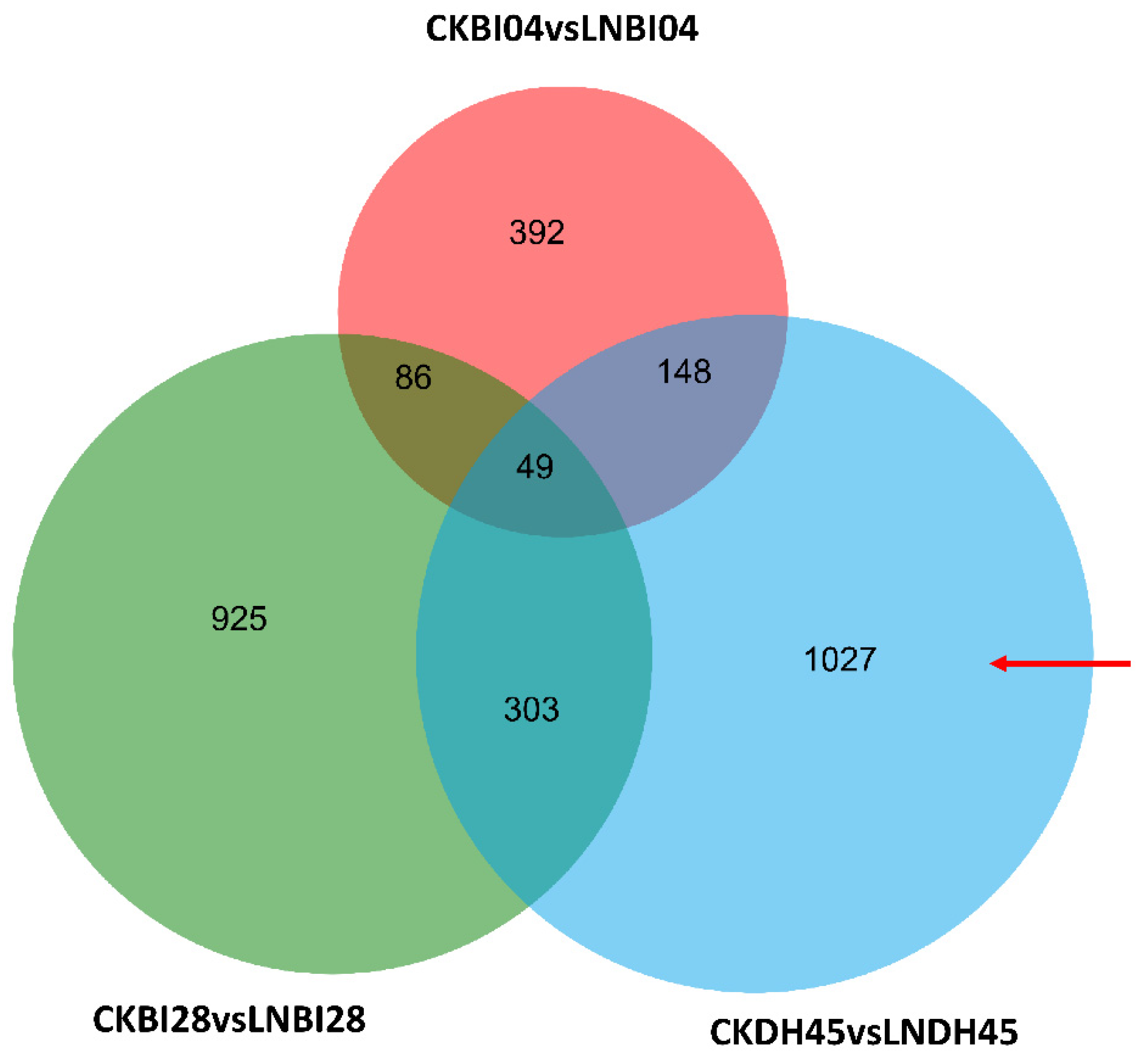
2.5. Identification of Specific Differentially Expressed Genes (DEGs) in DH45 under LN Conditions
2.6. Functional Classification of the 1027 DEGHP in DH45 by GO and KEGG Enrichment Analysis
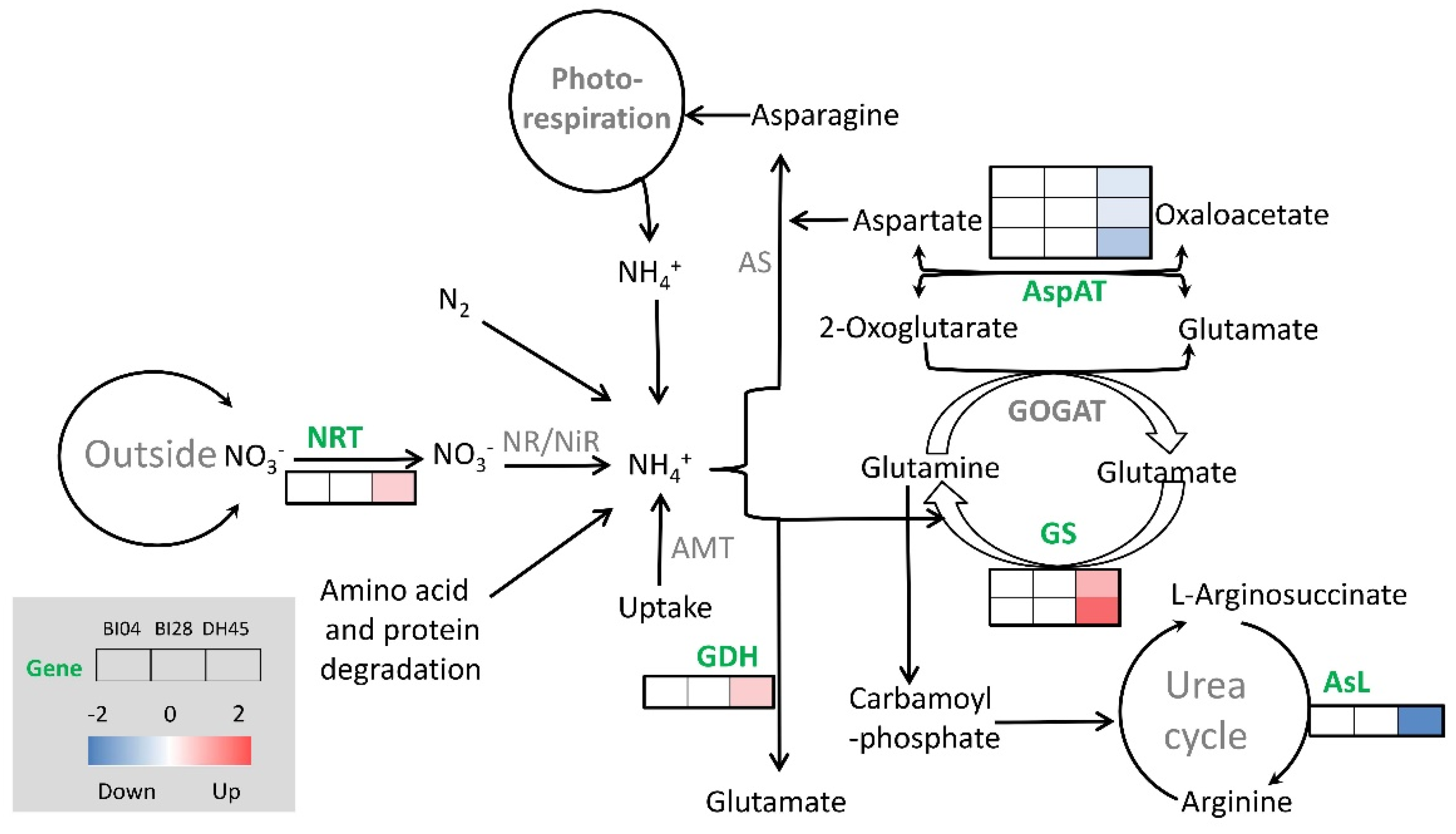
2.7. Expression Profiles of Eight Specific DEGHP Involved in Nitrogen Metabolism
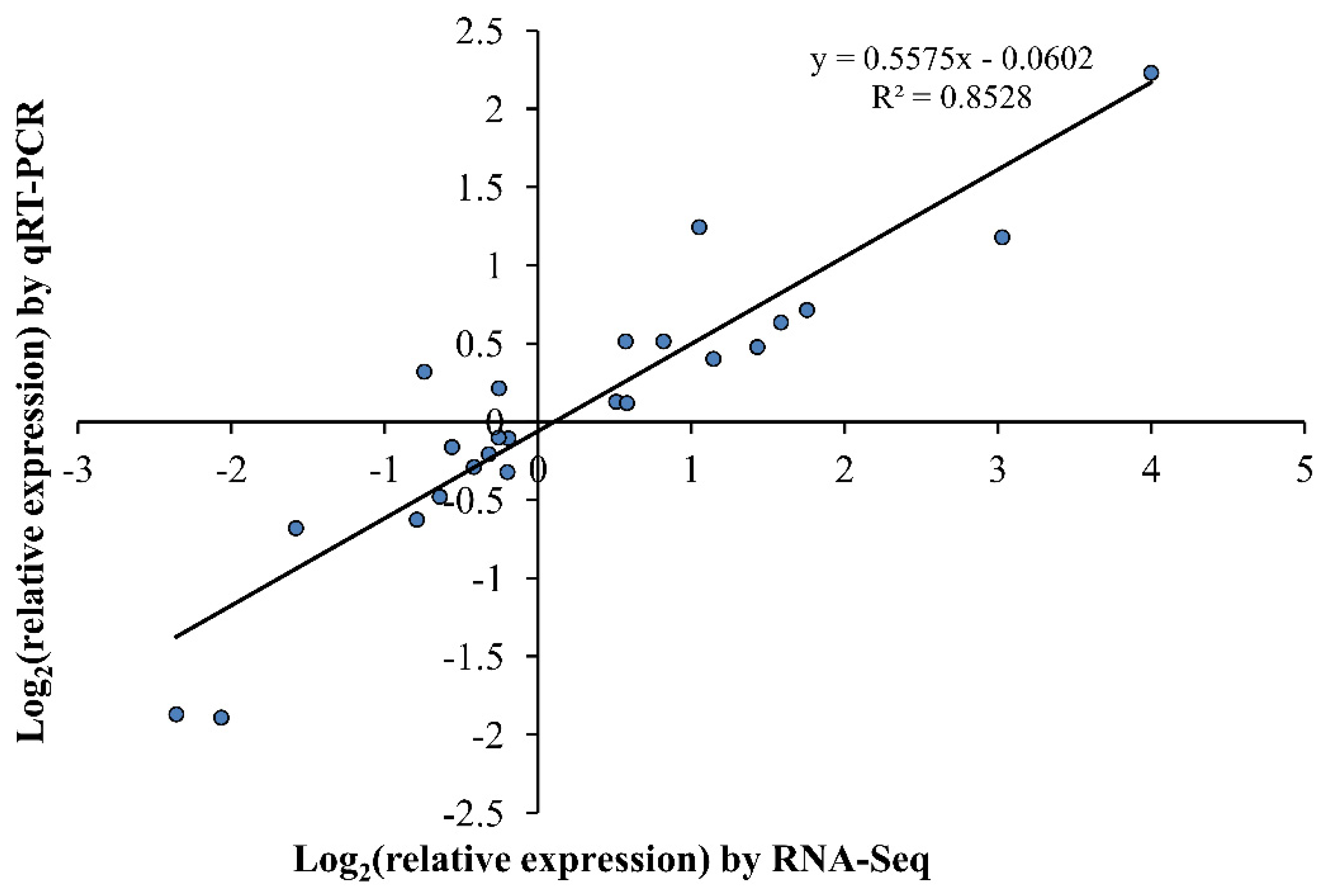
2.8. Validation of the Expression Profiles of Eight DEGHP Involved in Nitrogen Metabolism by qRT-PCR
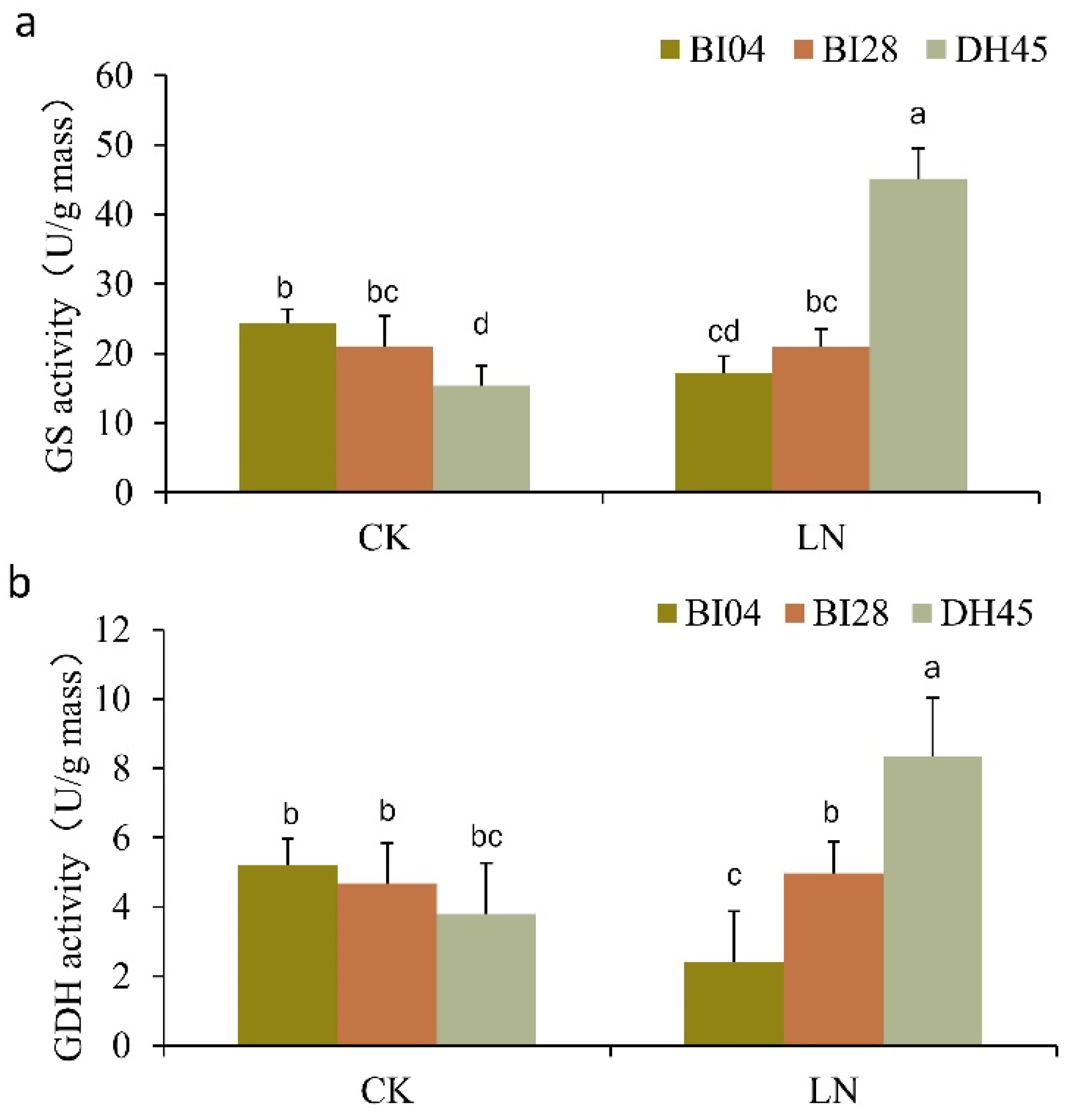
2.9. GS and GDH Activities of DH45 and Its Parents
3. Discussion
3.1. Production of Homozygous Lines for NUE Improvement by Crossing Combined with F1 Microspore Embryogenesis
3.2. Biological Response to Low-Nitrogen Stress in the Superior Line DH45
3.3. Exploration of Genes Potentially Responsible for NUE Improvement in the Superior Line DH45
4. Materials and Methods
4.1. Plant Materials
4.2. Isolated Microspore Culture
4.3. Field and Hydroponic Culture Screening
4.4. Measurement of Biomass and NUE
4.5. RNA Sampling, Sequencing, and Reads Mapping
4.6. Identification of DEGs and Functional Analysis
4.7. Validation of RNA-Seq by qRT-PCR
4.8. Quantification of GS and GDH Activities
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CK | Control |
| LN | Low nitrogen |
| GY | Grain yield per plant |
| NUE | Nitrogen use efficiency |
| DH | Doubled haploid |
| HPD | Over best parent value |
| MPD | Over middle parent value |
| GO | Gene Ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| DEGs | Differentially expressed genes |
| NRQ | Normalized relative quantity |
References
- Stahl, A.; Vollrath, P.; Samans, B.; Frisch, M.; Wittkop, B.; Snowdon, R.J. Effect of breeding on nitrogen use efficiency-associated traits in oilseed rape. J. Exp. Bot. 2019, 70, 1969–1986. [Google Scholar] [CrossRef]
- Jez, J.M.; Lee, S.G.; Sherp, A.M. The next green movement: Plant biology for the environment and sustainability. Science 2016, 353, 1241–1244. [Google Scholar] [CrossRef]
- Ali, J.; Jewel, Z.A.; Mahender, A.; Anandan, A.; Hernandez, J.; Li, Z. Molecular genetics and breeding for nutrient use efficiency in rice. Int. J. Mol. Sci. 2018, 19, 1762. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Xu, S.; Jiang, C.; Sun, H.; Feng, S.; Zhou, S.; Zhuang, G.; Bai, Z.; Zhuang, X. Transcriptomic sequencing and coexpression network analysis on key genes and pathways regulating nitrogen use efficiency in Myriophyllum aquaticum. Int. J. Mol. Sci. 2019, 20, 1587. [Google Scholar] [CrossRef] [PubMed]
- Tilman, D.; Reich, P.B.; Knops, J.; Wedin, D.; Mielke, T.; Lehman, C. Diversity and productivity in a long-term grassland experiment. Science 2001, 294, 843–845. [Google Scholar] [CrossRef] [PubMed]
- Ray, D.K.; Ramankutty, N.; Mueller, N.D.; West, P.C.; Foley, J.A. Recent patterns of crop yield growth and stagnation. Nat. Commun. 2012, 3, 1293. [Google Scholar] [CrossRef]
- Rothstein, S.J. Returning to our roots: Making plant biology research relevant to future challenges in agriculture. Plant Cell 2007, 19, 2695–2699. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, G.; Ding, G.; Li, L.; Cai, H.; Ye, X.; Zou, J.; Xu, F. Identification and characterization of improved nitrogen efficiency in interspecific hybridized new-type Brassica napus. Ann. Bot. 2014, 114, 549–559. [Google Scholar] [CrossRef]
- Huang, A.; Sang, Y.; Sun, W.; Fu, Y.; Yang, Z. Transcriptomic analysis of responses to imbalanced carbon: Nitrogen availabilities in rice seedlings. PLoS ONE 2016, 11, e0165732. [Google Scholar]
- Ren, J.; Wu, P.; Trampe, B.; Tian, X.; Chen, S. Novel technologies in doubled haploid line development. Plant Biotechnol. J. 2017, 15, 1361–1370. [Google Scholar] [CrossRef]
- Ma, H.; Li, G.; Wurschum, T.; Zhang, Y.; Zheng, D.; Yang, X.; Li, J.; Liu, W.; Yan, J.; Chen, S. Genome-wide association study of haploid male fertility in maize (Zea Mays L.). Front. Plant Sci. 2018, 9, 974. [Google Scholar] [CrossRef]
- Castillo, A.M.; Cistué, L.; Vallés, M.P.; Sanz, J.M.; Romagosa, I.; Molina-Cano, J.L. Efficient production of androgenic doubled-haploid mutants in barley by the application of sodium azide to anther and microspore cultures. Plant Cell Rep. 2001, 20, 105–111. [Google Scholar] [CrossRef]
- Szarejko, I.; Forster, B.P. Doubled haploidy and induced mutation. Euphytica 2007, 158, 359–370. [Google Scholar] [CrossRef]
- Soriano, M.; Li, H.; Boutilier, K. Microspore embryogenesis: Establishment of embryo identity and pattern in culture. Plant Reprod. 2013, 26, 181–196. [Google Scholar] [CrossRef]
- Gao, R.; Guo, G.; Fang, C.; Huang, S.; Chen, J.; Lu, R.; Huang, J.; Fan, X.; Liu, C. Rapid generation of barley mutant lines with high nitrogen uptake efficiency by microspore mutagenesis and field screening. Front. Plant Sci. 2018, 9, 450. [Google Scholar] [CrossRef]
- Dwivedi, S.L.; Britt, A.B.; Tripathiet, L.; Sharma, S.; Upadhyaya, H.; Ortiz, R. Haploids: Constraints and opportunities in plant breeding. Biotechnol. Adv. 2015, 33, 812–829. [Google Scholar] [CrossRef]
- Han, M.; Wong, J.; Su, T.; Beatty, P.H.; Good, A.G. Identification of nitrogen use efficiency genes in barley: Searching for QTLs controlling complex physiological traits. Front. Plant Sci. 2016, 7, 1587. [Google Scholar] [CrossRef]
- Mascher, M.; Gundlach, H.; Himmelbach, A.; Beier, S.; Twardziok, S.O.; Wicker, T.; Radchuk, V.; Dockter, C.; Hedley, P.E.; Russell, J.; et al. Chromosome conformation capture ordered sequence of the barley genome. Nature 2017, 544, 427–433. [Google Scholar] [CrossRef] [PubMed]
- Eudes, F.; Shim, Y.S.; Jiang, F. Engineering the haploid genome of microspores. Biocatal. Agric. Biotechnol. 2014, 3, 20–23. [Google Scholar] [CrossRef]
- Beaith, M.E.; Fletcher, R.S.; Kott, L.S. Reduction of saturated fats by mutagenesis and heat selection in Brassica napus L. Euphytica 2005, 144, 1–9. [Google Scholar] [CrossRef]
- Quan, X.; Zeng, J.; Chen, G.; Zhang, G. Transcriptomic analysis reveals adaptive strategies to chronic low nitrogen in Tibetan wild barley. BMC Plant Biol. 2019, 19, 68. [Google Scholar] [CrossRef]
- Li, H.; Hu, B.; Chu, C. Nitrogen use efficiency in crops: Lessons from Arabidopsis and rice. J. Exp. Bot. 2017, 68, 2477–2488. [Google Scholar] [CrossRef] [PubMed]
- Bi, Y.; Meyer, A.; Downs, G.S.; Shi, X.; EI-kereamy, A.; Lukens, L.; Rothstein, S.J. High throughput RNA sequencing of a hybrid maize and its parents shows different mechanisms responsive to nitrogen limitation. BMC Genom. 2014, 15, 77. [Google Scholar] [CrossRef]
- Goel, P.; Sharma, N.K.; Bhuria, M.; Sharma, V.; Chauhan, R.; Pathania, S.; Swarnkar, M.K.; Chawla, V.; Acharya, V.; Shankar, R.; et al. Transcriptome and co-expression network analyses identify key genes regulating nitrogen use efficiency in Brassica juncea L. Sci. Rep. 2018, 8, 7451. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Osuna, A.; Calatrava, V.; Galvan, A.; Fernandez, E.; Liamas, A. Identification of the MAPK cascade and its relationship with nitrogen metabolism in the green Alga Chlamydomonas reinhardtii. Int. J. Mol. Sci. 2020, 21, 3417. [Google Scholar] [CrossRef] [PubMed]
- Ho, C.-H.; Lin, S.; Hu, H.; Tsay, Y. CHL1 Functions as a Nitrate Sensor in Plants. Cell 2009, 138, 1184–1194. [Google Scholar] [CrossRef]
- Wang, P.; Du, Y.; Li, Y.; Ren, D.; Song, C. Hydrogen peroxide–mediated activation of MAP Kinase 6 modulates nitric oxide biosynthesis and signal transduction in Arabidopsis. Plant Cell 2010, 22, 2981–2998. [Google Scholar] [CrossRef] [PubMed]
- Forde, B.G.; Cutler, S.R.; Zaman, N.; Krysan, P.J. Glutamate signalling via a MEKK1 kinase-dependent pathway induces changes in Arabidopsis root architecture. Plant J. 2013, 75, 1–10. [Google Scholar] [CrossRef]
- Siddappaa, S.; Marathe, G.K. What we know about plant arginases? Plant Physiol. Biochem. 2020, 156, 600–610. [Google Scholar] [CrossRef]
- Ca’novas, F.M.; Avila, C.; Cantón, F.R.; Rafael, A.; Cañas, R.A.; Torre, F.D.L. Ammonium assimilation and amino acid metabolism in conifers. J. Exp. Bot. 2007, 59, 2307–2318. [Google Scholar] [CrossRef] [PubMed]
- Llácer, J.L.; Fita, I.; Rubio, V. Arginine and nitrogen storage. Curr. Opin. Struct. Biol. 2008, 18, 673–681. [Google Scholar] [CrossRef]
- Seebauer, J.R.; Moose, S.P.; Fabbri, B.J.; Crossland, L.D.; Below, F.E. Amino acid metabolism in maize earshoots. Implications for assimilate preconditioning and nitrogen signaling. Plant Physiol. 2004, 136, 4326–4334. [Google Scholar] [CrossRef] [PubMed]
- Quan, X.; Zeng, J.; Ye, L.; Chen, G.; Han, Z.; Shah, J.M.; Zhang, G. Transcriptome profiling analysis for two tibetan wild barley genotypes in responses to low nitrogen. BMC Plant Biol. 2016, 16, 30. [Google Scholar] [CrossRef] [PubMed]
- Gelli, M.; Mitchell, S.E.; Liu, K.; Clemente, T.E.; Weeks, D.P.; Zhang, C.; Holding, D.R.; Dweikat, I.M. Mapping QTLs and association of differentially expressed gene transcripts for multiple agronomic traits under different nitrogen levels in soghum. BMC Plant Biol. 2016, 16, 16. [Google Scholar] [CrossRef]
- Forde, B.G.; Lea, P.J. Glutamate in plants: Metabolism, regulation, and signalling. J. Exp. Bot. 2007, 58, 2339–2358. [Google Scholar] [CrossRef] [PubMed]
- Lezhneva, L.; Kiba, T.; Feria-Bourrellier, A.B.; Lafouge, F.; Boutet-Mercey, S.; Zoufan, P.; Sakakibara, H.; Daniel-Vedele, F.; Krapp, A. The Arabidopsis nitrate transporter NRT2.5 plays a role in nitrate acquisition and remobilization in nitrogen-starved plants. Plant J. 2014, 80, 230–241. [Google Scholar] [CrossRef]
- Xu, G.; Fan, X.; Miller, A.J. Plant nitrogen assimilation and use efficiency. Annu. Rev. Plant Biol. 2012, 63, 153–182. [Google Scholar] [CrossRef]
- Miflin, B.J.; Habash, D.Z. The role of glutamine synthetase and glutamate dehydrogenase in nitrogen assimilation and possibilities for improvement in the nitrogen utilization of crops. J. Exp. Bot. 2002, 53, 979–987. [Google Scholar] [CrossRef] [PubMed]
- Gaufichon, L.; Rothstein, S.J.; Suzuki, A. Asparagine Metabolic Pathways in Arabidopsis. Plant Cell Physiol. 2016, 57, 675–689. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Liu, C.; Wang, Y.; He, T.; Gao, R.; Xu, H.; Guo, G.; Li, Y.; Zhou, L.; Lu, R.; et al. Expression analysis of nitrogen metabolism-related genes reveal differences in adaptation to low-nitrogen stress between two different barley cultivars at seedling stage. Int. J. Genom. 2018, 2018, 8152860. [Google Scholar] [CrossRef]
- Xu, H.; Liu, C.; Lu, R.; Guo, G.; Chen, Z.; He, T.; Gao, R.; Li, Y.; Huang, J. The difference in responses to nitrogen deprivation and re-supply at seedling stage between two barley genotypes differing nitrogen use efficiency. Plant Growth Regul. 2016, 79, 119–126. [Google Scholar] [CrossRef]
- Lu, R.; Wang, Y.; Sun, Y.; Shan, L.; Chen, P.; Huang, J. Improvement of isolated microspore culture of barley (Hordeum vulgare L.): The effect of floret co-culture. Plant Cell Tiss Organ Cult. 2008, 93, 21–27. [Google Scholar] [CrossRef]
- Liu, C.; Lu, R.; Guo, G.; He, T.; Li, Y.; Xu, H.; Gao, R.; Chen, Z.; Huang, J. Transcriptome analysis reveals translational regulation in barley microspore-derived embryogenic callus under salt stress. Plant Cell Rep. 2016, 35, 1719–1728. [Google Scholar] [CrossRef]
- De Macale, M.A.R.; Velk, P.L.G. The role of Azolla covers in improving the nitrogen use efficiency of lowland rice. Plant Soil 2004, 263, 311–321. [Google Scholar] [CrossRef]
- Moll, R.H.; Kamprath, E.J.; Jackson, W.A. Analysis and interpretation of factors which contribute to efficiency of nitrogen utilization1. Agron. J. 1982, 74, 562–564. [Google Scholar] [CrossRef]
- Zhai, R.; Feng, Y.; Wang, H.; Zhan, X.; Shen, X.; Wu, W.; Zhang, Y.; Chen, D.; Dai, G.; Yang, Z.; et al. Transcriptome analysis of rice root heterosis by RNA-Seq. BMC Genom. 2013, 14, 19. [Google Scholar] [CrossRef]
- Monat, C.; Padmarasu, S.; Lux, T.M.; Wicker, T.; Gundlach, H.; Himmelbach, A.; Ens, J.; Li, C.; Muehlbauer, G.J.; Schulman, A.H.; et al. TRITEX: Chromosome-scale sequence assembly of Triticeae genomes with open-source tools. Genome Biol. 2019, 20, 284. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Li, Y.; Liu, C.; Wang, Y.; He, T.; Guo, G.; Fang, C.; Gao, R.; Xu, H.; Zhou, L.; et al. Reference gene selection for quantitative RT-PCR normalisation in barley under low-nitrogen stress, based on RNAseq data. J. Cereal Sci. 2018, 82, 213–215. [Google Scholar] [CrossRef]



| Genotype | NUE/(g.g−1) | Relative Value of NUE | |
|---|---|---|---|
| Control (CK) | Low Nitrogen (LN) | ||
| BI-04 | 23.95 ± 0.83 ab | 45.8 ± 4.16 a | 1.91 |
| BI-28 | 24.49 ± 0.09 a | 45.27 ± 0.72 ab | 1.84 |
| BI-35 | 23.34 ± 0.45 b | 41.17 ± 1.53 bc | 1.76 |
| BI-45 | 23.05 ± 0.34 b | 37.83 ± 1.20 c | 1.64 |
| Treatment and Line | 2016–2017 | 2017–2018 | 2018–2019 | ||||
|---|---|---|---|---|---|---|---|
| GY/g | NUE/g·g−1 | GY/g | NUE/g·g−1 | GY/g | NUE/g·g−1 | ||
| Control (CK) | BI04 | 14.5 ± 1.53 ab | 46.24 ± 4.88 d | 8.03 ± 2.55 ab | 25.61 ± 8.13 c | 20.4 ± 0.51 a | 65.05 ± 13.72 b |
| BI28 | 16.08 ± 2.16 a | 51.27 ± 6.88 cd | 9.59 ± 2.38 a | 30.57 ± 7.60 c | 16.27 ± 0.72 a | 51.87 ± 6.92 b | |
| DH45 | 14.24 ± 2.58 ab | 45.4 ± 8.22 d | 9.64 ± 1.69 a | 30.73 ± 5.40 c | 18.84 ± 0.86 a | 60.06 ± 12.43 b | |
| Low nitrogen (LN) | BI04 | 6.11 ± 1.72 d | 69.26 ± 19.45 c | 4.17 ± 1.60 c | 47.27 ± 10.16 b | 6.34 ± 0.57 c | 71.86 ± 15.61 b |
| BI28 | 10.43 ± 1.60 c | 118.23 ± 18.08 b | 4.24 ± 0.95 c | 48.06 ± 10.79 b | 5.52 ± 0.53 c | 62.57 ± 12.27 b | |
| DH45 | 13.22 ± 1.86 b | 149.86 ± 21.09 a | 5.67 ± 1.41 bc | 64.27 ± 16.03 a | 10.00 ± 0.62 b | 113.36 ± 26.41 a | |
| Traits | Nitrogen Regimes | Mean ± SD | MPD (%) | HPD (%) | ||
|---|---|---|---|---|---|---|
| BI04 | BI28 | DH45 | ||||
| Biomass/g | Control (CK) | 0.35±0.02 a | 0.36±0.03 a | 0.39±0.03 a | 8.97 | 7.69 |
| Low nitrogen (LN) | 0.14±0.02 c | 0.16±0.02 c | 0.27±0.01 b | 80.00 * | 68.75 * | |
| NUE/g·g-1 | Control (CK) | 7.92±1.72 C | 8.32±1.53 C | 9.00±1.63 C | 10.84 | 8.17 |
| Low nitrogen (LN) | 32.00±1.74 B | 37.23±1.63 B | 61.31±1.33 A | 77.11 * | 64.68 * | |
| Term ID | Description | −Log10 (p Value) | ||
|---|---|---|---|---|
| BI04 | BI28 | DH45 | ||
| GO:0006468 | protein phosphorylation | 2.91 | 6.14 | 9.09 |
| GO:0048544 | recognition of pollen | 7.23 | ||
| GO:0015696 | ammonium transport | 3.46 | ||
| GO:0005975 | carbohydrate metabolic process | 5.72 | 2.22 | |
| GO:0006355 | regulation of transcription, DNA-templated | 10.99 | 2.18 | |
| GO:0006644 | phospholipid metabolic process | 1.67 | ||
| GO:0006665 | sphingolipid metabolic process | 1.67 | ||
| GO:0006807 | nitrogen compound metabolic process | 1.61 | ||
| GO:0016042 | lipid catabolic process | 1.46 | ||
| GO:0006817 | phosphate ion transport | 1.37 | ||
| Pathway ID | Description | −Log10 (p Value) | ||
|---|---|---|---|---|
| BI04 | BI28 | DH45 | ||
| ko04016 | MAPK signaling pathway-plant | 1.66 | 4.04 | |
| ko00250 | Alanine, aspartate and glutamate metabolism | 3.97 | ||
| ko00052 | Galactose metabolism | 1.95 | 3.55 | |
| ko00220 | Arginine biosynthesis | 3.18 | ||
| ko00910 | Nitrogen metabolism | 0.81 | 2.59 | |
| ko00940 | Phenylpropanoid biosynthesis | 2.58 | ||
| ko00330 | Arginine and proline metabolism | 2.48 | ||
| ko04014 | Ras signaling pathway | 0.75 | 2.48 | |
| ko00500 | Starch and sucrose metabolism | 1.95 | 2.40 | |
| ko04075 | Plant hormone signal transduction | 1.31 | 2.13 | |
| ko00360 | Phenylalanine metabolism | 2.11 | ||
| ko04010 | MAPK signaling pathway | 1.96 | ||
| ko04141 | Protein processing in endoplasmic reticulum | 1.05 | 1.85 | |
| ko00592 | Alpha-Linolenic acid metabolism | 0.66 | 1.70 | |
| ko00511 | Other glycan degradation | 1.50 | ||
| ko00565 | Ether lipid metabolism | 1.47 | ||
| ko00270 | Cysteine and methionine metabolism | 1.16 | 1.44 | |
| ko00590 | Arachidonic acid metabolism | 0.94 | 1.43 | |
| ko00564 | Glycerophospholipid metabolism | 1.35 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, H.; Li, Y.; Gao, R.; Xu, R.; Guo, G.; Lu, R.; Halford, N.G.; Chen, Z.; Liu, C. Rapid Generation and Analysis of a Barley Doubled Haploid Line with Higher Nitrogen Use Efficiency Than Parental Lines by F1 Microspore Embryogenesis. Plants 2021, 10, 1588. https://doi.org/10.3390/plants10081588
Xu H, Li Y, Gao R, Xu R, Guo G, Lu R, Halford NG, Chen Z, Liu C. Rapid Generation and Analysis of a Barley Doubled Haploid Line with Higher Nitrogen Use Efficiency Than Parental Lines by F1 Microspore Embryogenesis. Plants. 2021; 10(8):1588. https://doi.org/10.3390/plants10081588
Chicago/Turabian StyleXu, Hongwei, Yingbo Li, Runhong Gao, Rugen Xu, Guimei Guo, Ruiju Lu, Nigel G. Halford, Zhiwei Chen, and Chenghong Liu. 2021. "Rapid Generation and Analysis of a Barley Doubled Haploid Line with Higher Nitrogen Use Efficiency Than Parental Lines by F1 Microspore Embryogenesis" Plants 10, no. 8: 1588. https://doi.org/10.3390/plants10081588
APA StyleXu, H., Li, Y., Gao, R., Xu, R., Guo, G., Lu, R., Halford, N. G., Chen, Z., & Liu, C. (2021). Rapid Generation and Analysis of a Barley Doubled Haploid Line with Higher Nitrogen Use Efficiency Than Parental Lines by F1 Microspore Embryogenesis. Plants, 10(8), 1588. https://doi.org/10.3390/plants10081588









